Abstract
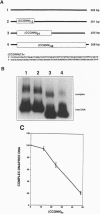
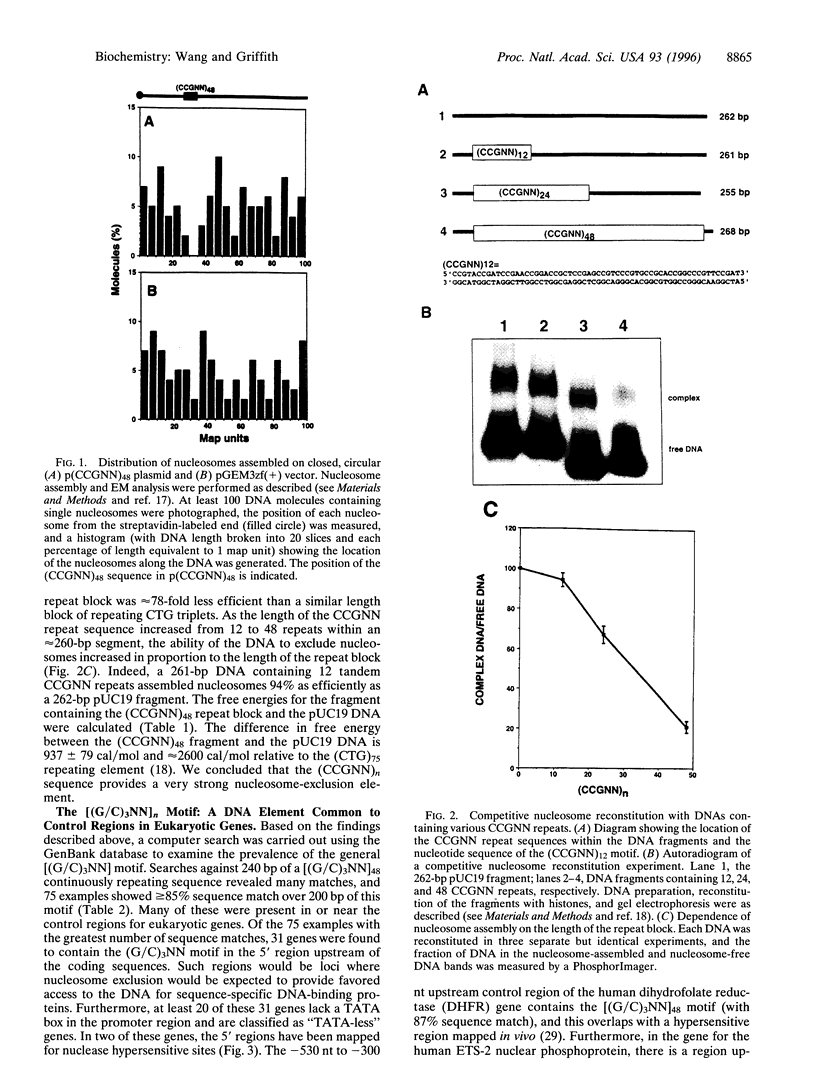
Nucleosomes, the basic structural elements of chromosomes, consist of 146 bp of DNA coiled around an octamer of histone proteins, and their presence can strongly influence gene expression. Considerations of the anisotropic flexibility of nucleotide triplets containing 3 cytosines or guanines suggested that a [5'(G/C)3 NN3']n motif might resist wrapping around a histone octamer. To test this, DNAs were constructed containing a 5'-CCGNN-3' pentanucleotide repeat with the Ns varied. Using in vitro nucleosome reconstitution and electron microscopy, a plasmid with 48 contiguous CCGNN repeats strongly excluded nucleosomes in the repeat region. Competitive reconstitution gel retardation experiments using DNA fragments containing 12, 24, or 48 CCGNN repeats showed that the propensity to exclude nucleosomes increased with the length of the repeat. Analysis showed that a 268-bp DNA containing a (CCGNN)48 block is 4.9 +/- 0.6-fold less efficient in nucleosome assembly than a similar length pUC19 fragment and approximately 78-fold less efficient than a similar length (CTG)n sequence, based on results from previous studies. Computer searches against the GenBank database for matches with a [(G/C)3NN]48 sequence revealed numerous examples that frequently were present in the control regions of "TATA-less" genes, including the human ETS-2 and human dihydrofolate reductase genes. In both cases the (G/C)3NN repeat, present in the promoter region, co-maps with loci previously shown to be nuclease hypersensitive sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azizkhan J. C., Jensen D. E., Pierce A. J., Wade M. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit Rev Eukaryot Gene Expr. 1993;3(4):229–254. [PubMed] [Google Scholar]
- Dunn K., Griffith J. D. The presence of RNA in a double helix inhibits its interaction with histone protein. Nucleic Acids Res. 1980 Feb 11;8(3):555–566. doi: 10.1093/nar/8.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesfeld J. M. DNA sequence-directed nucleosome reconstitution on 5S RNA genes of Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1612–1622. doi: 10.1128/mcb.7.5.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. D., Christiansen G. Electron microscope visualization of chromatin and other DNA-protein complexes. Annu Rev Biophys Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. Chromatin structure: deduced from a minichromosome. Science. 1975 Mar 28;187(4182):1202–1203. doi: 10.1126/science.187.4182.1202. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Iyer V., Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995 Jun 1;14(11):2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. L., Archer T. K. Nucleosome-mediated disruption of transcription factor-chromatin initiation complexes at the mouse mammary tumor virus long terminal repeat in vivo. Mol Cell Biol. 1994 Jan;14(1):32–41. doi: 10.1128/mcb.14.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrothalassitis G. J., Watson D. K., Papas T. S. The human ETS-2 gene promoter: molecular dissection and nuclease hypersensitivity. Oncogene. 1990 Sep;5(9):1337–1342. [PubMed] [Google Scholar]
- McPherson C. E., Shim E. Y., Friedman D. S., Zaret K. S. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell. 1993 Oct 22;75(2):387–398. doi: 10.1016/0092-8674(93)80079-t. [DOI] [PubMed] [Google Scholar]
- Nelson D. L. The fragile X syndromes. Semin Cell Biol. 1995 Feb;6(1):5–11. doi: 10.1016/1043-4682(95)90009-8. [DOI] [PubMed] [Google Scholar]
- Nickol J., Behe M., Felsenfeld G. Effect of the B--Z transition in poly(dG-m5dC) . poly(dG-m5dC) on nucleosome formation. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1771–1775. doi: 10.1073/pnas.79.6.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten A. D., Tapscott S. J. Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5465–5469. doi: 10.1073/pnas.92.12.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay N. Deletion analysis of a DNA sequence that positions itself precisely on the nucleosome core. J Mol Biol. 1986 May 5;189(1):179–188. doi: 10.1016/0022-2836(86)90389-x. [DOI] [PubMed] [Google Scholar]
- Rhodes D. Structural analysis of a triple complex between the histone octamer, a Xenopus gene for 5S RNA and transcription factor IIIA. EMBO J. 1985 Dec 16;4(13A):3473–3482. doi: 10.1002/j.1460-2075.1985.tb04106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schild C., Claret F. X., Wahli W., Wolffe A. P. A nucleosome-dependent static loop potentiates estrogen-regulated transcription from the Xenopus vitellogenin B1 promoter in vitro. EMBO J. 1993 Feb;12(2):423–433. doi: 10.1002/j.1460-2075.1993.tb05674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T., Inokuchi K., Nienhuis A. W. Chromatin structure of the human dihydrofolate reductase gene promoter. Multiple protein-binding sites. J Biol Chem. 1986 Jan 25;261(3):1445–1452. [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Artificial nucleosome positioning sequences. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrader T. E., Crothers D. M. Effects of DNA sequence and histone-histone interactions on nucleosome placement. J Mol Biol. 1990 Nov 5;216(1):69–84. doi: 10.1016/S0022-2836(05)80061-0. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Künzler P. Cromatin and core particles formed from the inner histones and synthetic polydeoxyribonucleotides of defined sequence. Nucleic Acids Res. 1979 Apr;6(4):1387–1415. doi: 10.1093/nar/6.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland G. R., Richards R. I. The molecular basis of fragile sites in human chromosomes. Curr Opin Genet Dev. 1995 Jun;5(3):323–327. doi: 10.1016/0959-437x(95)80046-8. [DOI] [PubMed] [Google Scholar]
- Thoma F. Nucleosome positioning. Biochim Biophys Acta. 1992 Feb 28;1130(1):1–19. doi: 10.1016/0167-4781(92)90455-9. [DOI] [PubMed] [Google Scholar]
- Timchenko L., Monckton D. G., Caskey C. T. Myotonic dystrophy: an unstable CTG repeat in a protein kinase gene. Semin Cell Biol. 1995 Feb;6(1):13–19. doi: 10.1016/1043-4682(95)90010-1. [DOI] [PubMed] [Google Scholar]
- Trifonov E. N. Curved DNA. CRC Crit Rev Biochem. 1985;19(2):89–106. doi: 10.3109/10409238509082540. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Amirhaeri S., Kang S., Wells R. D., Griffith J. D. Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science. 1994 Jul 29;265(5172):669–671. doi: 10.1126/science.8036515. [DOI] [PubMed] [Google Scholar]
- Wang Y. H., Griffith J. Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics. 1995 Jan 20;25(2):570–573. doi: 10.1016/0888-7543(95)80061-p. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Transcription: in tune with the histones. Cell. 1994 Apr 8;77(1):13–16. doi: 10.1016/0092-8674(94)90229-1. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Kingston R. E. Nucleosome core displacement in vitro via a metastable transcription factor-nucleosome complex. Science. 1992 Dec 11;258(5089):1780–1784. doi: 10.1126/science.1465613. [DOI] [PubMed] [Google Scholar]
- Workman J. L., Roeder R. G. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell. 1987 Nov 20;51(4):613–622. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]