Abstract
BMP4 is one of the transforming growth factor-β superfamily, which can participate in adipogenesis. Gene encoding BMP4 is acknowledged as a convincing candidate that may contribute to both glucose and lipid metabolism. In this paper, we aimed to test the impacts of BMP4 variants on type 2 diabetes in a large sample of Chinese population. We genotyped 10 tagging single nucleotide polymorphisms within the BMP4 region in 6822 participants and acquired detailed clinical investigations and biochemistry measurements. We found that BMP4 rs8014363 showed nominal association towards type 2 diabetes, with the T allele conferring a high risk of type 2 diabetes (OR = 1.108, 95%CI 0.999–1.229, P = 0.051 for allele; OR = 1.110, 95%CI 1.000–1.231, P = 0.050 for genotype), but it was no longer statistically significant after adjusting for multiple testing (empirical P = 0.3689 for allele based on 10,000 permutations). Moreover, we observed a significant association of rs8014363 with triglyceride level and a trend towards association with high-density lipoprotein cholesterol after adjusting for age, gender, and BMI (P = 0.035 and 0.068, resp.). Our data suggested that the genetic variants of BMP4 may not play a dominant role in glucose metabolism in Chinese Han population, but a minor effect cannot be ignored.
1. Background
According to the IDF Diabetes Atlas 2012, there are more than 371 million diabetic patients worldwide and 4.8 million patients died due to diabetes [1]. With the staggering increase of diabetes pandemic creating an overwhelming array of serious complications and high mortality rate, exploring the etiology behind diabetes is of great essentiality. Although environmental factors contribute significantly to diabetes, it is generally considered that the importance of genetic factors cannot be ignored as well [2, 3]. Recently along with the powerful genome-wide association study, the candidate gene approach can also guide a better understanding of the pathophysiology of complex diseases. Up to now, more than 60 loci have been confirmed to confer susceptibility to type 2 diabetes [4, 5] however, they are still not enough to interpret the genetic mechanism of the disease. As a consequence, it is worthy for further research.
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-β superfamily, which is now acknowledged to be involved in regulating embryonic development and differentiation as well as cellular function [6–8]. Among them, BMP4 has been suggested to play an important role in adipogenesis, especially the white adipocyte differentiation through interaction with BMP receptor (BMPR) and subsequently activating the Smad signaling pathways [9–13]. White adipocyte tissues (WAT) are originally recognized as the primary site of triglycerides storage; however, accumulating evidences indicate that WAT is an endocrine organ that participates in the whole body energy metabolism and is highly associated with the risk of developing metabolic syndrome [14, 15]. Thus, BMP4, which is considered as a convincing candidate gene that may contribute substantially to both glucose and lipid metabolism, should arise more attention. Nevertheless, up to now, there is no report focusing on the genetic association studies of BMP4 with type 2 diabetes and related clinical traits in East Asian population. In view of this, the aim of this current study was to test for the possible correlation between them in a Chinese Han population.
2. Methods
2.1. Ethics Statement
The study was approved by the institution review board of Shanghai Jiao Tong University Affiliated Sixth People's Hospital in accordance with the principle of the Helsinki Declaration II. Written informed consent was obtained from each participant.
2.2. Subjects
A total of 6822 participants of Han ancestry residing in Shanghai were recruited, including 3410 cases with type 2 diabetes and 3412 controls. Detailed information concerning this study population has been described elsewhere [16, 17]. In brief, all cases were unrelated type 2 diabetes patients defined according to 1999 WHO criteria and were recruited from the clinical inpatient database of Shanghai Diabetes Institute [18]. The controls were enrolled from community-based random sample epidemiological studies of diabetes and related metabolic diseases. All of them were unrelated subjects with normal glucose tolerance as assessed by 75 g oral glucose tolerant tests (OGTTs) and with negative family history of diabetes. The clinical characteristics of all participants were shown in Table 1.
Table 1.
Clinical characteristics of the study sample.
| Cases | Controls | |
|---|---|---|
| Samples (n) | 3410 | 3412 |
| Male/female (n) | 1812/1597 | 1364/2048 |
| Age (years) | 60.33 ± 12.49 | 51.41 ± 14.39 |
| BMI (kg/m2) | 24.20 (22.00, 26.60) | 23.23 (21.27, 27.68) |
| Fasting plasma glucose (mmol/L) | 12.78 (9.00, 16.00) | 5.02 (4.70, 5.40) |
| 2 h plasma glucose (mmol/L) | 17.00 (13.00, 22.00) | 5.42 (4.60, 6.30) |
| Total cholesterol (mmol/L) | 4.70 (4.00, 5.50) | 4.70 (4.04, 5.35) |
| Triglyceride (mmol/L) | 1.49 (0.99, 2.18) | 1.25 (0.87, 1.82) |
| HDL-C (mmol/L) | 1.11 (0.94, 1.33) | 1.33 (1.13, 1.51) |
| LDL-C (mmol/L) | 2.97 (2.42, 3.57) | 3.04 (2.49, 3.61) |
Data are shown as n or median (interquartile range).
BMI: body mass index; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol.
2.3. Clinical Measurements
All participants underwent a detailed clinical investigation as described previously [19]. Briefly, anthropometric parameters such as height, weight, blood pressure, and waist and hip circumference were measured. Body mass index (BMI) was calculated as weight in kilometers divided by height in meters squared. For the controls, OGTTs which were assessed by standard 75 g glucose in the morning after an overnight fast were performed. And blood samples were obtained at the fasting and 2 h during OGTTs. Plasma glucose, serum insulin and lipid profile were measured. Homeostasis model assessment (HOMA), which was calculated by fasting plasma glucose and insulin, was used for estimating insulin resistance index and β cell function [20]. In addition, Insulin sensitivity from the OGTT was also estimated according to the insulin sensitivity index (ISI) proposed by Gutt et al. [21]. ISI = [75000 + (fasting plasma glucose − 2 h plasma glucose) × 0.19 × weight]/120/mean plasma glucose/log10 mean insulin.
2.4. Single Nucleotide Polymorphism (SNP) Selection and Genotyping
In the present study, 10 tagging SNPs were selected according to the HapMap Phase III (release 27) Han Chinese database using the threshold of r 2 ≥ 0.8, which stretched 9 kb in the upstream to 9 kb in the downstream of the BMP4 gene region. The 10 tagging SNPs could tag 73% SNPs (14 SNPs out of 19 SNPs in the HapMap Chinese Han sample) with a minor allele frequency (MAF) of >0.05. All the SNPs were genotyped using the primer extension of multiplex products with detection by matrix-assisted laser desorption ionization-time of flight mass spectroscopy using a MassARRAY Compact Analyzer (Sequenom, San Diego, CA, USA) and the overall call rate was 98.7%.
2.5. Statistical Analysis
The Hardy-Weinberg equilibrium test was performed in the cases and controls separately for each variant before association analysis. SNPs that failed this test (P < 0.01 in the controls) should be excluded. Pairwise linkage disequilibrium including |D′| and r 2 was estimated using Haploview (version 4.2). Allele and genotype distributions between the patients and control subjects were compared with χ 2 test or logistic regression [22], and odds ratios (ORs) with 95% confidence intervals (CIs) were presented. All skewly distributed quantitative traits (including fasting plasma glucose, 2 h plasma glucose, fasting insulin, 2 h insulin, triglycerides, total cholesterol, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), estimated ISI, HOMA for β-cell function, and insulin resistance) were logarithmically transformed to approximate univariate normality. Quantitative traits were analyzed in the control group by linear regression under an additive genetic model adjusted for age, gender, and BMI as confounding factors. Correction of multiple testing on allele association was performed using Haploview (version 4.2) through 10,000 permutations that randomly permutated the case/control status independently of genotypes. The statistical analyses were performed using SAS for Windows (version 8.0; SAS Institute, Cary, NC, USA). A two-tailed P value of 0.05 was considered statistically significant.
The statistic power was calculated under an additive model based on the allele frequency observed in our samples. Upon the assumption that the population risk was 9.6% and two-side α of 0.05, for SNPs with the minor allele frequency over 0.2, our case-control sample size has over 80% power to detect the minimum OR of 1.15.
3. Results
The genotype distributions of all SNPs were in Hardy-Weinberg equilibrium. Pairwise linkage disequilibrium indicated that these 10 SNPs were in modest linkage disequilibrium and formed 3 haplotype blocks in this region (Figure 1).
Figure 1.
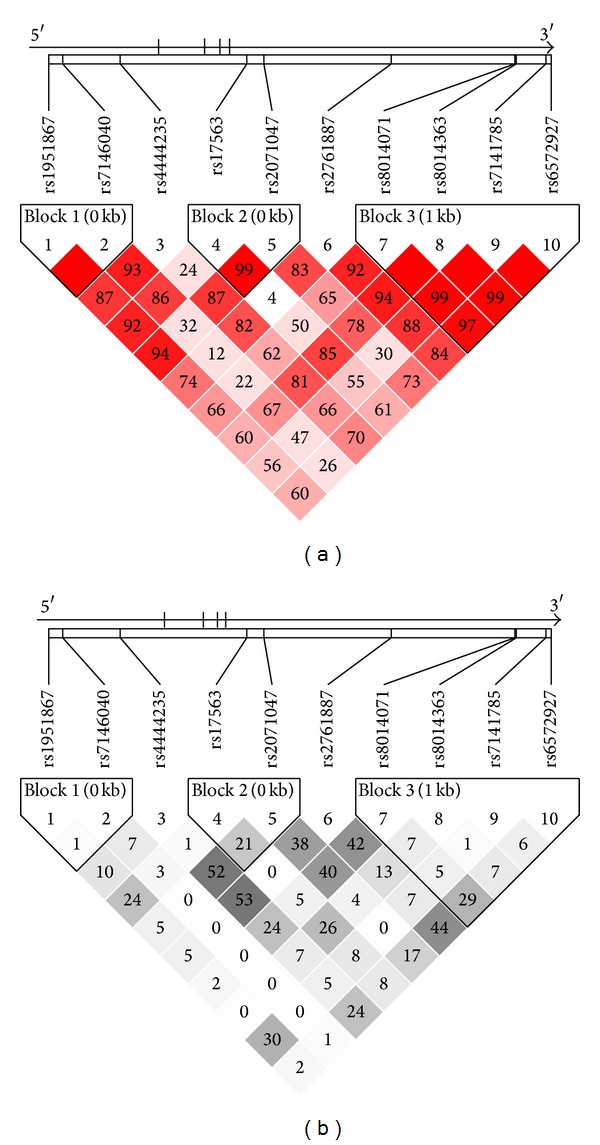
Linkage disequilibrium maps for SNPs genotyped in BMP4 region. (a) Shades of red demonstrate the strength of the pairwise linkage disequilibrium based on D′ and numbers represent the value of D′ expressed as a percentage. (b) Shades of grey show the strength of the pairwise linkage disequilibrium based on r 2 and numbers indicate the value of r 2 expressed as a percentage.
We firstly examined the associations of these 10 SNPs with type 2 diabetes. Nominal evidence of the association was observed only for rs8014363, with the T allele conferring a high risk of type 2 diabetes (OR = 1.108, 95%CI 0.999 − 1.229, P = 0.051 for allele; OR = 1.110, 95%CI 1.000 − 1.231, P = 0.050 for genotype, Table 2). However, the result was no longer significant after correction for multiple testing by 10,000 permutations (empirical P = 0.3689). For the haplotype analysis, by comparing the frequencies between the cases and control subjects, we found that haplotype ACCT in block 3 comprised by rs8014071-rs8014363-rs7141785-rs6572927 indicated a marginal association with type 2 diabetes (P = 0.056, Table 3). After adjusting for multiple testing, the statistical significance was remarkably attenuated (empirical P = 0.4263).
Table 2.
Associations of BMP4 SNPs with type 2 diabetes.
| SNP | Chr. position (Build 37.3) | Major/minor allele | Risk allele | Cases (n = 3410) | Controls (n = 3412) | OR for risk allele (95% CI) | P value for risk allele | OR for genotype (95% CI) | P value for genotype (empirical P value) | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk allele frequencies | Genotype count 11/12/22# |
Risk allele frequencies | Genotype count 11/12/22# |
||||||||
| rs1951867 | 54407192 | G/C | G | 0.903 | 2748/586/35 | 0.896 | 2673/617/36 | 1.073 (0.959, 1.201) | 0.220 | 1.071 (0.957, 1.198) | 0.233 |
| rs7146040 | 54407920 | A/G | G | 0.097 | 2755/586/36 | 0.096 | 2768/572/38 | 1.017 (0.908, 1.140) | 0.767 | 0.983 (0.878, 1.101) | 0.769 |
| rs4444235 | 54410919 | T/C | C | 0.459 | 996/167/717 | 0.452 | 1034/1593/716 | 1.026 (0.959, 1.098) | 0.461 | 0.975 (0.912, 1.043) | 0.465 |
| rs17563 | 54417522 | T/C | T | 0.720 | 1800/1319/273 | 0.717 | 1733/1375/269 | 1.042 (0.967, 1.124) | 0.280 | 1.042 (0.967, 1.123) | 0.283 |
| rs2071047 | 54418411 | C/T | T | 0.368 | 1380/1522/485 | 0.355 | 1428/1527/444 | 1.056 (0.985, 1.133) | 0.126 | 0.948 (0.885, 1.016) | 0.131 |
| rs2761887 | 54425052 | C/A | A | 0.519 | 793/1675/920 | 0.511 | 795/1674/866 | 1.033 (0.965, 1.105) | 0.348 | 0.968 (0.905, 1.036) | 0.348 |
| rs8014071 | 54431500 | A/G | G | 0.350 | 1426/1572/404 | 0.341 | 1462/1539/386 | 1.039 (0.968, 1.115) | 0.290 | 0.962 (0.896, 1.033) | 0.287 |
| rs8014363 | 54431575 | T/C | T | 0.884 | 2617/690/45 | 0.873 | 2574/774/45 | 1.108 (0.999, 1.229) | 0.051 | 1.110 (1.000, 1.231) | 0.050 (0.3689) |
| rs7141785 | 54433114 | C/T | C | 0.904 | 2740/572/36 | 0.902 | 2727/594/32 | 1.028 (0.916, 1.154) | 0.639 | 1.022 (0.912, 1.146) | 0.705 |
| rs6572927 | 54433390 | T/A | T | 0.633 | 1345/1534/456 | 0.632 | 1327/1533/455 | 1.008 (0.940, 1.082) | 0.816 | 1.008 (0.939, 1.081) | 0.834 |
P values <0.1 were shown in bold.
The additive model was used in the association analyses between genotype and type 2 diabetes.
#11: major allele homozygotes; 12: heterozygotes; 22: minor allele homozygotes. Empirical P values are for the alleles based on 10,000 permutations.
Table 3.
Associations of three haplotypes in BMP4 region with type 2 diabetes.
| Haplotype | Haplotype frequencies | P value (empirical P value) | |
|---|---|---|---|
| Cases | Controls | ||
| Block1 (rs1951867-rs7146040) | |||
| GA | 0.805 | 0.801 | 0.522 |
| CA | 0.097 | 0.103 | 0.248 |
| GG | 0.097 | 0.096 | 0.755 |
| Block2 (rs17563-rs2071047) | |||
| TT | 0.368 | 0.355 | 0.116 |
| TC | 0.358 | 0.361 | 0.688 |
| CC | 0.274 | 0.284 | 0.210 |
| Block3 (rs8014071-rs8014363-rs7141785-rs6572927) | |||
| ATCA | 0.365 | 0.366 | 0.960 |
| GTCT | 0.348 | 0.338 | 0.223 |
| ACCT | 0.117 | 0.127 | 0.056 (0.4263) |
| ATTT | 0.096 | 0.098 | 0.705 |
| ATCT | 0.073 | 0.068 | 0.247 |
P values <0.1 were shown in bold.
Empirical P values are for the haplotypes based on 10,000 permutations.
In addition, we further analyzed the effect of the SNP rs8014363 on clinical characteristics in the control group under an additive model. As shown in Table 4, rs8014363 was significantly associated with triglyceride level, with carriers of more type 2 diabetes risk alleles (T) exhibiting higher values of triglyceride than CC homozygotes after adjusting for age, gender and BMI as confounding variants (P = 0.035). Moreover, we also observed a trend towards association between rs8014363 and HDL-C, with carriers of the risk alleles (T) demonstrating lower values (P = 0.068). However, no conspicuous associations were detected in other lipid profile indexes.
Table 4.
Association analyses of the rs8014363 genotype with clinical characteristics in the normal glucose tolerant group.
| CC (n = 45) | CT (n = 774) | TT (n = 2574) | β | SE | P | P* | |
|---|---|---|---|---|---|---|---|
| Age (years) | 48.33 ± 14.93 | 51.02 ± 14.44 | 51.58 ± 14.39 | 0.7892 | 0.5317 | 0.138 | / |
| BMI (kg/m2) | 22.98 (21.48, 24.65) | 23.26 (21.12, 25.53) | 23.23 (21.30, 25.51) | 0.0003 | 0.0022 | 0.884 | / |
| Fasting plasma glucose (mmol/L) | 5.06 (4.60, 5.40) | 5.00 (4.63, 5.39) | 5.03 (4.70, 5.40) | 0.0015 | 0.0017 | 0.371 | 0.462 |
| 2 h plasma glucose (mmol/L) | 5.09 (4.50, 6.14) | 5.44 (4.70, 6.30) | 5.43 (4.60, 6.34) | 0.0017 | 0.0035 | 0.637 | 0.888 |
| Fasting insulin (mU/L) | 5.41 (3.39, 6.52) | 6.00 (4.23, 8.34) | 6.23 (4.39, 8.72) | 0.0181 | 0.0115 | 0.114 | 0.127 |
| 2 h insulin (mU/L) | 29.36 (12.43, 51.29) | 28.34 (14.07, 45.17) | 27.81 (16.10, 47.10) | 0.0182 | 0.0158 | 0.249 | 0.365 |
| Total cholesterol (mmol/L) | 4.75 (4.04, 5.25) | 4.64 (4.01, 5.32) | 4.70 (4.05, 5.35) | 0.0034 | 0.0036 | 0.341 | 0.643 |
| Triglyceride (mmol/L) | 1.11 (0.74, 1.71) | 1.24 (0.84, 1.82) | 1.26 (0.88, 1.83) | 0.0188 | 0.0087 | 0.031 | 0.035 |
| HDL-C (mmol/L) | 1.38 (1.19, 1.58) | 1.34 (1.14, 1.52) | 1.33 (1.12, 1.51) | −0.0054 | 0.0036 | 0.134 | 0.068 |
| LDL-C (mmol/L) | 3.00 (2.45, 3.62) | 3.03 (2.45, 3.60) | 3.04 (2.50, 3.62) | 0.0031 | 0.0049 | 0.527 | 0.846 |
| HOMA-IR | 1.13 (0.79, 1.56) | 1.27 (0.91, 1.89) | 1.34 (0.93, 1.91) | 0.0201 | 0.0119 | 0.092 | 0.104 |
| HOMA-B | 79.81 (47.82, 128.65) | 89.25 (61.45, 137.90) | 90.30 (62.92, 135.68) | 0.0115 | 0.0128 | 0.369 | 0.382 |
| Gutt-ISI | 104.36 (83.45, 138.38) | 100.24 (81.62, 131.43) | 99.56 (80.97, 127.80) | −0.0095 | 0.0068 | 0.160 | 0.220 |
Data are shown as mean ± SD or median (interquartile range).
BMI: body mass index; HDL-C: high-density lipoprotein cholesterol; LDL-C: low-density lipoprotein cholesterol; HOMA-IR: homeostasis assessment model of insulin resistance; HOMA-B: homeostasis assessment model of β-cell function; Gutt-ISI: insulin sensitivity index proposed by Gutt.
P values <0.1 were shown in bold.
*Adjusted for age, gender, and BMI.
4. Discussion
In the current study, we tested the effects of 10 tagging SNPs in BMP4 region on type 2 diabetes in a Chinese population. We identified modest effects of BMP4 variants on the risk of type 2 diabetes.
Over the past years, it has become increasingly obvious that obesity is a major independent risk factor for developing type 2 diabetes [23, 24]. In accompany with obesity, there is always increase of adipose tissues results from both adipocyte hypertrophy and hyperplasia [25]. BMP4 is considered generally to induce commitment of pluripotent stem cells to the white adipocyte lineage [9–11]. In our present analysis, we observed that the triglyceride level significantly increased with the increasing number of risk allele in rs8014363. Nonetheless, an opposite situation between HDL-C levels and the number of risk allele in rs8014363 could also be seen. They are consistent with that WAT is a depot which is not only specialized in storing energy in the form of triglyceride but also a crucial target organ for insulin action. While HDL-C is mainly responsible for reverse-transporting cholesterol to the liver, it serves as a beneficial effect toward atherosclerosis, which is in contrast with triglyceride. That is to say, BMP4 pose an influence on the glucose and lipid metabolism, which may through participating in WAT differentiation.
Furthermore, One functional analysis using multiple genetic approaches showed that BMP4 and its affinity BMPR1A were both expressed in β cells; besides, results not only in vitro but also in vivo all provided consistent evidences that BMP4 in β cells was required for insulin secretion and advantageous to ameliorate glucose tolerance through significantly stimulating glucose-stimulated insulin secretion (GSIS) [26]. However, in our analysis, we failed to find any association with β cell function for variants of BMP4. Accordingly, the exact mechanisms behind these results should be warranted for further investigation.
Some limitations should be considered in the present study. Firstly, as the genetic effect of BMP4 variants on type 2 diabetes was mild, our samples may not have enough power to detect the association. Secondly, because the patients we enrolled were not newly diagnosed and they were treated with antidiabetic drugs and/or insulin. The effect of BMP4 variants on type 2 diabetes which is definitely through impact on impaired insulin secretion or insulin sensitivity is rather ambiguous. Thirdly, the status of taking lipid lowering medication in the control subjects was not clear. Nonetheless, the condition is very rare. Finally, although we found modest correlation between BMP4 variant and type 2 diabetes and related clinical characteristics, we did not perform a replicated research in another independent sample to confirm these results. Thus, it is imperative to further replicate the influence of the variant of BMP4 on type 2 diabetes and metabolic traits in other Chinese samples.
5. Conclusion
Our data suggested that the genetic variants of BMP4 may not play a dominant role in glucose metabolism in Chinese Han population, but a minor effect cannot be ignored. Further investigations are of great necessity to confirm our observations and elucidate the unequivocal mechanisms underlying such association.
Conflict of Interests
The authors declare that they have no conflict of interests.
Authors' Contribution
Shanshan Tang and Rong Zhang contributed equally to this paper.
Acknowledgments
This work was funded by Grants from the Key Program of the Shanghai Municipality for Basic Research (11JC1409600), the National 863 program (2012AA02A509), the 973 program (2011CB504001), the National Science Foundation of China (81200582), the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (2011ZX09307-001-02), and the Key Discipline of Public Health of Shanghai (12GWZX0104). The authors appreciate all the participants of this research. The authors gratefully acknowledge the skillful technical support of all nursing and medical staff at Shanghai Clinical Center for Diabetes.
References
- 1. http://www.idf.org/diabetesatlas/5e/Update2012.
- 2.Florez JC, Hirschhorn J, Altshuler D. The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annual Review of Genomics and Human Genetics. 2003;4:257–291. doi: 10.1146/annurev.genom.4.070802.110436. [DOI] [PubMed] [Google Scholar]
- 3.O’Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: the end of the beginning? Science. 2005;307(5708):370–373. doi: 10.1126/science.1104346. [DOI] [PubMed] [Google Scholar]
- 4.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddi AH. BMPs: from bone morphogenetic proteins to body morphogenetic proteins. Cytokine and Growth Factor Reviews. 2005;16(3):249–250. doi: 10.1016/j.cytogfr.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Wagner DO, Sieber C, Bhushan R, Börgermann JH, Graf D, Knaus P. BMPs: from bone to body morphogenetic proteins. Science Signaling. 2010;3(107, article mr1) doi: 10.1126/scisignal.3107mr1. [DOI] [PubMed] [Google Scholar]
- 8.Kim M, Choe S. BMPs and their clinical potentials. BMB Reports. 2011;44(10):619–634. doi: 10.5483/BMBRep.2011.44.10.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulz TJ, Tseng Y-H. Emerging role of bone morphogenetic proteins in adipogenesis and energy metabolism. Cytokine and Growth Factor Reviews. 2009;20(5):523–531. doi: 10.1016/j.cytogfr.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto TC, Lane MD. Adipose development: from stem cell to adipocyte. Critical Reviews in Biochemistry and Molecular Biology. 2005;40(4):229–242. doi: 10.1080/10409230591008189. [DOI] [PubMed] [Google Scholar]
- 11.Tang Q-Q, Otto TC, Lane MD. Commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(26):9607–9611. doi: 10.1073/pnas.0403100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tseng Y-H, Kokkotou E, Schulz TJ, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454(7207):1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowers RR, Kim JW, Otto TC, Lane MD. Stable stem cell commitment to the adipocyte lineage by inhibition of DNA methylation: role of the BMP-4 gene. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(35):13022–13027. doi: 10.1073/pnas.0605789103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gesta S, Tseng Y-H, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131(2):242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 15.Kissebah AH, Krakower GR. Regional adiposity and morbidity. Physiological Reviews. 1994;74(4):761–811. doi: 10.1152/physrev.1994.74.4.761. [DOI] [PubMed] [Google Scholar]
- 16.Hu C, Wang C, Zhang R, et al. Association of genetic variants of NOS1AP with type 2 diabetes in a Chinese population. Diabetologia. 2010;53(2):290–298. doi: 10.1007/s00125-009-1594-2. [DOI] [PubMed] [Google Scholar]
- 17.Hu C, Zhang R, Wang C, et al. A genetic variant of G6PC2 is associated with type 2 diabetes and fasting plasma glucose level in the Chinese population. Diabetologia. 2009;52(3):451–456. doi: 10.1007/s00125-008-1241-3. [DOI] [PubMed] [Google Scholar]
- 18.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic Medicine. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 19.Jia WP, Pang C, Chen L, et al. Epidemiological characteristics of diabetes mellitus and impaired glucose regulation in a Chinese adult population: the Shanghai diabetes studies, a cross-sectional 3-year follow-up study in Shanghai urban communities. Diabetologia. 2007;50(2):286–292. doi: 10.1007/s00125-006-0503-1. [DOI] [PubMed] [Google Scholar]
- 20.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 21.Gutt M, Davis CL, Spitzer SB, et al. Validation of the insulin sensitivity index (ISI0,120): comparison with other measures. Diabetes Research and Clinical Practice. 2000;47(3):177–184. doi: 10.1016/s0168-8227(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan JM, Rimm EB, Colditz GA, Stampfer MJ, Willett WC. Obesity, fat distribution, and weight gain as risk factors for clinical diabetes in men. Diabetes Care. 1994;17(9):961–969. doi: 10.2337/diacare.17.9.961. [DOI] [PubMed] [Google Scholar]
- 24.Poirier P, Giles TD, Bray GA, et al. Obesity and cardiovascular disease: pathophysiology, evaluation, and effect of weight loss: an update of the 1997 American Heart Association Scientific Statement on obesity and heart disease from the Obesity Committee of the Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2006;113(6):898–918. doi: 10.1161/CIRCULATIONAHA.106.171016. [DOI] [PubMed] [Google Scholar]
- 25.Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. Journal of Biological Chemistry. 1993;268(30):22243–22246. [PubMed] [Google Scholar]
- 26.Goulley J, Dahl U, Baeza N, Mishina Y, Edlund H. BMP4-BMPR1A signaling in β cells is required for and augments glucose-stimulated insulin secretion. Cell Metabolism. 2007;5(3):207–219. doi: 10.1016/j.cmet.2007.01.009. [DOI] [PubMed] [Google Scholar]


