Abstract
Laboratory mice carry 3 host range groups of gammaretroviruses all of which are linked to leukemia induction. Although polytropic mouse leukemia viruses (P-MLVs) are generally recognized as the proximate cause of MLV-induced leukemias in laboratory mice, wild mice that carry only endogenous P-MLVs do not produce infectious virus and are not prone to disease; these mice carry the permissive XPR1 retroviral receptor and an attenuated variant of the retroviral restriction factor, APOBEC3. In contrast, Eurasian mice carrying ecotropic and xenotropic MLVs have evolved multiple restrictive XPR1 variants, other factors that interfere with MLV entry, and more effectively antiviral variants of APOBEC3. These different antiviral restrictions in Mus musculus subspecies suggest that the different virus types found in these natural populations may pose different but largely uncharacterized survival risks in their host subspecies.
Keywords: mouse gammaretroviruses, XPR1 retrovirus receptor, retrovirus restriction, mouse APOBEC3
Introduction
The mouse leukemia viruses (MLVs) are simple retroviruses in the gammaretrovirus genus. They are broad host range viruses that lack accessory factors and use ubiquitously expressed host genes as receptors for entry. Inbred strains of laboratory mice and wild mouse species differ in their susceptibility to mouse gammaretrovirus infection and to virus-induced diseases, and they also differ in the types of MLVs they carry. Three MLV host range subgroups contribute to neoplastic disease in laboratory mice. As originally defined, ecotropic (E-MLVs) infect only mouse cells, xenotropic (X-MLVs) infect cells of non-rodent species, and polytropic (P-MLVs) infect both mouse and non-rodent cells [1]. All 3 of these virus types are found as endogenous retroviruses (ERVs) in the germlines of laboratory mice [2–4], and all 3 contribute to the generation of the recombinant infectious P-MLV viruses that have been identified as the proximate cause of disease in laboratory mice [5]. ERVs of the 3 virus types involved in the disease process are found in different subspecies of wild house mice [6] (Figure 1), and this segregation provides the opportunity to assess their separate roles in pathogenesis and the evolution of the antiviral responses of their mouse hosts that interfere with replication.
Figure 1.
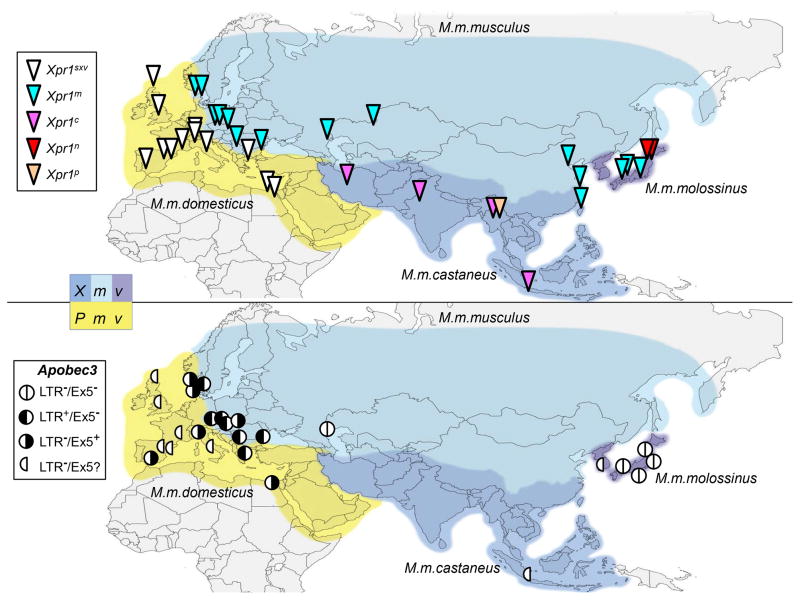
Distribution of endogenous MLVs and allelic variants of Xpr1 and Apobec3 in M. musculus subspecies and common strains of laboratory mice. The permissive allele was termed sxv for susceptibility to xenotropic virus and the other alleles were named for the cells/species in which they were discovered: n, NIH 3T3; m, M. m. musculus and M. m. molossinus; c, M. m. castaneus, p, M. pahari.
Virus-infected mice carry numerous constitutively expressed antiviral factors that target the various stages of the retroviral life cycle (reviewed for example in [7]). This review will focus on those factors that inhibit early stages of virus replication, and the distribution of restrictive and nonrestrictive genes and allelic variants in virus-infected wild mouse populations that carry different MLVs.
At the level of entry, there are two types of resistance genes that target the receptor-virus interaction. Resistance can result from polymorphisms of the cell surface receptors, or from the presence of ERVs that produce receptor-blocking viral envelope glycoprotein (Env). After the gammaretrovirus enters the receptive cell, reverse transcription and translocation to the nucleus can be inhibited or altered by the virus resistance factors Fv1, APOBEC3, and TRIM5α. Two of these factors, Fv1 and APOBEC3, are encoded in the mouse genome and are known to have anti-gammaretroviral activity in mice, but no mouse TRIM5α ortholog with virus restriction activity has been identified [8].
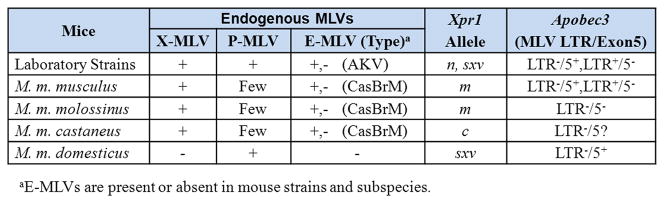
Analyses of these MLVs and host restriction factors in the four house mouse subspecies demonstrate that different restriction genes and alleles evolved in house mice that carry P-MLVs (Mus musculus domesticus), and in mice harboring X-MLVs and E-MLVs (M. m. molossinus, M. m. castaneus, M. m. musculus) (Figure 1, 2).
Figure 2.
Distribution of functionally distinct alleles of the Xpr1 receptor and Apobec3 in Eurasian mice. Blue blocks represent the ranges of Mus subspecies carrying X-MLV ERVs (Xmvs) and yellow represents the range of M. m. domesticus, which carries P-MLV ERVs (Pmvs). Top panel: The ancestral permissive Xpr1sxv allele is in white; different colors indicate the 4 restrictive alleles. Bottom panel: The ancestral Apobec3 allele, in white, lacks the MLV LTR insertion and Exon 5, features acquired by different Musmusculus populations.
Restriction of virus entry
Two gammaretrovirus receptors, CAT-1 and XPR1, are used by the three MLV host range groups. Both of these receptors have naturally occurring variants that are responsible for different virus restriction phenotypes. There are two functional variants of the CAT-1 receptor used by E -MLVs [9,10]and 5 variants of the XPR1 receptor for the X -MLVs and P-MLVs, all of which have mutations that alter the virus binding site [2]. These variants are not only important host factors that can restrict infection, but they can also alter virus/host interactions in ways that influence virus-induced pathology.
The XPR1 receptor for X-MLVs and P-MLVs
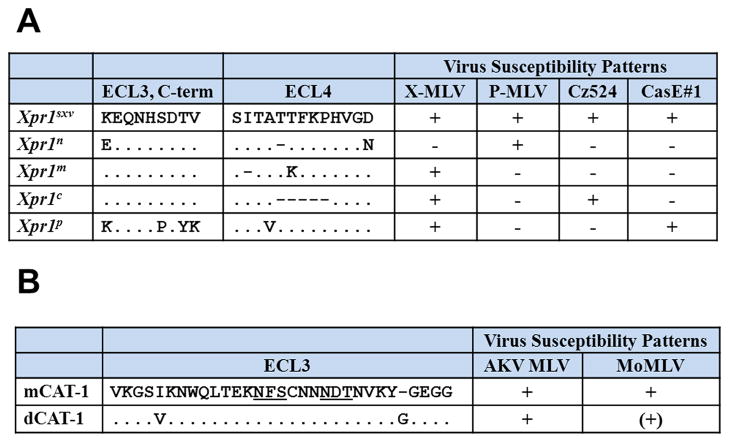
X-MLVs and P-MLVs are viruses capable of infecting cells of non-rodent species and were originally distinguished as separate subgroups because of their different species tropism, their non-reciprocal interference patterns, and by the pathogenicity of P-MLVs [11–16]. The XPR1 receptor for this family of viruses (XP-MLVs) is a multipass transmembrane protein [17–19]. Sequence polymorphisms in the virus envelope and in XPR1 are responsible for the various viral tropisms. The 5 known Mus Xpr1 variants differentially mediate entry of XP-MLV host range variants due to mutations in two XPR1 extracellular loops (Figure 3A) [11,20–22], and these virus host range variants differ in the receptor determining VRA region of Env [23].
Figure 3.
Functional polymorphisms in the receptor determining regions of the MLV receptors, CAT-1 and XPR1, in Mus species. A) Variation in the C-terminal region of the 3rd extracellular loop (ECL3) and in ECL4 of Xpr1. B) Variation in the third extracellular loop of CAT-1 (gene name, Slc7a1). Consensus sites for N-linked glycosylation are underlined. Susceptibility was determined by infection of fibroblasts with infectious viruses or pseudoviruses carrying the LacZ reporter. Log10 titer: −, no detectable infection; (+), 1–2; +, 3–6.
The laboratory mouse carries two of the 5 Mus Xpr1 alleles, the restrictive Xpr1n and the permissive Xpr1sxv; 3 additional restrictive alleles are found in various wild mouse populations and their distribution is subspecies specific (Figure 1). M. m. domesticus, which harbors only P-MLVs, carries the permissive Xpr1sxv allele, whereas the 4 receptor variants that restrict entry of one or more XP-MLVs are found in house mouse subspecies carrying predominantly X-MLVs or in a species (M. pahari, which carries Xpr1p) that is sympatric with X-MLV infected mice [11,22,24] (Figure 2, 3A).
E-MLVs and their CAT-1 receptor
Infectious E-MLVs of 3 different subtypes have been described: AKV MLV in Asian M. m. castaneus and M. m. molossinus, HoMLV in M. spicilegus of Europe, and CasBrM/Fv4 MLV in Asia and in areas of California where Asian mice were introduced by passive transport by humans [6,25,26]. No endogenous or infectious E-MLVs have been found in European M. m. domesticus. One of these three subtypes, AKV MLV, is endogenous in many strains of laboratory mice [3].
The E-MLVs use the CAT-1 receptor [9] which encodes a glycoprotein that functions as a cationic amino acid transporter. It is a multipass transmembrane protein with 14 predicted transmembrane domains. The sites in the mouse protein critical for virus entry lie in the putative third extracellular loop along with two consensus recognition sites for N-linked glycosylation, both of which carry N-glycans [27–29] (Figure 3B).
There are 2 functionally distinct CAT-1 sequence variants in Mus (Figure 3B), and both receptors are recognized by the three E-MLV subtypes. The receptor identified in cells from M. dunni mice, dCAT-1, differs from the laboratory mouse receptor, mCAT-1, in that it restricts infection by an AKV-type MLV, Moloney MLV, although these cells are fully susceptible to other E-MLV isolates [10]. dCAT-1 differs from mCAT-1 by 4 residues, two of which are in the receptor determining third extracellular loop [10] (Figure 3B). Susceptibility differences are due to polymorphisms in env as well as receptor, and these mutations are also associated with cytopathicity [30–32]. Several cytopathic E-MLVs induce syncytia and cell death; in some cases, this response is receptor-specific, and in all 3 cases has been attributed to mutations that alter 2 of the 3 amino acid residues in Env that form the receptor binding pocket [1,33].
Although naturally occurring receptor polymorphisms thus alter both infectivity and cytopathicity, there is no evidence of CAT-1 receptor variation in E-MLV-infected house mouse populations, all of which carry the laboratory mouse variant, mCAT-1 [1].
Interference Genes
Retrovirus infection can be restricted by specific ERVs that express Env glycoprotein. Resistance genes that function through such an Env-mediated interference mechanism have been identified in chickens, cats and sheep, as well as mice, and thus represent a common survival mechanism in natural populations exposed to endemic infections. In mice, an E-MLV ERV associated with the Fv4 resistance locus, is a truncated provirus which encodes Env [34]. Fv4 is found in California and Asian wild mice [35]. These mice also carry E-MLV ERVs that produce at least one pathogenic virus, CasBrM [36]. The Fv4 Env has a defect in the TMenv fusion peptide, and its incorporation into virions reduces their infectivity [37]. There is also a family of genes, termed Rmcf, linked to resistance to XP-MLVs. These genes represent defective Env-producing XP-MLV ERVs and have been found in laboratory strains and in M. m. castaneus [38,39]. The prototype Rmcf gene effectively blocks diseases that involve formation of recombinant P-MLVs [40].
Thus, 2 types of entry level restrictions have evolved in mice carrying MLVs. Mice carrying X-MLVs are protected from exogenous infection by multiple restrictive XPR1 receptor variants as well as by interfering Envs. While polymorphic CAT-1 variants are not found in E-MLV infected mice, resistance at entry results from receptor blocks by the widely disseminated Env-producing ERV, Fv4.
Restriction of post-entry replication
Mice carry two restriction genes responsible for post-entry inhibition of MLV replication, Fv1 and mouse Apobec3 (mA3), both of which have allelic variants with different antiviral phenotypes. Fv1 is a mouse specific gene that targets the virus capsid, and the allelic variants found in laboratory mice block different subgroups of mouse-tropic MLVs after reverse transcription and before integration [41]. mA3 encodes a cytidine deaminase that can be packaged in virions and can restrict virus either by causing G to A hypermutation in viral (+) strands during reverse transcription [42], or through a nonenzymatic mechanism involving inhibition of early reverse transcription [43]. There are two allelic variants of mA3 in laboratory mice, and the C57BL mA3 is more effectively antiviral than the BALB/c allele [44]. Both Fv1 and mA3 have been under positive selection in Mus confirming their defensive roles [45,46], but while there are many Fv1 sequence variants in wild mice [45], there is little information on the antiviral activity of these variants and their distribution in virus-infected wild mouse populations.
Adaptive evolution of the virus resistance gene Apobec3 in Mus
The BALB/c and C57BL mA3 alleles differ in expression level, protein sequence and splicing pattern, all of which contribute to antiviral activity. Increased mA3 expression level in C57BL has been associated with insertion of the long terminal repeat (LTR) of an X-MLV [46], suggesting involvement of the LTR transcriptional enhancer.
Multiple replacement mutations distinguish BALB/c and C57BL mA3. Many of these codons are under strong positive selection in Mus; 6 of these selected residues lie in two clusters in the N-terminal catalytically active deaminase domain of mA3, the region that has also been implicated in virus resistance [46,47]. A structural model of mA3 positioned these two clusters of positively selected residues on opposite sides of the substrate groove where they have the potential to affect substrate interactions [46,48].
C57BL and BALB/c mice also express mA3 transcripts with different splicing patterns: C57BL mice preferentially express exon 5-deficient (5−) mA3 mRNA, while the major mA3 transcript in BALB/c mice contains exon 5 (5+). mA3 exon 5 influences protein synthesis at a post-transcriptional level, and inclusion of this exon is effected by two critical polymorphisms: the number of TCCT repeats upstream of exon 5 and a single nucleotide polymorphism within exon 5 [49].
Analysis of wild mouse mA3 genes for the presence of the LTR, for coding sequence variation and for the splicing signals show that the ancestral mA3 lacked both the MLV LTR and the polymorphisms that determine exon 5 inclusion[46,49]. These two genetic features were acquired independently at about the time of the house mouse radiation 0.5 million years ago but are found in different Mus populations. The more antiviral LTR+Ex5− mA3 is found largely in X -MLV infected Eastern European M. m. musculus, while the 5+ mA3 allele originated and became fixed in Western European M. m. domesticus [49](Figure 1, 2).
The fixation of 5+ mA3 is surprising, because this allele would seem to be evolutionarily deleterious, with its reduced level of mA3 expression and lower antiviral activity. The subspecies with this mA3, M. m. domesticus, carries P-MLV ERVs, which are not known to produce infectious virus and do not cause disease in these populations. However, these P-MLV ERVs have been edited by mA3 [50], and these P-MLV ERVs do not encode the glycosylated Gag variant (glycogag) carried by all infectious MLVs and most endogenous X-MLVs that is known to antagonize mA3 [51,52]. This suggests that the 5+ variant of the potentially genotoxic mA3 enzyme may provide sufficient antiviral benefits in M. m. domesticus.
Conclusions
Acquired retroviral ERVs can alter gene function and genome structure as well as cause disease in their new hosts, but these elements also form stable associations with their hosts over evolutionary time. Host antiviral factors co-evolve with these pathogens, and this “arms race” is responsible for the acquisition of novel antiviral genes and alleles. While most studies on resistance to MLV-induced disease have focused on laboratory mouse strains, the more interesting question is how natural populations deal with endemic infection. Studies on inbred strains of laboratory mice have highlighted the role of P-MLVs in disease-induction, but examination of wild mice produces a different picture in which different antiviral responses have evolved in mice with different ERV profiles (Figure 1,2).
Eurasian mice
The 3 Eurasian house mouse subspecies carry active E- and X-MLV ERVs that produce infectious virus of both types, but virus spread and virus-induced disease are restricted by several host factors, including restrictive XPR1 receptors, interfering Env genes like Fv4 and Rmcf2, and the more antiviral LTR+/Ex5− mA3 subtype.
Western European mice
Wild mice that carry ERVs related to the leukemogenic P-MLVs, namely M. m. domesticus, have low tumor incidence. These mice carry the less antiviral mA3 gene, have fully functional MLV receptors, and are not known to encode interfering Envs, suggesting that these mice are either not in need of protection or rely on unknown antiviral factors. P-MLV ERVs do not produce infectious virus and, in laboratory mice and wild mice, involvement of P-MLVs in disease is dependent on their rescue by infection with E-MLVs [53,54]. No E-MLVs have been found in wild M. m. domesticus; it is tempting to speculate that this absence may be the result of purifying selection.
Laboratory mice
The classical strains of laboratory mice were created by interbreeding 3 Mus musculus subspecies [55]. This interbreeding introduced the 3 MLV types into the same breeding lines, and also removed some natural host restriction factors. Many of the resulting fancy mouse colonies and inbred strains are plagued with tumors that are uncommon in their wild mouse progenitors [56,57].
The fact that multiple types of antiviral factors have evolved in X-MLV, but not P-MLV infected mice suggests that X-MLVs may be more deleterious than appreciated. The pathogenic properties of these viruses are not easily studied, since they are noninfectious in most common strains of laboratory mice [58,59]. The observations detailed here suggest that these viruses may deserve a second look. Further studies on these infected wild mice may also provide insight into the host factors that most influence cross-species and cross-subspecies virus transmission.
Highlights.
Wild mice carrying XP-MLVs rarely develop virus-induced disease.
Different antiviral host factors are found in mice with X-MLVs and P-MLVs.
Mice with X-MLVs have defective or blocked receptors and a restrictive APOBEC3.
Mice with P-MLVs have a permissive receptor, a weaker APOBEC3, but make no virus.
Acknowledgments
This work was supported by the Intramural Program of the NIAID, NIH, Bethesda, MD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1*.Kozak CA. Naturally occurring polymorphisms of the mouse gammaretrovirus receptors CAT-1 and XPR1 alter virus tropism and pathogenicity. Adv Virol. 2011:975801. doi: 10.1155/2011/975801. Discussion of complementary polymorphisms in virus Env glycoprotein and in their receptors, and the consequences for tropism and cytopathicity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2*.Kozak CA. The mouse “xenotropic” gammaretroviruses and their XPR1 receptor. Retrovirology. 2010;7:101. doi: 10.1186/1742-4690-7-101. Comprehensive review of the distribution, receptor requirements and host restriction of X-MLV replication in lab strains and wild mouse species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Neill RR, Khan AS, Hoggan MD, Hartley JW, Martin MA, Repaske R. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J Virol. 1986;58:359–366. doi: 10.1128/jvi.58.2.359-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoye JP, Moroni C, Coffin JM. Virological events leading to spontaneous AKR thymomas. J Virol. 1991;65:1273–1285. doi: 10.1128/jvi.65.3.1273-1285.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozak CA, O’Neill RR. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stocking C, Kozak C. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tareen SU, Sawyer SL, Malik HS, Emerman M. An expanded clade of rodent Trim5 genes. Virology. 2009;385:473–483. doi: 10.1016/j.virol.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albritton LM, Tseng L, Scadden D, Cunningham JM. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 10.Eiden MV, Farrell K, Warsowe J, Mahan LC, Wilson CA. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y, Liu Q, Wollenberg K, Martin C, Buckler-White A, Kozak CA. Evolution of functional and sequence variants of the mammalian XPR1 receptor for mouse xenotropic gammaretroviruses and the human-derived XMRV. J Virol. 2010;84:11970–11980. doi: 10.1128/JVI.01549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cloyd MW, Hartley JW, Rowe WP. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980;151:542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chesebro B, Wehrly K. Different murine cell lines manifest unique patterns of interference to superinfection by murine leukemia viruses. Virology. 1985;141:119–129. doi: 10.1016/0042-6822(85)90188-6. [DOI] [PubMed] [Google Scholar]
- 14.Hartley JW, Wolford NK, Old LJ, Rowe WP. New class of murine leukemia-virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci U S A. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982;120:251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 16.Cloyd MW, Thompson MM, Hartley JW. Host range of mink cell focus-inducing viruses. Virology. 1985;140:239–248. doi: 10.1016/0042-6822(85)90362-9. [DOI] [PubMed] [Google Scholar]
- 17.Tailor CS, Nouri A, Lee CG, Kozak C, Kabat D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc Natl Acad Sci U S A. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battini J-L, Rasko JEJ, Miller AD. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: possible role in G protein-coupled signal transduction. Proc Natl Acad Sci U S A. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y-L, Guo L, Xu S, Holland CA, Kitamura T, Hunter K, Cunningham JM. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Liu Q, Kozak CA. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology. 2009;6:87. doi: 10.1186/1742-4690-6-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marin M, Tailor CS, Nouri A, Kozak SL, Kabat D. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J Virol. 1999;73:9362–9368. doi: 10.1128/jvi.73.11.9362-9368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Y, Knoper RC, Kozak CA. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J Virol. 2007;81:10550–10557. doi: 10.1128/JVI.00933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battini JL, Heard JM, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bamunusinghe D, Liu Q, Lu X, Oler A, Kozak CA. Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor. J Virol. 2013 doi: 10.1128/JVI.01264-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaguma Y, Miyashita N, Moriwaki K, Huai WC, Jin ML, He XQ, Ikeda H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J Virol. 1991;65:1796–1802. doi: 10.1128/jvi.65.4.1796-1802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voytek P, Kozak CA. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology. 1989;173:58–67. doi: 10.1016/0042-6822(89)90221-3. [DOI] [PubMed] [Google Scholar]
- 27.Albritton LM, Kim JW, Tseng L, Cunningham JM. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JW, Cunningham JM. N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. J Biol Chem. 1993;268:16316–16320. [PubMed] [Google Scholar]
- 30.Park BH, Matuschke B, Lavi E, Gaulton GN. A point mutation in the env gene of a murine leukemia virus induces syncytium formation and neurologic disease. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung YT, Kozak CA. Generation of novel syncytium-inducing and host range variants of ecotropic Moloney murine leukemia virus in Mus spicilegus. J Virol. 2003;77:5065–5072. doi: 10.1128/JVI.77.9.5065-5072.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung YT, Wu T, Kozak CA. Novel host range and cytopathic variant of ecotropic Friend murine leukemia virus. J Virol. 2004;78:12189–12197. doi: 10.1128/JVI.78.22.12189-12197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davey RA, Zuo Y, Cunningham JM. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda H, Laigret F, Martin MA, Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55:768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gardner MB, Kozak CA, O’Brien SJ. The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet. 1991;7:22–27. doi: 10.1016/0168-9525(91)90017-k. [DOI] [PubMed] [Google Scholar]
- 36.Gardner MB, Klement V, Rongey RR, McConahey P, Estes JD, Huebner RJ. Type C virus expression in lymphoma-paralysis-prone wild mice. J Natl Cancer Inst. 1976;57:585–590. doi: 10.1093/jnci/57.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor GM, Gao Y, Sanders DA. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J Virol. 2001;75:11244–11248. doi: 10.1128/JVI.75.22.11244-11248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartley JW, Yetter RA, Morse HC. A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J Exp Med. 1983;158:16–24. doi: 10.1084/jem.158.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lyu MS, Nihrane A, Kozak CA. Receptor-mediated interference mechanism responsible for resistance to polytropic leukemia viruses in Mus castaneus. J Virol. 1999;73:3733–3736. doi: 10.1128/jvi.73.5.3733-3736.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruscetti S, Davis L, Feild J, Oliff A. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J Exp Med. 1981;154:907–920. doi: 10.1084/jem.154.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartley JW, Rowe WP, Huebner RJ. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J Virol. 1970;5:221–225. doi: 10.1128/jvi.5.2.221-225.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 43.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282:2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 44.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y, Buckler-White A, Wollenberg K, Kozak CA. Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus. Proc Natl Acad Sci U S A. 2009;106:3259–3263. doi: 10.1073/pnas.0900181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Sanville B, Dolan MA, Wollenberg K, Yan Y, Martin C, Yeung ML, Strebel K, Buckler-White A, Kozak CA. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 2010;6:e1000974. doi: 10.1371/journal.ppat.1000974. Demonstration that P-MLV genomes are preferentially packaged in E-MLV virions, explaining how ERVs that fail to produce virus can be amplified. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakata Y, Landau NR. Reversed functional organization of mouse and human APOBEC3 cytidine deaminase domains. J Biol Chem. 2006;281:36624–36631. doi: 10.1074/jbc.M604980200. [DOI] [PubMed] [Google Scholar]
- 48.Kohli RM, Abrams SR, Gajula KS, Maul RW, Gearhart PJ, Stivers JT. A portable hot spot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J Biol Chem. 2009;284:22898–22904. doi: 10.1074/jbc.M109.025536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J, Hakata Y, Takeda E, Liu Q, Iwatani Y, Kozak CA, Miyazawa M. Two genetic determinants acquired late in Mus evolution regulate the inclusion of exon 5, which alters mouse APOBEC3 translation efficiency. PLoS Pathog. 2012;8:e1002478. doi: 10.1371/journal.ppat.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jern P, Stoye JP, Coffin JM. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:e183. doi: 10.1371/journal.pgen.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc Natl Acad Sci U S A. 2013;110:9078–9083. doi: 10.1073/pnas.1217399110. Demonstration that the MLV glygogag renders the MLV RT complex resistant to APOBEC3 and that glycogag mutants revert in wild type but not mA3 KO mice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J Virol. 2010;84:10933–10936. doi: 10.1128/JVI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung YT, Wu T, Kozak CA. Characterization of recombinant nonecotropic murine leukemia viruses from the wild mouse species Mus spretus. J Virol. 2003;77:12773–12781. doi: 10.1128/JVI.77.23.12773-12781.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosenke K, Lavignon M, Malik F, Kolokithas A, Hendrick D, Virtaneva K, Peterson K, Evans LH. Profound amplification of pathogenic murine polytropic retrovirus release from coinfected cells. J Virol. 2012;86:7241–7248. doi: 10.1128/JVI.00225-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Wang JR, Didion JP, Buus RJ, Bell TA, Welsh CE, Bonhomme F, Yu AH, Nachman MW, Pialek J, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lathrop AE, Loeb L. Further investigations on the origin of tumors in mice: V. the tumor rate in hybrid strains. J Exp Med. 1918;28:475–500. doi: 10.1084/jem.28.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lilly F, Pincus T. Genetic control of murine viral leukemogenesis. Advances in Cancer Research. 1973;17:231–277. [Google Scholar]
- 58*.Sakuma T, Tonne JM, Squillace KA, Ohmine S, Thatava T, Peng KW, Barry MA, Ikeda Y. Early events in retrovirus XMRV infection of the wild-derived mouse Mus pahari. J Virol. 2011;85:1205–1213. doi: 10.1128/JVI.00886-10. The transmission and disease potential of X-MLVs cannot be evaluated in laboratory strains due to a defective receptor. This study makes use of a wild mouse species carrying a functional X-MLV receptor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59*.Baliji S, Liu Q, Kozak CA. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J Virol. 2010;84:12841–12849. doi: 10.1128/JVI.01863-10. Demonstration that some inbred strains are actually susceptible to X-MLVs, originally defined as being unable to infect inbred mice. [DOI] [PMC free article] [PubMed] [Google Scholar]