Abstract
Pan1 is a multi-domain scaffold that enables dynamic interactions with both structural and regulatory components of the endocytic pathway. Pan1 is composed of Eps15 Homology (EH) domains which interact with adaptor proteins, a central region that is responsible for its oligomerization and C-terminal binding sites for Arp2/3, F-actin, and type-I myosin motors. In this study, we have characterized the binding sites between Pan1 and its constitutive binding partner End3, another EH domain containing endocytic protein. The C-terminal End3 Repeats of End3 associate with the N-terminal part of Pan1’s central coiled-coil region. These repeats appear to act independently of one another as tandem, redundant binding sites for Pan1. The end3-1 allele was sequenced, and corresponds to a C-terminal truncation lacking the End3 Repeats. Mutations of the End3 Repeats highlight that those residues which are identical between these repeats serve as contact sites for the interaction with Pan1.
Keywords: Pan1, End3, Endocytosis, EH domain, Amphipathic helix, End3-1
Endocytosis occurs via sequential membrane and protein interaction events that cluster plasma membrane cargo for incorporation into an internalized vesicle in the cytosol. While many of the protein components that are recruited to an endocytic site have been identified, in most cases their stoichiometry and mechanism of association have been poorly described. Based on work in fission yeast, it is expected that thousands of individual proteins are recruited to each developing endocytic site, but how they interact and function in such a confined space is unclear [1]. Many of these endocytic proteins contain well characterized domains and motifs that mediate the various protein-protein or protein-lipid interactions necessary for progression of endocytosis, such as Eps15 Homology (EH) domains which interact with NPF tripeptide motifs and SH3 domains with their cognate proline rich domain binding partners [2,3]. Contact sites between some endocytic proteins are less clearly characterized, as they are lacking the well studied and conserved domains or may contain small motifs that are difficult to define. A specific example of this is the poorly described interaction between Pan1 and End3, which is surprising given the large literature describing these proteins as constitutive binding partners that are key regulators of endocytosis.
Pan1 is thought to act as a scaffold that contributes to the proper progression of protein recruitment and activation at endocytic sites. The N-terminus of Pan1 binds early-acting endocytic proteins such as adaptors, whereas the C-terminus interacts with late-acting factors that regulate actin polymerization. The N-terminus contains two “long repeat” regions, (LR1 and LR2): each containing an EH domain that mediates binding to the adaptors Ent1/2 and Yap1801/2 (Figure 1) [4,5]. The LR1 has also been shown to associate with the C-terminal portion of another endocytic adaptor protein Sla1 [6]. The central region of Pan1 defines the oligomerization domain that contains predicted coiled-coils that contribute to formation of homodimers and higher order oligomers of Pan1, as well as association with Sla2, the yeast homolog of the endocytic protein Hip1R [2,7]. The C-terminal domain contains regions that bind Arp2/3, F-actin, and the type-I myosins Myo3/5; these proteins regulate actin polymerization during the internalization and scission steps of endocytosis [8–10]. Through the spatio-temporal regulation of endocytosis, Pan1 may control the progression toward vesicle internalization by monitoring proper protein recruitment and interactions. Importantly, Pan1 activity and localization must be tightly regulated to control the timely coordination of Arp2/3 stimulation at the later stages of endocytosis as well as its removal from an internalized vesicle. This control is mediated through phosphorylation of Pan1 by Prk1, an endocytic regulatory kinase. Pan1 contains 19 consensus Prk1 sites (L/IxxQxTG), 18 of which lie in its N-terminal LR1 and LR2 regions [11]. The literature suggests that phosphorylation of Pan1 terminates its Arp2/3 activation, disrupts its association with Sla1, and returns Pan1 to the cytosol where it is predicted to be dephosphorylated [7,9,12,13].
Figure 1. End3 binds to the Central Domain of Pan1.
A) Schematics of Pan1, End3, and end3-1 protein. The thick bars above Pan1 and End3 represent the yeast two-hybrid interacting regions. Domains and motifs important for each protein are indicated. EH, Eps15 Homology Domain; LR, Long Repeat; A, Acidic motif; PP, polyproline; 1, End3 Repeat 1; 2, End3 Repeat 2. The green bars represent predicted coiled-coils within Pan1 and End3. The sequence alignment of End3 with end3-1 is shown below the schematics and begins at the final residue of the second EH domain. end3-1 contains a frame shift due to deletion of nucleotide 715 and produces 33 non-native amino acids before termination. The underlined residues are the End3 Repeats at the C-terminus of End3, and the bolded and italicized sequence is the non-native end3-1 protein sequence. B) End3 binds to the coiled-coil region of Pan1. Recombinant protein-binding assay with normalized protein amounts of GST, GST-LR2360–690, or GST-CC792–1252 bound to glutathione agarose beads incubated with lysates from bacteria expressing His6-tagged full length End3. His6-End lysate input, aliquots of each supernatant (S) and washed pellet (P) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-His6 or anti-GST antibodies, as indicated. The percentages of End3 protein bound were calculated from anti-His6 pellet band intensity relative to the supernatant lane from the same sample.
Most of Pan1’s interactions with its endocytic partners occur at either the N- or C-terminus of Pan1 [5–8,10]. In contrast, we know much less about the significance of Pan1’s central oligomerization domain, in part because many fewer of the binding partners for this domain have been identified.
The association between Pan1 and End3 has previously been shown to be required to mediate proper Pan1 dephosphorylation to prevent excess Arp2/3 stimulation [13]. The End3 C-terminus was originally shown to bind Pan1 through a directed yeast two-hybrid assay. The binding site in Pan1 was at first presumed to be localized to the LR2 region, but was later revised to the coiled-coil region of Pan1, somewhere C-terminal to the second EH domain of Pan1 (EH2) domain, and thus not part of LR2 [13,14]. However, the data upon which the new binding site was based are unpublished, so the precise binding site on Pan1 has not been clearly defined. All Pan1-End3 protein work published since then has used either full length Pan1 protein or amino acids 384–846, which include both the entire LR2 domain as well as a significant portion of the coiled-coil domain [6,7,12,13]. Thus, no analysis has been performed to test where the End3 binding site lies within the coiled-coil region of Pan1.
End3 and Pan1 are considered a constitutive complex as they are reported to copurify with a 1:1 stoichiometry, yet the binding interfaces required for their association have never been characterized [7]. In this study we demonstrate that End3 contains two binding sites for Pan1 that lie in tandem about 40 amino acids apart in its C-terminus. These tandem sites were previously named “End3 Repeats” (E3R) due to their conserved motifs that lie within predicted coiled-coils (Figure 1) [15]. We have generated E3R mutants that are unable to bind to Pan1 in vitro. These E3R mutants phenocopy end3Δ mutant phenotypes, including accumulation of hyper-phosphorylated Pan1 species and severe disruption of endocytosis. This suggests that the End3 Repeats mediate functionally important interactions with Pan1’s central coiled-coil domain to maintain endocytic homeostasis. Our findings contribute to a better understanding of how these two, key endocytic proteins bind to one another to promote progression of an endocytic site.
Results
A Yeast two-Hybrid Screen with the Pan1 Central Domain
The EH2 domain and the first half of the central region of Pan1 are required to maintain viability in the absence of endogenous PAN1 [16]. A PCR-based mutagenesis screen of a PAN1 segment encoding amino acids (aa) 527–918, which includes both of these regions, identified temperature sensitive alleles with mutations in the coiled-coil-containing central region of Pan1 rather than the EH2 domain that binds to adaptors AP1801/2 and Ent1/2 [5] (Nick Miliaras and BW, unpublished data). This highlighted that the central coiled-coil containing region of Pan1 was a ‘hotspot’ for temperature sensitive alleles, and thus indicated that this central region is functionally important.
In order to identify important binding partners within this mutationally sensitive region of Pan1, we undertook a yeast two-hybrid (Y2H) screen using aa792–948 as the bait. From this screen we identified novel potential binding partners of interest, Bbc1, Cna1, and Kel2 as well as the known Pan1 binding partner End3 (Supplemental Table 1).
End3 Binds the Central Domain of Pan1
Our yeast two-hybrid interaction between Pan1 and End3 agreed with the unpublished data from the Cai lab, suggesting that the central domain of Pan1 contains the binding site for the C-terminus of End3 [13]. We next validated the yeast two-hybrid results by testing if End3 binds to the coiled-coil region of Pan1, rather than the LR2 region, using a pulldown with the recombinant End3 protein mixed with either GST, GST-LR2, or GST-Coiled-coil (Figure 1B). His6-End3 associated with glutathione beads pre-bound with GST-Coiled-coil, and showed only low level background association with LR2-bound beads or GST-only beads. This validated and confirmed our yeast two-hybrid results and the unpublished data from the Cai lab that the binding site of End3 does not lie within LR2, but rather is in the central region following EH2 [13].
To narrow down the binding site within Pan1, we paired End3 with different Pan1 coiled-coil containing yeast two-hybrid constructs. Results from the yeast two-hybrid reporter assay in Table 1 showed that End3 bound to the entire Pan1 central region (aa687–1190), as well as to another region of Pan1 (aa687–846) that overlaps with the original Y2H bait (aa792–948). These two fragments of the central domain of Pan1 contain aa792–846; thus, we concluded that the minimal binding region for End3 lies within this sequence of Pan1. Testing the direct association between Pan1 aa792–846 and End3 via yeast two-hybrid or recombinant binding was not possible due to technical difficulties expressing this small Pan1 fragment. In an attempt to define the minimal binding site for End3 within the Pan1 aa792–846 region, we performed alanine scanning mutagenesis. However, while some Pan1 alanine mutations showed reduced End3 binding by Y2H assays, none of them disrupted the Pan1-End3 interaction when tested by pulldown of recombinant proteins.
Table 1.
Minimal Binding Region in Pan1 for End3
| DNA-binding domain |
Activation domain |
Miller Units | BD-pan1 | AD-End3 |
|---|---|---|---|---|
| Pan1762–948 | End3176–349 | 99.74±4.30 |  |
 |
| Pan1687–846 | End3176–349 | 80.10±6.04 | ||
| Pan1687–1190 | End3176–349 | 412.28±107.73 | ||
| vector | End3176–349 | 0.16±0.04 | ||
| Pan1792–948 | vector | 0.23±0.02 | ||
| Pan1687–846 | vector | 3.20±0.68 | ||
| Pan1687–1190 | vector | 0.19±0.02 | ||
| vector | vector | 0.45±0.28 |
Superscript numbers indicate amino acids. Standard deviation of mean values are given
Further recombinant binding experiments using various fragments of Pan1 central region confirmed the yeast two-hybrid data in Table 1 that the region responsible for binding to End3 lies within the more N-terminal portion of the central coiled-coil (data not shown). Future efforts will be necessary to narrow down the sequence responsible for this interaction.
Pan1 Binds to the C-terminal End3 Repeats
To test which portion of the C-terminus of End3 is required for Pan1 binding, we performed a recombinant pull down assay using C-terminal fragments of End3. This region of End3 is comprised of three predicted coiled-coils, two of which contribute to the E3Rs (Figure 1A). Pan1aa687–1190-TAP or TAP protein was purified from yeast cells and immobilized on calmodulin-sepharose beads (the central region of Pan1 must be purified from yeast due to its toxicity in bacteria) [16]. Bacterial lysates containing fragments of His6-End3 were incubated with the Pan1- or TAP-bound beads, washed, and analyzed by western blot (Figure 2). Full length End3 and aa176–349 (the Y2H prey fragment) each bound the beads with the Pan1 central region but not TAP-only beads. End3’s EH domains do not contribute to this association, as aa228–349, which contain only the coiled-coil sequence after the second End3 EH domain, bound to Pan1 with similar efficiency as aa176–349. Each End3 Repeat can independently bind Pan1, as seen for fragments aa176–303 and aa304–349. While the final End3 coiled-coil is not required for binding since aa176–303 bound Pan1, when tested on its own, this final coiled-coil, aa304–349, showed just a slight enrichment on Pan1-TAP bound beads that was undetectable on the TAP beads. The apparent weakness of the aa304–349 binding is likely due to the poorly folded nature of this predicted coiled-coil when expressed on its own. That two independent coiled-coils of End3 are capable of interacting with Pan1 indicated that there are likely two binding sites for Pan1 on a single End3 protein.
Figure 2. Pan1 Binds to the C-terminal End3 Repeats.
Each End3 Repeat can independently associate with Pan1. Recombinant protein-binding assay with TAP-alone or Pan1687–1190-TAP bound to calmodulin sepharose beads, incubated with lysates from bacteria expressing His6-tagged fragments of End3 as indicated in the schematics. The End3 Repeats are underlined beneath the full length schematic and labelled E3R1 and E3R2. The purple box at the C-terminus of end3-1 corresponds to the non-native amino acid sequence due to the frame shift in this mutant. Aliquots of each supernatant (S) and washed pellet (P) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-His6 antibody. On the second panel, the TAP protein band resolves slightly larger than His6-End3176–349 (20.4kD and 21.9kD, respectively), thus the second lane to the right only contains TAP protein and no detectable His6-End3176–349. The TAP tag contains Protein A which reacts with the secondary antibody (anti-mouse) and is thus detected in the anti-His6 blot.
We next asked if end3-1 binds to Pan1. end3-1 has been widely used as an endocytic mutant because it was one of the first endocytosis mutants discovered, and it shows internalization defects at the lower temperature of 24°C, but the mutation of this allele is uncharacterized [14,15,17]. We were curious if sequencing the end3-1 allele could define those resides crucial for association with Pan1, and identified a deletion of nucleotide 715 that causes a frame shift at aa238, after the second EH domain, followed by 33 non-native amino acids (Figure 1A). end3-1 protein has both N-terminal EH domains intact, but is truncated within the first coiled-coil of the C-terminus; thus, end3-1 is lacking the final two coiled-coils which contain the End3 Repeats first identified by the Riezman lab [15].
Based upon this sequence information as well as our own and previously published yeast two-hybrid results, we predicted that end3-1 would be unable to associate with Pan1 as it is lacking the binding site(s) necessary for this interaction [14]. As expected, end3-1 protein showed no enrichment for binding to Pan1-TAP over TAP-only beads, confirming that end3-1 protein is unable to bind Pan1 in this in vitro assay (Figure 2).
Mutation of the End3 Repeats
Two independent binding sites for Pan1 within the C-terminal coiled-coils of End3 highlighted the potential functional significance of the End3 Repeats (E3Rs) that were first identified by Riezman et al [15]. The E3Rs are 50% identical to one another and 75% similar. These repeats are predicted to lie within coiled-coils based upon COILS analysis (http://www.ch.embnet.org/software/COILS_form.html). When threaded through a helical wheel projection, hydrophobic residues lie within the a- and d-positions of the heptad repeat, and the remaining residues fall on the hydrophilic face of this predicted coiled-coil (Figure 3). Those residues that are identical between E3R1 and E3R2 lie on the hydrophilic face of the repeats, suggesting that this face contains conserved binding sites that are present in tandem on End3. In contrast, the hydrophobic face of the helical wheel projections show that the hydrophobic residues are conserved but not identical between E3R1 and E3R2. This indicates that this face of the coiled-coil is required for a hydrophobic homo- or intramolecular interaction, but that a conserved, tandem site for interactions with other proteins may not exist within this face.
Figure 3. End3 Repeat Mutants.
A) Helical Wheel representation of the End3 Repeats, E3R1 and E3R2. These repeats fall within predicted coiled-coil sequences, and have been projected around a helical wheel to visualize which face of the alpha-helical coil contains residues that are identical between E3R1 and E3R2 (Boxed). The amphipathic nature of the helix is indicated by the grey dotted curve with hydrophobic residues on the right and hydrophilic residues on the left. Alignment of the repeats are below with the colon (:) marking similar residues and an asterisk (*) marking identical residues. Four classes of mutants are shown below the wildtype sequence, with the mutated amino acids shown in red. B) Testing complementation of temperature sensitivity of end3Δ cells with E3R mutants. Serial dilutions were spotted onto selective -Ura medium and incubated at 30°C or 38°C for 3 days. All plasmids are single copy plasmids in the pRS.416 backbone. C) Similar assay as shown in 3B with high copy (indicated as .426) and single copy plasmids (.416) of Hζ and single E3R mutants. HΦR high copy and single repeat mutants displayed identical growth as shown here for the Hζ mutants.
In order to test if the E3Rs are binding sites for Pan1, mutations were introduced into conserved residues on either the hydrophobic or hydrophilic faces of both repeats. Each mutant maintained high probability predicted coiled-coils based on analysis with the COILS software, and was confirmed to maintain helical nature through circular dichroism analysis of expressed mutant protein samples (data not shown). Two types of hydrophilic mutants were made, one with mutations in residues that are identical between E3R1 and E3R2 (Hζ) as well as another mutant that focused on residues that are conserved between fungal homologs of End3 (FH). Both hydrophilic mutants were generated to account for two possibilities: 1) the binding site was conserved as a repeating, identical motif within the End3 protein, or 2) the important residues have been conserved evolutionarily among fungal homologs of End3, but may not be identical on each repeat. Two types of hydrophobic mutants were also made in the a- or d- positions, one with residues mutated to alanine to remove the hydrophobic character (HΦA) and a second mutant with arginine insertions (HΦR) to add a bulkier, charged residue that would sterically interfere with a hydrophobic binding pocket. Figure 3A presents each of these mutants. The greek symbols used in this study represent the commonly accepted short-hand for hydrophilic (ζ) or hydrophobic (Φ) [18].
Characterization of End3 Repeat Mutants
Each of the four E3R mutants were introduced into both End3 Repeats within a single End3 protein and then assayed for temperature sensitivity when expressed in end3Δ cells (Figure 3B). The FH mutant was temperature resistant, indicating that the fungally conserved hydrophilic residues of the E3R was not important for growth at 38°C. The Hζ and HΦR mutants were unable to grow at 38°C, similar to empty vector in end3Δ cells, suggesting that these two mutants have disrupted an important End3 function, while the HΦA mutation is less disruptive. Single E3R mutants, where one repeat is wildtype while the other is mutant, did not exhibit temperature sensitivity, suggesting that a single E3R is sufficient for function (as also reported by the Riezman lab) [15] (Figure 3C).
To test stability of the mutant proteins at low as well as high temperature, we probed cell extracts grown at both 30°C and 37°C with an anti-End3 antibody (Supplemental Figure 1). We confirmed that the protein levels were within the linear range of the detection limits for this antibody. Wildtype and End3 mutant proteins were stable after one hr at 37°C, indicating that protein instability at high temperature does not contribute to the temperature sensitive growth of the E3R mutants. Hζ and HΦR showed reduced protein levels as compared to wildtype and HΦA, but had similar expression levels as compared to the single E3R mutants. This suggests that the temperature sensitivity of Hζ and HΦR mutants cannot be solely attributed to their slightly lowered expression levels. Instead, these mutants must have disrupted function, and likely have reduced binding to Pan1.
The FH mutant was not further analyzed since the strain behaved as wildtype, even though recombinant FH mutant protein was difficult to purify. The temperature resistant HΦA mutant was used as a control in all further assays.
end3Δ cells expressing high copy Hζ and HΦR were temperature resistant, suggesting that higher levels of mutant protein can overcome reduced Pan1 binding. We predicted that higher levels of Hζ and HΦR mutant protein might restore temperature resistance due to a bridging interaction between End3 and Pan1. Sla1 is the only known binding partner common to both End3 and Pan1, and previous studies have shown that the C-terminus of Sla1 associates with the N-termini of both Pan1 and End3 [6]. Since Pan1 and End3 bind to Sla1 with regions distinct from the Pan1-End3 interaction, it is possible that Sla1 forms a bridging interaction to bring Pan1 and End3 mutant proteins together that otherwise cannot directly associate with one another. A sla1ΔCt mutant was generated previously that displayed temperature resistance at elevated temperatures, so this strain background could be used to test for growth at 36°C [19]. A sla1ΔCt truncation lacking the entire C-terminal repeat region of Sla1 was generated in an end3Δ strain, and the high copy E3R mutants were tested for temperature resistance. We found that sla1ΔCt interfered with the temperature resistant phenotype of the high copy E3R mutants, while wildtype End3 protein remained temperature resistant (Figure 4). Thus, Sla1 is predicted to act as a bridge between End3 and Pan1 when they are unable to directly associate with one another. This bridging interaction may be facilitated by the high levels of mutant End3 protein.
Figure 4. sla1ΔCt Disrupts the Temperature Resistance of High Copy E3R Mutants.
Testing complementation of High Copy Hζ E3R in an end3Δsla1ΔCt strain. Serial dilutions were spotted onto selective -Ura medium and incubated at 30°C or 36°C for 3 days. Single copy wildtype End3 was chosen as a positive control in this assay to complement the end3Δ temperature sensitivity. HΦR high copy and single repeat mutants displayed identical growth as shown here for the Hζ mutant.
End3 Repeat Mutants have Reduced Binding to Pan1
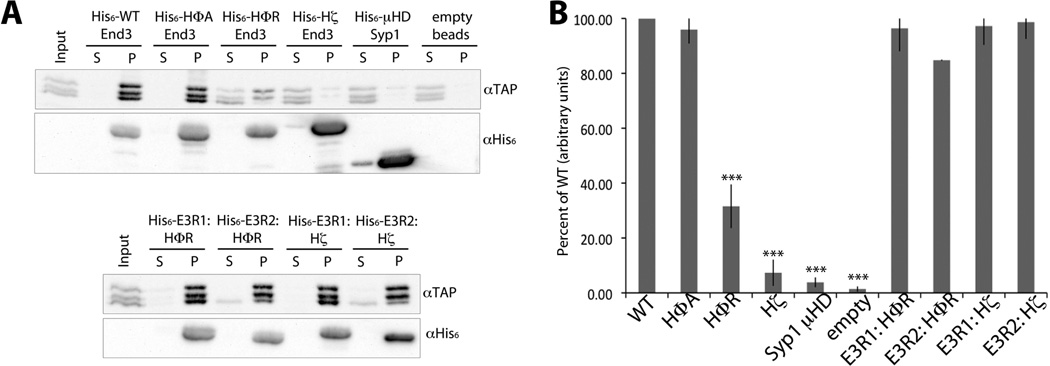
In order to directly test the ability of the E3R mutants to bind to the central region of Pan1, recombinant His-tagged End3 mutant proteins were immobilized on Talon beads and then incubated with purified TAP-tagged Pan1 aa687–1190. Negative controls were the Syp1 µ homology domain (µHD), an endocytic adaptor protein which does not bind to the central domain of Pan1, and empty Talon beads with no pre-bound protein [20]. As shown in Figure 5A, wildtype and HΦA End3 bound Pan1, and also depleted the unbound lysate sample. HΦR had markedly reduced binding to Pan1 (~ 70% less than WT) (Figure 5B), while Hζ did not bind Pan1, similar to Syp1 µHD and the empty bead controls.
Figure 5. End3 Repeat Mutants do not Bind Pan1.
A) Recombinant protein-binding assay with equivalent His6-tagged End3 wildtype or mutant protein bound to Talon sepharose beads, incubated with purified Pan1687–1190-TAP. Aliquots of each supernatant (S) and washed pellet (P) were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-TAP and anti-His6 antibodies, as indicated. B) Quantified percent Pan1-TAP bound in each pellet lane, relative to wildtype His6-End3. HΦR, Hζ, Syp1, and empty samples are significant (p<0.001) relative to wildtype End3. Values are presented as a mean ±SD (n=3).
As expected from their ability to rescue end3Δ temperature sensitive growth, the single E3R mutants showed similar levels of Pan1 association as wildtype. This confirmed that only a single wildtype End3 Repeat is needed to bind to Pan1, and there does not seem to be preference between the first and second repeats for this interaction in vitro.
The Pan1aa687–1190-TAP protein fragment was purified from yeast and showed significant N-terminal degradation (Figure 5). The protein appeared as a triplet with an intact C-terminal tag that was detectable by western analysis. Interestingly, the largest of the triplet bands showed a reproducible preference for binding to HΦR, suggesting that the smallest of the three bands may contain a poorly presented End3 binding site in the absence of more N-terminal protein sequence. This supports the hypothesis that the End3 binding site lies in the N-terminal portion of Pan1’s central region.
Pan1 is Hyperphosphorylated in E3R Mutant Strains
End3 is necessary to recruit Scd5, the targeting factor for the Glc7 phosphatase, to Pan1; thus, Pan1 is hyperphosphorylated in an end3Δ strain [13]. It follows that E3R mutants that are unable to bind Pan1 should accumulate hyperphosphorylated Pan1. To test this hypothesis, whole cell extracts were probed with a Pan1 antiserum. This experiment revealed a slower mobility band in the temperature sensitive Hζ and HΦR strains, similar to what is seen for end3Δ (Figure 6A). Since Prk1 is the major protein kinase that acts on Pan1 and has a close homolog Ark1, the double deletion control strain prk1Δark1Δ showed mostly dephosphorylated Pan1 protein. The single E3R mutants that bind Pan1 showed the expected result; Pan1 protein was dephosphorylated, similar to end3Δ strains complemented with wildtype End3.
Figure 6. Pan1 is Hyperphosphorylated in the E3R Mutants.
A) Immunoblots of cell extracts from wildtype cells, prk1Δark1Δ cells, and end3Δ with wildtype, empty and various E3R mutant vectors, probed with anti-Pan1 antibody. B) Immunoblots of cell extracts treated either with phosphatase inhibitors (−) or calf intestinal phosphatase (CIP) (+) for 30 min at 30°C before resolving by SDS-PAGE, transferring to nitrocellulose, and probing with anti-Pan1 antibody.
To confirm that these mobility shifts were due to phosphorylation rather than another post-translational modification, the extracts were treated with Calf Intestinal Phosphatase, resolved by SDS-PAGE, and again probed with anti-Pan1 serum (Figure 6B). All the slower mobility species collapsed to a single band that represents unphosphorylated Pan1 protein. We conclude that Pan1 is hyperphosphorylated in the Hζ and HΦR mutants strains because End3 cannot properly bind Pan1 to recruit the phosphatase complex.
E3R Mutant Strains Exhibit Reduced Endocytosis
Since the End3 Repeat mutants are unable to bind Pan1 and thus lead to a build up of hyperphosphorylated Pan1, it was expected that the efficiency of endocytosis of these strains would be impaired. In order to quantify the endocytic efficiency of the E3R mutant strains, the fluorescence intensity of the endocytic cargo Mup1-pHluorin was monitored over time [21]. When tagged with the pH-sensitive pHluorin protein, the fluorescence of this methionine permease accumulates at the cell surface under methionine starvation; upon methionine feeding, fluorescence quickly declines as the Mup1-pHluorin protein is internalized and quenched upon reaching the acidic vacuole. When this fluorescence is quantified over time, a wildtype strain shows ~80–90% decreased fluorescence within an hour (Figure 7A and B). The endocytic mutant strains end3Δ and end3-1 are less efficient at Mup1 internalization and only show ~30% reduction during the same time.
Figure 7. E3R Mutant Strains show Reduced Endocytic Efficiency.
A) Wildtype, end3Δ and end3-1 cells expressing Mup1-pHluorin were grown in the absence of methionine before addition of methionine at time zero. Total cellular fluorescence was measured at the given intervals. Fluorescence intensities were normalized to the first time-point of the series, and values are presented as a mean ±SD (n=10). B) Representative images of the change in fluorescence intensity quantified in 7A. Scale bar, 10 microns. C) Total cellular fluorescence after 45 min incubation with methionine, relative to fluorescence intensity at time zero. Supplemental Figure 2 includes representative images from this experiment. end3Δ cells with various E3R mutant vectors were quantified and compared to wildtype, end3Δ and end3-1 strains with empty pRS.416. The colored columns represent phenotypic classes that are statistically similar. The blue columns represent wildtype Mup1-pHluorin fluorescence intensity 45 minutes after methionine addition. The red columns represent defective Mup1 internalization which is significantly different (p<0.001) relative to the wildtype and complemented end3Δ strain (blue columns). The grey columns display an intermediate Mup1 internalization phenotype that is statistically significant (p<0.001) relative to the defective phenotypes (red columns) as well as the wildtype internalization (blue columns). All values are presented as a mean ±SD (n=60).
Like the end3Δ and end3-1 strains, HΦR and both low and high copy Hζ (red columns in Figure 7C) have similarly reduced endocytic Mup1 internalization (30–50% reduced from initial fluorescence intensity), suggesting that the function of the End3 Repeats contributes to the efficiency of endocytosis. Unlike previous assays, where HΦA and the single E3R mutants (grey columns in Figure 7C) showed wildtype phenotypes, this quantitative assay revealed an intermediate rescue of the endocytic efficiency in these mutants (~65% loss of Mup1-pHluorin fluorescence, compared to 85% for wildtype and END3 complemented end3Δ cells, blue columns). This suggests that while only a single E3R is needed to associate with Pan1 in vitro, both repeats may be required for full function in vivo. Since Pan1 and End3 are known to form a constitutive complex at a developing endocytic site, it is not surprising that specifically reducing this interaction reveals significant endocytic delays.
E3R Mutant Strains Display Increased Pan1-GFP Patch Lifetimes
Since the E3R mutants exhibited disrupted endocytic progression through the Mup1 experiment as well as disrupted Pan1 association, this suggested that Pan1 lifetime at the endocytic patch is likely also affected. The patch lifetime of genomically-expressed Pan1-GFP was calculated from various endΔ strains, and most of the strains showed patch lifetimes comparable to the wildtype strain (Figure 8, blue columns). HΦR, Hζ, and Hζ.426 displayed lengthened Pan1-GFP patch lifetimes, but none of these are as severe as seen with end3Δ strains expressing empty vector. This suggests that the N-terminus of End3 provides some temporal control of Pan1 patch lifetime, presumably through Sla1.
Figure 8. E3R Mutant Strains Display Increased Pan1-GFP Patch Lifetimes.
A) Quantified Pan1-GFP patch lifetimes from midlog cultures grown at 30°C and imaged at 30°C. The colored columns represent the same phenotypic classes discussed in Figure 7, with similar statistical significance. All values are presented as a mean ±SD (n=30). B) Representative kymograph images of wildtype, end3Δ, and end3Δ with the three mutant vectors (HΦR, Hζ, and Hζ.426) expressing GFP-tagged Pan1. Images were collected every 2.5 seconds for 72 frames (180 seconds total).
In agreement with the Mup1-pHluorin uptake experiment, the Hζ.426 strain was unable to restore Pan1-GFP patch lifetime to wildtype behavior. This is consistent with high copy Hζ plasmid not fully rescuing endocytosis (Figure 7C), even though the temperature-dependent growth is restored.
Discussion
The findings in this study contribute to a better understanding of how Pan1 and End3 bind to one another, which in turn yields a greater appreciation of the overall molecular mechanisms that underlie endocytosis. This interaction was first identified and a coarse-scale map of the binding sites was determined through a directed yeast two-hybrid analysis [14]. This approach led to some incorrect conclusions about the binding interfaces; most notably, that the second long repeat (LR2) of Pan1 binds to End3. This was determined based on the abolished interaction when the N-terminal portion of LR2 was removed from the Y2H construct. This conclusion was also supported by the finding that a construct containing the entire Pan1 coiled-coil plus its C-terminus was unable to bind to End3 in the Y2H experiment. Based on the data shown in our study here, it is unclear why the coiled-coil plus C-terminus construct of Pan1 that contained the binding site for End3 we defined did not yield a positive interaction in the prior study. A second incorrect conclusion from the prior yeast two-hybrid analysis was that both End3 Repeats are required to bind Pan1, as it was reported that removing the first E3R abolished the interaction. This finding has now been directly tested in vitro through a recombinant pulldown experiment with End3 fragments, as well as through mutational analysis of E3R1 or E3R2. Both of these experiments confirm that the End3 Repeats are functionally redundant binding sites for Pan1 and that either one can contribute to the interaction. One explanation for the differences in the results in the first analysis is that the protein expression could have been unreliable among the various yeast two-hybrid constructs tested, or perhaps the truncations produced poorly folded proteins that were unable to form functional binding sites.
Binding Partners for the Central Region of Pan1
Our Y2H assay using the central coiled-coil region of Pan1 revealed novel potential binding partners. The first was the conserved C-terminal domain of Bbc1, a protein that regulates Las17-dependent actin nucleation and colocalizes with Myo3 and Myo5 [22,23]. A second novel binding partner was the auto-inhibitory domain of Cna1, the catalytic subunit of yeast calcineurin, a Ca2+/Calmodulin-dependent phosphatase, suggesting that Cna1 may act as a phosphatase for Pan1; however, our preliminary attempts to test this idea did not support it [24] (data not shown). The final novel binding partner was the C-terminal coiled-coils of Kel2, a Kelch-repeat protein that is implicated in cell polarity [25]. A similar interaction was observed between Kel1 and End3 coiled-coils, indicating that a complex of Pan1-End3 and the Kelch proteins may have an unidentified function [26]. These new potential binding partners suggest that the central domain of Pan1 is not simply a flexible linker between its functionally distinct N- and C-termini, but that this region contains binding sites that may contribute to endocytosis as well as other, unexplored functions for Pan1.
The End3 Binding Site on Pan1 is Complex
We confirmed that End3 does not bind Pan1 LR2 region. Data provided in Table 1, along with our recombinant binding experiments, confirm that the binding site for End3 lies within aa687–846. Identification of the precise residues that interact with End3’s E3Rs will require significantly more work. Our preliminary evidence supports the idea that at least two binding sites exist (KW and BW, unpublished).
The Hydrophilic face of the E3Rs Represent the Pan1 Binding Site
In an effort to define the residues within the E3Rs responsible for binding to Pan1, two mutants have reduced (HΦR) or abolished (Hζ) binding to Pan1. Since these mutations lie on opposite faces of the predicted coiled-coil of the E3Rs, this raises the question: which face represents the binding site for Pan1? The Hζ mutant shows a complete loss of Pan1 binding, confirming that this face of the coiled-coil E3R contains the Pan1 binding site, suggesting that these hydrophilic residues show high similarity between the two End3 Repeats because there is selective pressure to maintain both Pan1 binding sites. The HΦR mutant, on the other hand, was still able to bind to Pan1 with about 70% reduced affinity relative to wildtype. This suggests that the binding site is still intact in HΦR but has lowered affinity due to the mutations on the hydrophobic face of the helix, perhaps caused by allosteric changes or altered flexibility of the coiled-coil that change the Pan1 binding pocket. It is also possible that an intramolecular interaction between the hydrophobic faces of the E3Rs could contribute to the proper presentation of the Pan1 binding sites on the hydrophilic faces of the coiled-coil. The HΦR mutant would likely be more disruptive of such an intramolecular association than the HΦA mutant, and would also be more likely to induce allosteric structural changes in the shape of the coiled-coil and the Pan1 binding site. Due to these changes, the reduced binding of HΦR to Pan1 still correlated with strong phenotypes, similar to the Hζ mutant.
The E3Rs are Required for Phosphoregulation of Pan1
Support for the phosphoregulation of Pan1 being the function of End3 comes from genetic analyses that end3Δ temperature sensitivity can be rescued by deleting the kinase Prk1 [11]. End3 is thought to control Pan1 activity by recruiting Scd5 and the Glc7 phosphatase [13]. The dephosphorylation of Pan1 is dependent upon an End3-Pan1 interaction via the End3 Repeats, as the E3R mutant cells accumulate phosphorylated Pan1. Only when E3R mutant proteins are overexpressed is Pan1 found as a primarily dephosphorylated species, likely due to the overabundance of available End3-Scd5 complexes in the vicinity of Pan1. Since Sla1 can act as a bridge between Pan1 and End3, this Sla1 bridging factor could also bring the End3-Scd5 complexes in close proximity to Pan1.
end3-1 Encodes a Truncated Protein Lacking the E3Rs
Our sequence analysis of the original end3-1 mutant strain shows that this allele does not contain End3 Repeat motifs. As predicted, the end3-1 protein is unable to bind Pan1 in a recombinant pulldown assay. This information allows us to now re-evaluate the mechanism contributing to the phenotypes of the end3-1 strain. This mutant, among the first characterized endocytosis mutants in yeast, has been widely used due to its endocytic deficiency at low temperatures (24°C) as well as depolarized actin cytoskeleton and temperature sensitivity at 37°C, and it appears to act like an end3Δ mutant [15,27–45]. The mechanism of the end3-1 allele phenotype is most likely explained by this mutant protein being unable to bind Pan1 and properly regulate its phosphorylation and activity, thus leading to deficiencies in endocytosis and the actin cytoskeleton.
The E3Rs are Functionally Redundant Binding Sites
Our data suggest that the End3 Repeats are redundant binding sites for Pan1. Based upon our recombinant pulldown experiments with End3 truncations and the single E3R mutants, we can conclude that these repeats are each capable of binding to Pan1, with no observable preference for E3R1 or E3R2 when tested in the context of full length End3 protein. The Hζ and HΦR E3R mutants showed either abolished or reduced ability to bind Pan1 in vitro, respectively, and were unable to restore temperature resistance in an end3Δ strain. In agreement with the in vitro binding data, these mutants are also unable to mediate proper Pan1 dephosphorylation in vivo [13].
Single E3R mutants, on the other hand, are able to bind to Pan1 and complement the temperature sensitivity and Pan1 hyperphosphorylation phenotypes of end3Δ. This agrees with previously published data that a single E3R is necessary for full End3 function, as measured by growth at 37°C and endocytosis at 24°C [15]. The Hζ and HΦR mutants showed reduced endocytic rates that were very similar to end3Δ and end3-1 cells, while the single E3R mutants showed an intermediate rescue of Mup1-pHluorin internalization. This suggests that there might be a requirement for both repeats for normal in vivo function, in spite of a single E3R being sufficient for binding Pan1 in vitro.
Why does End3 contain two, redundant binding sites for Pan1? There must be an advantage that two repeats provide to a yeast cell. The presence of duplicate, conserved motifs/domains is seen throughout fungal species, and there is also selective pressure for these motifs/domains to retain identical residues between them. One possible advantage is that a single End3 protein can bind to more than one Pan1 protein at a time. However, this does not seem likely, as Pan1 and End3 form a 1:1 stoichiometric complex when co-purified from yeast lysates, as well as when recombinantly purified and then mixed in vitro [7] (unpublished data). However, it is possible that End3 uses one E3R to form a stable complex with Pan1 that cycles on and off the membrane while the other E3R remains free for a second, independent function. It is also possible that there are other binding partners for the C-terminus of End3 that have not yet been characterized that contribute to the in vivo endocytic delay observed in single E3R mutants. If there are other, significant interactions that End3 must mediate with these repeats, then perhaps having two repeats allows End3 to overcome the competition of two distinct proteins binding to the same site. Alternatively, perhaps the efficient recruitment and complex molecular structure of the various proteins at the growing endocytic site depend on the presence of two tandem End3 Repeats that can mediate interactions with Pan1. It is possible that these repeats increase the local avidity between Pan1 and End3.
Based on studies in fission yeast, we know that hundreds of individual endocytic proteins are recruited to a developing endocytic patch, but we have little understanding of how these proteins pack together into a functional coat that coordinates cargo selection, membrane deformation, and inward movement of a coated pit [1]. The fact that End3 has redundant binding sites may allow it to form stable, discrete complexes that aid in the formation of large protein networks at an endocytic site.
This work contributes to the understanding of how two endocytic proteins interact, but it also raises more questions about the architecture and organization of an endocytic patch. Due to the restricted environment in the vicinity of a clathrin-coated pit, are specific protein-protein interactions required, or is a high local concentration of binding partners sufficient for proper activity? Are multiple, redundant binding sites used to gather smaller protein complexes into an overlapping meshwork of protein interactions to promote vesicle internalization? Ongoing studies will eventually address these remaining questions.
Materials and Methods
Strains, plasmids and general methods
The yeast strains and plasmids used in this study are listed in Tables 2 and 3.
Table 2.
Yeast Strains Used
| Strain | Genotype | Source |
|---|---|---|
| AH109 | Yeast two-hybrid screen strain | ClonTech |
| SFY526 | lacZ Reporter Strain | ClonTech |
| DDY1810 | MATa leu2-3,112 trp1-Δ901 ura3-52 prb1-1122 pep4-3 prc1-407 gal2 | Drubin Lab |
| SEY6210 | his3-Δ200 trp1-Δ901 leu2-3, 112 ura3-52 lys2-801 suc2-Δ9 BAR1 | Laboratory strain |
| BWY1346 | MATα trp1 leu2 ura3 lys2 end3-1 | [23] |
| BWY1830 | MATα ark1 Δprkl Δ | Laboratory strain |
| BWY2068 | MATα his3-Δ200 trp 1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 end3∷G418 | This Study |
| BWY2459 | MATα his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 pan 1-GFP∷G418 | This Study |
| BWY4255 | MATa his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 MUP1-pHluorin∷NAT | This Study |
| BWY4847 | MATa his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 end3∷G418 pan1-GFP∷G418 | This Study |
| BWY4849 | MATa his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 end3∷G418 Mup1-pHluorin∷NAT | This Study |
| BWY4853 | MATa trp1 leu2 ura3 lys2 end3-1 Mup1-pHluorin∷NAT | This Study |
| BWY5370 | MATa his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 end3∷G418 Mup1-pHluorin∷NAT SLA1 ΔCterm∷HIS (aa1-855 expressed) | This Study |
| BWY5376 | MATa his3-Δ200 trp1-Δ901 leu2-3,112 ura3-52 lys2-801 suc2-Δ9 BAR1 Mup1-pHluorin∷NAT SLA1ΔCterm∷HIS (aa1-855 expressed) | This Study |
Table 3.
Plasmids Used
| Plasmid | Description | Details | Source |
|---|---|---|---|
| pRS.416 | CEN, URA3 | Laboratory plasmid | |
| pRS.426 | 2µ, URA3 | Laboratory plasmid | |
| pBW0622 | GST-PAN1 CC | pEGKT∷GST-PAN1, aa792–1252 | Laboratory plasmid |
| pBW0623 | DBD-PAN1792–948 | pGBKT7∷Gal4 DBD-PAN1, aa 792–948 | Laboratory plasmid |
| pBW0910 | END3 | pRS.416∷END3 | Laboratory plasmid |
| pBW0923 | GST | pEGKT∷GST (CEN, URA3) | Laboratory plasmid |
| pBW1505 | HIS6-SYP1 µHD | pET28 CEN/URA∷HIS6-SYP1 µHD | Laboratory plasmid |
| pBW1649 | PAN1687–1190-TAP | pEGKT∷PAN1-TAP, aa687–1190 | Laboratory plasmid |
| pBW1659 | pGST-PAN1 LR2 | pEGKT∷GST-PAN1, aa360–690 | Laboratory plasmid |
| pBW1673 | DBD-PAN1687–1190 | pGBKT7∷Gal4 DBD-PAN1, aa687–1190 | Laboratory plasmid |
| pBW1680 | AD-BBC1 | pACTII∷Gal4 AD-BBC1, aa1096–1157 | This Study |
| pBW1681 | AD-CNA1 | pACTII∷Gal4 AD-CNA1 aa428–493 | This Study |
| pBW1684 | AD-END3 | pACTII∷Gal4 AD-END3, aa176–349 | This Study |
| pBW1685 | AD-KEL2 | pACTII∷Gal4 AD-KEL2, aa605–748 | This Study |
| pBW1858 | DBD-PAN1687–846 | pGBKT7∷Gal4 DBD-PAN1, aa687–846 | This Study |
| pBW1891 | HIS6-END3176–349 | pET28 CEN/URA∷HIS6-END3, aa176–349 | This Study |
| pBW1892 | HIS6-END3176–303 | pET28 CEN/URA∷HIS6-END3 aa176–303 | This Study |
| pBW1893 | HIS6-END3228–349 | pET28 CEN/URA∷HIS6-END3 aa228–349 | This Study |
| pBW2032 | HIS6-HΦA | pET28 CEN/URA∷HIS6-END3 HΦA | This Study |
| pBW2034 | HIS6-HΦR | pET28 CEN/URA∷HIS6-END3 HΦR | This Study |
| pBW2036 | HIS6-Hζ | pET28 CEN/URA∷HIS6-END3 Hζ | This Study |
| pBW2038 | HIS6-FH | pET28 CEN/URA∷HIS6-END3 FH | This Study |
| pBW2039 | HΦA | pRS.416∷END3 HΦA | This Study |
| pBW2040 | HΦR | pRS.416∷END3 HΦR | This Study |
| pBW2041 | Hζ | pRS.416∷END3 Hζ | This Study |
| pBW2042 | FH | pRS.416∷END3 FH | This Study |
| pBW2044 | HIS6-END31–349 | pET28 CEN/URA∷HIS6-END3, aa1–349 | This Study |
| pBW2046 | HIS6end3-1 | pET28 CEN/URA∷HIS6-end3–1 | This Study |
| pBW2199 | HIS6-E3R1:HΦR | pET28 CEN/URA∷HIS6-END3 E3R1:HΦR | This Study |
| pBW2201 | HIS6-E3R2:HΦR | pET28 CEN/URA∷HIS6-END3 E3R2:HΦR | This Study |
| pBW2203 | HIS6-E3R1:Hζ | pET28 CEN/URA∷HIS6-END3 E3R2:Hζ | This Study |
| pBW2205 | HIS6-E3R2:Hζ | pET28 CEN/URA∷HIS6-END3 E3R2:Hζ | This Study |
| pBW2207 | Hζ426 | pRS.426∷END3 Hζ | This Study |
| pBW2208 | E3R1:HΦR | pRS.416∷END3 E3R1:HΦR | This Study |
| pBW2209 | E3R2:HΦR | pRS.416∷END3 E3R2:HΦR | This Study |
| pBW2210 | E3R1:Hζ | pRS.416∷END3 E3R1:Hζ | This Study |
| pBW2211 | E3R2:Hζ | pRS.416∷END3 E3R2:Hζ | This Study |
| pBW2212 | HIS6-END3304–349 | pET28 CEN/URA∷HIS6-END3, aa304–349 | This Study |
| pBW2270 | HΦR.426 | pRS.426∷END3 HΦR | This Study |
| pBW2271 | E3R1:HΦR.426 | pRS.426∷END3 E3R1:HΦR | This Study |
| pBW2272 | E3R2:HΦR.426 | pRS.426∷END3 E3R2:HΦR | This Study |
| pBW2273 | E3R1:Hζ426 | pRS.426∷END3 E3R1:Hζ | This Study |
| pBW2274 | E3R2:Hζ426 | pRS.426∷END3 E3R2:Hζ | This Study |
Genomic deletions or integrations were generated by chromosomal integration of PCR products (Longtine et al., 1998). Amino acid substitutions for E3R mutants were made using homologous recombination with oligonucleotides from Operon (Huntsville, AL).
Yeast two-hybrid screening and quantitative β-galactosidase analysis
A two-hybrid screen was performed by transforming the FRYL library (Fromont-Racine et al., 1997) into AH109 cells (Clontech, Mountain View, CA) containing pBW623, a plasmid of Gal4-DNA-binding domain fusion of Pan1 (aa 792–948), and grown on YNB-TRP-LEU-HIS-ADE media. From ~750,000 transformants, 32 positive clones were obtained, and the activation domain library plasmid was rescued and confirmed by retransforming into AH109 with pBW623 before being sequenced. For quantitative analysis, SFY526 cells transformed with pBW623 and the FRYL prey plasmid (or empty, negative control vectors pACT2 and pGBKT7) were grown in selective liquid culture to mid-log phase and assayed for the expression of Galp-LacZ as described previously (Jarvis et al., 1988). Each value represents the average and SD for three independent quantifications.
Protein purification
His6-tagged proteins were purified from Rosetta cells (Novagen, Darmstadt, Germany) transformed with pET28a-derived plasmids. Cells were induced with 0.5 mM isopropyl β-D-thiogalactoside (IPTG) for 5 hours at 30°C, harvested, frozen (−80°C), and resuspended in 20 mM Tris Base pH 7.5, 200 mM NaCl 0.1% Tween 20 (with AEBSF (0.1 mM) and complete EDTA-free protease inhibitor cocktail (Roche, Pleasanton, CA)). Lysates produced by lysozyme treatment and sonication were centrifuged at 4°C for 20 min at 26,000 g. Protein lysates were incubated with Talon metal affinity resin (Clontech, Mountain View, CA) with 5mM imidazole for 2 h at 4°C. The beads were washed with increasing concentrations of imidazole (15–30mM) and proteins eluted with 500mM imidazole in 20mM Tris Base pH 7.5, 200mM NaCl. As necessary, purified proteins were concentrated using 10 000 or 30 000 MWCO Amicon Ultra centrifugal filter devices (Millipore, Billerica, MA), or buffer exchanged using protein desalting spin columns (Pierce, Rockford, IL).
GST- and TAP-tagged proteins were purified from yeast DDY1810 cells (Drubin laboratory) transformed with pEGKT-derived plamids. Cells were induced grown for two nights in one liter volume in synthetic media minus Leu and Ura (for plasmid selection) and 2% raffinose as the sole carbon source. Cells were induced with 2% Galactose, 10g peptone and 20g yeast extract overnight, harvested, frozen (−80°C), and resuspended in 20 mM Tris Base pH 7.5, 200 mM NaCl 0.1% Tween 20 (with AEBSF (0.1 mM) and complete EDTA-free protease inhibitor cocktail (Roche, Pleasanton, CA)). Lysates produced by sonication were centrifuged at 4°C for 20 min at 26,000 g. TAP-tagged protein lysates were incubated with Calmodulin-affinity sepharose beads with 4mM CaCl2 for approximately 16 hours at 4°C. The beads were washed with buffer prior to elutions with 5mM EGTA in 20mM Tris Base pH 7.5, 200mM NaCl. GST-tagged protein lysates were incubated with glutathione agarose beads (Molecular Probes, Grand Island, NY) for approximately 16 hours at 4°C. As necessary, purified proteins were concentrated using 10 000 or 30 000 MWCO Amicon Ultra centrifugal filter devices (Millipore), or buffer exchanged using protein desalting spin columns (Pierce).
Whole cell extracts were prepared by TCA precipitation. Cells grown to mid-logarithmic phase were incubated with 10% TCA on ice for 10 minutes, washed with 4°C acetone, and then lysed with 0.4–0.6um glass beads in the presence of protein sample buffer. Total protein lysate was resolved by SDS–PAGE and detected by western immunoblotting with rabbit anti-End3, rabbit anti-actin, or and rabbit anti-Pan1 antibodies.
Recombinant binding assays
For Figure 1, GST, GST-LR2, or GST-CC were bound to glutathione agarose beads (Molecular probes), then incubated with Rosetta protein lysates expressing His6-tagged Full length End3 protein at 4°C for 2 hours. For Figure 2, empty TAP or Pan1aa687–1190 (pBW1649) were bound to Calmoduline-sepharose beads, then incubated with Rosetta protein lysates expressing His6-tagged End3 truncation protein at 4°C for 2 hours. For Figure 2–4 and Figure 5, approximately 300ng of His6-End3 (Wildtype or E3R mutants), His6-Syp1 µHD, or buffer alone were bound to Talon metal affinity resin (Clontech, Mountain View, CA), then incubated with purified Pan1aa687–1190-TAP (pBW1649) at 4°C for 2 hours. After sedimenting the beads, an aliquot of supernatant was resuspended in SDS-PAGE sample buffer, and the beads were washed and then resuspended in SDS-PAGE sample buffer. Supernatant and pellet fraction were resolved by SDS–PAGE, transferred to nitrocellulose, and immunoblotted with mouse anti-His6 antibody (Clontech, Mountain View, CA) or Rabbit anti-TAP antibody (Pierce, Rockford, IL). Horseradish peroxidase secondary antibodies and the Supersignal Chemiluminescent detection system (Pierce, Rockford, IL) was used for visualization. Images were quantified using Image J v1.41n software.
Calf Intestinal Phosphatase treatment
Five OD600 of cells were harvested and lysed as described above using TCA. To test for phosphate modifications of Pan1, total was TCA pellets were resuspended in phosphatase buffer (0.01 M Tris, 0.01 M MgCl2, 1 M NaCl, 1 mM DTT, 0.1 mM AEBSF), and incubated with or without 1ul of Calf Intestinal Protein Phosphatase (CIP) (New England Biolabs, Beverly, MA). Two microliters of phosphatase inhibitors (250 mM NaF, 10 mM EDTA, 4 mM Na-orthovanadate, 0.2 mM cyclosporin, 2 mM AEBSF) were added to appropriate control tubes without CIP, and samples were incubated at 30°C for 30 min. Samples were mixed with SDS-PAGE protein sample buffer, resolved by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with rabbit anti-Pan1 antibody.
Fluorescence microscopy
Mup1-pHluorin images were collected using a Zeiss Axiovert 135TV inverted microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY) with a Sensicam QE CCD camera (Cooke, Romulus, MI), Zeiss 100× 1.4 NA Plan-Apochromat objective, motorized filter wheels, fluorescein isothiocyanate (FITC) and Texas Red filter sets (Semrock, Rochester, NY), and Slidebook 4.2 software (Intelligent Imaging Innovations, Denver, CO). Images were captured with 500ms exposure, identical binning, intensification and illumination intensity. For quantification of fluorescence intensity, 16-bit image files were analyzed in Image J v1.41n. Background subtraction was performed before measurement of integrated density, and values were corrected for cell size.
To perform kinetic analysis of endocytosis, cells expressing Mup1-pHluorin were grown overnight in synthetic YNB medium lacking methionine (YNB −Met). Cells were then diluted to a density of 0.3–0.4 OD/mL in YNB −Met, and were grown to a density of 0.7–0.8 OD/mL. Cells were then seeded onto concanavalin A-coated 8-well glass-bottomed chamber slides (LabTek, Scotts Valley, CA) containing YNB −Met and were allowed to settle before imaging. Immediately before imaging, methionine was added to a concentration of 20 µg/mL, and images were then captured at 5-min intervals for 45 min. During image acquisition, cells were maintained at a constant temperature of 30°C.
Pan1-GFP and end3ΔPan1-GFP strains were transformed with empty pRS.416 and/or End3 plasmids for quantification of Pan1 lifetimes. Strains were grown to midlog at 30°C and imaged at 30°C in an environmental chamber on the Zeiss Axiovert 135TV inverted microscope. Movies were captures with 50ms exposure, 2.5seconds/frame for 72 frames. Lifetimes of 30 individual patches were quantified for each strain and averaged together. Representative kymographs were generated with Image J v1.41n following background subtraction.
Statistical analysis
Statistical significance between populations was determined by one-way ANOVA followed by Tukey’s Multiple Comparison post hoc analysis.
Supplementary Material
Synposis.
Pan1 is an endocytic scaffold that interacts with structural and regulatory endocytic components. Pan1 is composed of Eps15 Homology (EH) domains that bind adaptor proteins, a central oligomerization domain and C-terminal binding sites for Arp2/3, F-actin, and type-I myosin motors. We show that the central domain of Pan1 binds its constitutive partner End3, another EH domain protein, via the two C-terminal End3 Repeats (which are absent in end3-1). The conserved hydrophilic faces of the End3 Repeats mediate Pan1 binding.
Acknowledgements
We would like to thank Howard Riezman for providing anti-End3 serum, Nathan Wright for technical support, and members of the Wendland laboratory for helpful discussions and comments. All microscopy was performed at the JHU Integrated Imaging Center. This work was funded by a grant from the National Institutes of Health (to B.W., NIH RO1 GM60979) and KW, MKB and NC were in part supported by an NIH T32 Training Grant (T32 007231-37).
Footnotes
The authors have no conflict of interest to declare.
References
- 1.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol Biol Cell. 2010;21:2894–2904. doi: 10.1091/mbc.E10-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miliaras NB, Wendland B. EH proteins: multivalent regulators of endocytosis (and other pathways) Cell Biochem Biophys. 2004;41:295–318. doi: 10.1385/CBB:41:2:295. [DOI] [PubMed] [Google Scholar]
- 3.Tonikian R, Xin X, Toret CP, Gfeller D, Landgraf C, Panni S, Paoluzi S, Castagnoli L, Currell B, Seshagiri S, Yu H, Winsor B, Vidal M, Gerstein MB, Bader GD, Volkmer R, Cesareni G, Drubin DG, Kim PM, Sidhu SS, Boone C. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 2009;7:e1000218. doi: 10.1371/journal.pbio.1000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voss H, Tamames J, Teodoru C, Valencia A, Sensen C, Wiemann S, Schwager C, Zimmermann J, Sander C, Ansorge W. Nucleotide sequence and analysis of the centromeric region of yeast chromosome IX. Yeast. 1995;11:61–78. doi: 10.1002/yea.320110109. [DOI] [PubMed] [Google Scholar]
- 5.Wendland B, Emr S. Pan1p, Yeast eps15, Functions as a Multivalent Adaptor That Coordinates Protein–Protein Interactions Essential for Endocytosis. The Journal of Cell Biology. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang HY, Xu J, Cai M. Pan1p, End3p, and Sla1p, Three Yeast Proteins Required for Normal Cortical Actin Cytoskeleton Organization, Associate with Each Other and Play Essential Roles in Cell Wall Morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toshima J, Toshima JY, Duncan MC, Cope MJTV, Sun Y, Martin AC, Anderson S, Yates JR, Mizuno K, Drubin DG. Negative regulation of yeast Eps15-like Arp2/3 complex activator, Pan1p, by the Hip1R-related protein, Sla2p, during endocytosis. Mol Biol Cell. 2007;18:658–668. doi: 10.1091/mbc.E06-09-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 9.Toshima J, Toshima JY, Martin AC, Drubin DG. Phosphoregulation of Arp2/3-dependent actin assembly during receptor-mediated endocytosis. Nat Cell Biol. 2005;7:246–254. doi: 10.1038/ncb1229. [DOI] [PubMed] [Google Scholar]
- 10.Barker SL, Lee L, Pierce BD, Maldonado-Báez L, Drubin DG, Wendland B. Interaction of the endocytic scaffold protein Pan1 with the type I myosins contributes to the late stages of endocytosis. Mol Biol Cell. 2007;18:2893–2903. doi: 10.1091/mbc.E07-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng G, Cai M. Regulation of the Actin Cytoskeleton Organization in Yeast by a Novel Serine/Threonine Kinase Prk1p. The Journal of Cell Biology. 1999;144:71–82. doi: 10.1083/jcb.144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng G, Yu X, Cai M. Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol Biol Cell. 2001;12:3759–3772. doi: 10.1091/mbc.12.12.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng G, Huang B, Neo SP, Wang J, Cai M. Scd5p mediates phosphoregulation of actin and endocytosis by the type 1 phosphatase Glc7p in yeast. Mol Biol Cell. 2007;18:4885–4898. doi: 10.1091/mbc.E07-06-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang HY, Munn A, Cai M. EH domain proteins Pan1p and End3p are components of a complex that plays a dual role in organization of the cortical actin cytoskeleton and endocytosis in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4294–4304. doi: 10.1128/mcb.17.8.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bénédetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sachs A, Deardorff J. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 17.Rohrer J, Bénédetti H, Zanolari B, Riezman H. Identification of a novel sequence mediating regulated endocytosis of the G protein-coupled alpha-pheromone receptor in yeast. Mol Biol Cell. 1993;4:511–521. doi: 10.1091/mbc.4.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aasland R, Abrams C, Ampe C, Ball LJ, Bedford MT, Cesareni G, Gimona M, Hurley JH, Jarchau T, Lehto VP, Lemmon MA, Linding R, Mayer BJ, Nagai M, Sudol M, Walter U, Winder SJ. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513:141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- 19.Gourlay C, Ayscough K. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- 20.Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J. 2009;28:3103–3116. doi: 10.1038/emboj.2009.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prosser DC, Whitworth K, Wendland B. Quantitative analysis of endocytosis with cytoplasmic pHluorin chimeras. Traffic. 2010;11:1141–1150. doi: 10.1111/j.1600-0854.2010.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- 23.Mochida J, Yamamoto T, Fujimura-Kamada K, Tanaka K. The novel adaptor protein, Mti1p, and Vrp1p, a homolog of Wiskott-Aldrich syndrome protein-interacting protein (WIP), may antagonistically regulate type I myosins in Saccharomyces cerevisiae. Genetics. 2002;160:923–934. doi: 10.1093/genetics/160.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cyert MS, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci USA. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philips J, Herskowitz I. Identification of Kel1p, a kelch domain-containing protein involved in cell fusion and morphology in Saccharomyces cerevisiae. The Journal of Cell Biology. 1998;143:375–389. doi: 10.1083/jcb.143.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman JR, Wolf E, Kim PS. A computationally directed screen identifying interacting coiled coils from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:13203–13208. doi: 10.1073/pnas.97.24.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau V, Galan JM, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. The Journal of Cell Biology. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horazdovsky BF, Busch GR, Emr SD. VPS21 encodes a rab5-like GTP binding protein that is required for the sorting of yeast vacuolar proteins. EMBO J. 1994;13:1297. doi: 10.1002/j.1460-2075.1994.tb06382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang A. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1 prevents mislocalization of mutant ATPase to the vacuole. The Journal of Cell Biology. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vida TA. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. The Journal of Cell Biology. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan P, Howard J, Payne G. The sequence NPFXD defines a new class of endocytosis signal in Saccharomyces cerevisiae. The Journal of Cell Biology. 1996;135:1789–1800. doi: 10.1083/jcb.135.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuang JS, Schekman RW. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. The Journal of Cell Biology. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayscough K, Stryker J, Pokala N. High Rates of Actin Filament Turnover in Budding Yeast and Roles for Actin in Establishment and Maintenance of Cell Polarity Revealed Using the Actin Inhibitor Latrunculin-A. The Journal of Cell Biology. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowles CR. Novel Golgi to vacuole delivery pathway in yeast: identification of a sorting determinant and required transport component. EMBO J. 1997;16:2769–2782. doi: 10.1093/emboj/16.10.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wemmie JA, Moye-Rowley WS. Mutational analysis of the Saccharomyces cerevisiaeATP-binding cassette transporter protein Ycf1p. Molecular Microbiology. 1997;25:683–694. doi: 10.1046/j.1365-2958.1997.5061868.x. [DOI] [PubMed] [Google Scholar]
- 37.Naqvi S, Zahn R, Mitchell D, Stevenson B, Munn A. The WASp homologue Las17p functions with the WIP homologue End5p/verprolin and is essential for endocytosis in yeast. Current Biology. 1998;8:959–962. doi: 10.1016/s0960-9822(98)70396-3. [DOI] [PubMed] [Google Scholar]
- 38.Swaminathan S, Amerik A, Hochstrasser M. The Doa4 Deubiquitinating Enzyme Is Required for Ubiquitin Homeostasis in Yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kruckeberg AL, Ye L, Berden JA, Van Dam K. Functional expression, quantification and cellular localization of the Hxt2 hexose transporter of Saccharomyces cerevisiae tagged with the green fluorescent protein. Biochemical Journal. 1999;339:299. [PMC free article] [PubMed] [Google Scholar]
- 40.Gagny B, Wiederkehr A, Dumoulin P, Winsor B, Riezman H, Haguenauer-Tsapis R. A novel EH domain protein of Saccharomyces cerevisiae, Ede1p, involved in endocytosis. J Cell Sci. 2000;113:3309–3319. doi: 10.1242/jcs.113.18.3309. [DOI] [PubMed] [Google Scholar]
- 41.Shih SC, Sloper-Mould KE, Hicke L. Monoubiquitin carries a novel internalization signal that is appended to activated receptors. EMBO J. 2000;19:187–198. doi: 10.1093/emboj/19.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaminska J, Gajewska B, Hopper A. Rsp5p, a New Link between the Actin Cytoskeleton and Endocytosis in the Yeast Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6946–6958. doi: 10.1128/MCB.22.20.6946-6958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soulet D, Covassin L, Kaouass M, Charest-Gaudreault R, Audette M, Poulin R. Role of endocytosis in the internalization of spermidine-C2-BODIPY, a highly fluorescent probe of polyamine transport. Biochem J. 2002;367:347. doi: 10.1042/BJ20020764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gourlay C, Ayscough K. Identification of an upstream regulatory pathway controlling actin-mediated apoptosis in yeast. J Cell Sci. 2005;118:2119–2132. doi: 10.1242/jcs.02337. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Chang A. Quality control of a mutant plasma membrane ATPase: ubiquitylation prevents cell-surface stability. J Cell Sci. 2006;119:360–369. doi: 10.1242/jcs.02749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.