Abstract
Drought is a major threat to agriculture production worldwide. Mitogen-activated protein kinases (MAPKs) play a pivotal role in sensing and converting stress signals into appropriate responses so that plants can adapt and survive. To examine the function of MAPKs in the drought tolerance of tomato plants, we silenced the SpMPK1, SpMPK2, and SpMPK3 genes in wild-type plants using the virus-induced gene silencing (VIGS) method. The results indicate that silencing the individual genes or co-silencing SpMPK1, SpMPK2, and SpMPK3 reduced the drought tolerance of tomato plants by varying degrees. Co-silencing SpMPK1 and SpMPK2 impaired abscisic acid (ABA)-induced and hydrogen peroxide (H2O2)-induced stomatal closure and enhanced ABA-induced H2O2 production. Similar results were observed when silencing SpMPK3 alone, but not when SpMPK1 and SpMPK2 were individually silenced. These data suggest that the functions of SpMPK1 and SpMPK2 are redundant, and they overlap with that of SpMPK3 in drought stress signaling pathways. In addition, we found that SpMPK3 may regulate H2O2 levels by mediating the expression of CAT1. Hence, SpMPK1, SpMPK2, and SpMPK3 may play crucial roles in enhancing tomato plants’ drought tolerance by influencing stomatal activity and H2O2 production via the ABA-H2O2 pathway.
Keywords: Solanum pimpinellifolium, protein kinase, stomata, drought tolerance, virus-induced gene silencing (VIGS)
1. Introduction
The tomato plant, Solanum lycopersicum, is extensively cultivated and consumed around the world and therefore constitutes a major agricultural industry. Adequate water supply is a major concern for this industry because drought conditions impair the quality and yield of tomatoes by hindering the growth and development of the seedlings. Many signaling pathways are involved in mediating plants’ responses to drought stress. Mitogen-activated protein kinase (MAPK) cascade, the foundation of signal transduction networks, plays crucial roles in the ability of plants to tolerate and adapt to stresses by regulating stress signal transduction and the expression of relevant genes.
The MAPK signaling pathway is a three-tiered phosphorelay cascade consisting of MAPKs (MPKs), which are activated by MAPK kinases (MPKK or MKKs), which in turn are activated by MAPKK kinases (MAPKKKs). As the last component of the cascade to be activated, MAPKs can phosphorylate specific serine/threonine residues on the target protein, thereby regulating a variety of cellular activities [1]. An increasing body of evidence suggests, that in plants, MAPK cascades are involved in numerous developmental processes as well as signaling networks associated with stress responses, including tolerance to certain stressors [2–6]. Lampard and Wang reported that the MAPK kinases AtMKK4, AtMKK5, AtMKK7, and AtMKK9 regulate stomatal development by phosphorylating AtMPK3, AtMPK6, and other unknown MAPK genes [7,8]. Because stomata are channels for gas exchange and water evaporation, their movement is crucial for plants to adapt to prevailing environmental conditions. Thus, the ability of MAPK cascades to regulate stomatal development implies that they may be involved in shaping plants’ stress tolerance. Several studies have found that MAPK cascades affect plants’ innate immunity to biotic stresses, such as bacteria, insects, and aphids [9–11]. Additional studies suggest that MAPK cascades are involved in processes regulated by the plant hormone, abscisic acid (ABA), when plants are exposed to various abiotic stressors[12].
ABA is a universal hormone in plants and plays a major role in plant growth and development, including embryo maturation, seed dormancy and germination, seedling establishment, and root branching. As a stress hormone, ABA regulates how plants respond to a variety of stresses by transmitting stress signals to the appropriate cells. These cells then activate the expression of stress-responsive genes and elicit other physiological responses, such as hydrogen peroxide (H2O2) production and stomatal movement induction. In Arabidopsis species, AtMPKK1, AtMPKK3, and AtMPKK9 as well as AtMPK1, AtMPK2, AtMPK3, and AtMPK6 play positive roles in ABA signaling during seed germination, and MdMKK1 and MdMPK1 function similarly in apples [13–19]. In addition, MAPKs have been implicated in ABA-induced antioxidant defenses. The AtMEKK1-AtMKK1/AtMKK2-AtMPK4 cascade and the AtMKK1-AtMPK6 signaling pathway play important roles in mitigating the effects of reactive oxygen species (ROS) [15,17,20–23]. In addition, previous studies have shown that the ABA-induced activity of guard cells, which are responsible for allowing or preventing gas exchange to occur through stomata, is mediated by MAPK cascades when abiotic stresses are present. In Arabidopsis, it was found that inhibiting AtMPK3 caused stomata to partially lose sensitivity to ABA [24,25]. Similar behavior was found in atmpk9/atmpk12 double mutants, which became insensitive to ABA-induced stomatal closure and ABA-inhibited stomatal opening [26]. Therefore, crosstalk exists between ABA signaling and MAPK cascades in response to various stressors, especially stressors that are closely related to stomatal movements, such as drought stress.
Although it has been shown that SpMPK1, SpMPK2, and SpMPK3 positively regulate the responses of tomato plants to many biotic stresses, such as insects and bacteria, their functions under abiotic stresses are poorly understood. Here, we studied the functions of SpMPK1, SpMPK2, and SpMPK3 in the drought tolerance of wild-type Solanum pimpinellifolium plants using the virus-induced gene silencing (VIGS) method. The loss-of-function studies indicate that SpMPK1, SpMPK2, and SpMPK3 may play positive roles in drought stress tolerance in tomato via controlling ABA-induced stomatal movements and H2O2 production.
2. Results
2.1. Silencing SpMPK1, SpMPK2, and SpMPK3 Reduced the Drought Tolerance of Tomato Plants
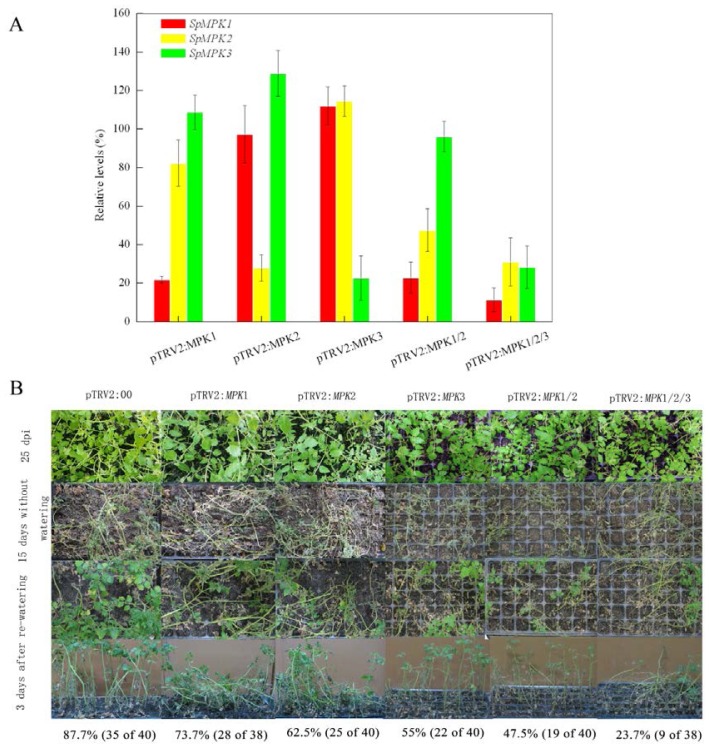
Gene-silenced plants were generated with VIGS constructs. VIGS efficiency was assessed using quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) by analyzing the transcription levels of SpMPK1, SpMPK2, and SpMPK3 in the gene-silenced plants compared with those in the control plants. After silencing single genes, the transcription levels of the MPK gene targets were reduced by 80% (SpMPK1), 73% (SpMPK2), and 78% (SpMPK3), but no reduction was observed in the transcription levels of other MPKs (Figure 1). For the co-silencing assays, the transcription levels of SpMPK1 and SpMPK2 in plants having both SpMPK1/SpMPK2 genes silenced were reduced by 77% and 65%, respectively. For plants with all three genes co-silenced, the transcription levels of SpMPK1, SpMPK2, and SpMPK3, were reduced by 89%, 70%, and 70%, respectively. These results suggest that endogenous SpMPK1, SpMPK2, and SpMPK3 genes were successfully silenced in the experimental plants.
Figure 1.
Silencing SpMPK1, SpMPK2, and SpMPK3 attenuated the drought tolerance of tomato plants. (A) Transcription levels of SpMPK1, SpMPK2, and SpMPK3 in each type of gene-silenced plants. Nine to 10-day-old plants were infiltrated with A. tumefaciens containing pTRV2:MPK1, pTRV2:MPK2, pTRV2:MPK3, pTRV2:MPK1/2, pTRV2:MPK1/2/3 and pTRV2:00 (control). SpMPKs gene silencing efficiency was analyzed for mRNA levels by quantitative RT-PCR at 25 dpi. The transcription levels in VIGS plants (n ≥ 50) were expressed as percentages of the mean levels in control plants, which were defined as 100%; (B) Drought sensitivity of gene-silenced plants. Gene-silenced plants at 25 to 30 dpi were further grown for 15 day without watering and consequently re-watered for 3 day, and then the surviving plants were counted; (C) Measurement of water loss of detached leaves. Detached leaves of gene-silenced plants were weighed at the indicated times after their excision. Water loss was calculated as the percentage of initial fresh weight. The data represent means ± SD of 10 leaves from each of three replicates.
To examine the drought tolerance of plants that underwent individual gene silencing (SpMPK1, SpMPK2, or SpMPK3) and combined gene silencing (SpMPK1/SpMPK2 or SpMPK1/SpMPK2/SpMPK3), those containing <50% of the target gene transcripts were grown for an additional 15 days without watering, until most of the gene-silenced plants wilted. These plants were then re-watered for three days and their survival rates were compared. After re-watering, the individually gene-silenced plants showed lower survival rates—73.7% (28 of 38), 62.5% (25 of 40), and 55% (22 of 40) for plants with the silenced SpMPK1, SpMPK2, and SpMPK3 genes—respectively-compared with the control plants, whose survival rate was 87.5% (35 of 40). In contrast, plants with co-silenced SpMPK1/SpMPK2 genes had a survival rate of only 47.5% (19 of 40), while those with co-silenced SpMPK1/SpMPK2/SpMPK3 genes had a survival rate of only 23.7% (9 of 38) after re-watering.
To further evaluate the responses of gene-silenced plants to drought stress, we examined water losses in detached leaves. As shown in Figure 1C, leaves from the individually gene-silenced plants lost more water than leaves of the control plants. After a 6-h incubation, control leaves lost 43% ± 4.4% of their initial weight, whereas leaves from the individually silenced SpMPK1, SpMPK2, and SpMPK3 plants lost 47% ± 0.5%, 54% ± 1.2%, and 55% ± 1.5%, respectively, of their initial weights. A similar outcome was seen for the leaves of SpMPK1/SpMPK2 co-silenced plants, which lost 57% ± 3.0% of their initial weight. Leaves from the SpMPK1/SpMPK2/SpMPK3 co-silenced plants lost water faster than the others, losing 31% ± 2.1% of their initial weight in the first hour after being detached; after 6 h of incubation, the leaves lost approximately 75% ± 4.5% of their initial weight, which was a significantly greater loss than was seen in the other experimental plants (p < 0.02). As a whole, detached leaves from gene-silenced plants lost more water than those of the control plants after 6 h of incubation.
2.2. Silencing SpMPK1, SpMPK2, and SpMPK3 Impaired Stomatal Closure in Response to Abscisic Acid (ABA) and Hydrogen Peroxide (H2O2)
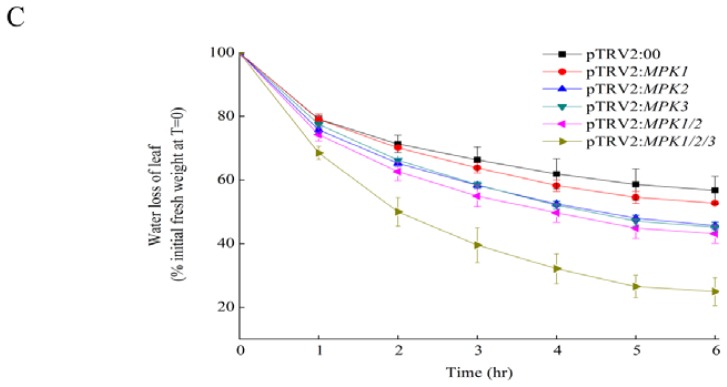
Stomata are pores formed by pairs of guard cells that are located in the epidermis. Plants can tolerate drought stress by regulating the aperture of the stomata to minimize water losses caused by transpiration. ABA-deficient plants or those with impaired ABA signaling are unable to adaptively regulate their stomatal apertures and are highly susceptible to drought stress [13,27,28]. To investigate whether SpMPK1, SpMPK2, and SpMPK3 genes function in ABA- and H2O2-sensitive stomatal responses, we examined the ABA-dependent stomatal activity in our gene-silenced plant models. Leaves from the control and experimental plants were submerged in stomatal opening solution and then treated with 50 μM ABA or 50 mM H2O2 for 2 h, and then stomatal apertures were measured. In the stomatal opening solution, all of the guard cells were fully opened prior to ABA and H2O2 exposure (Figure 2, CK). However, in the presence of ABA and H2O2, the control plants and the individually silenced SpMPK1 and SpMPK2 plants were highly sensitive to ABA and H2O2, which caused their stomata to close, as shown in Figure 2. In contrast, stomatal closure was markedly impaired in the leaves of SpMPK1/SpMPK2 co-silenced plants, as compared with that of the individual SpMPK1 and SPMPK2 silenced plants (p < 0.0001). These results suggest that SpMPK1 and SpMPK2 may redundantly regulate ABA- and H2O2-induced stomatal closure. In addition, individual SpMPK3 gene-silenced plants and SpMPK1/SpMPK2/SpMPK3 co-silenced plants exhibited the same stomatal behavior as SpMPK1/SpMPK2 co-silenced plants when exposed to ABA and H2O2. These results indicate that the function of SpMPK1 and SpMPK2 may overlap with that of SpMPK3 in regulating ABA- and H2O2-induced stomatal closure.
Figure 2.
Stomatal apertures (the ratio of width to length) of gene-silenced plants in response to ABA and H2O2. Leaves of control, individual gene-silenced (SpMPK1, SpMPK2, or SpMPK3) and combined gene-silenced (SpMPK1/SpMPK2 or SpMPK1/SpMPK2/SpMPK3) plants were incubated in stomatal opening solution for 2 h. Some of them were transferred to solutions containing 50 μM ABA or 50 mM H2O2 for 2 h and the others remained in the opening solution for control (CK). Stomata on the adaxial surface were observed by light microscopy. For each type of gene-silenced plants, at least 20 stomata from each sample of ten genes-silenced plants were measured. The data represent means ± SD. Asterisks indicate that the mean value is signficantly different from the control at p < 0.0001.
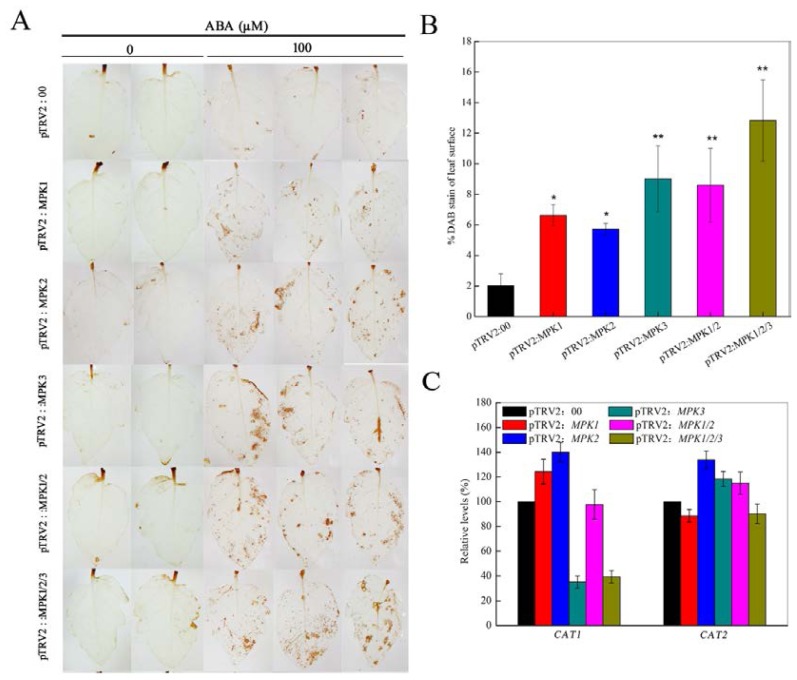
2.3. SpMPK3 is Involved in Abscisic Acid (ABA)-Induced Hydrogen Peroxide (H2O2) Production by Regulating the Expression of CAT1
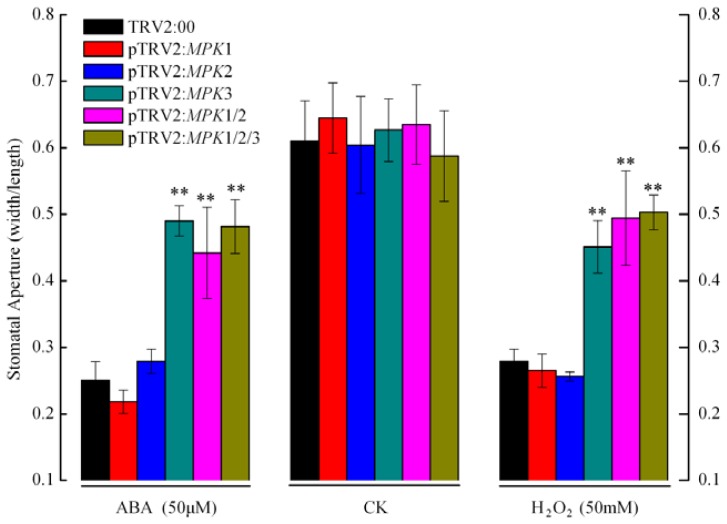
According to previous studies, ROS is also involved in the ABA signal transduction pathway that regulates guard cells [29,30]. The above results prompted us to wonder whether or not SpMPKs act on ABA-ROS signaling. Therefore we used our gene-silenced plant models to investigate how SpMPK1, SpMPK2, and SpMPK3 influence ABA-induced H2O2 production. To do this, the H2O2 levels in the gene-silenced plants were measured when plants were exposed to ABA. As shown in Figure 3A, only basal levels of H2O2 were detected in non-treated control plants, but H2O2 production increased substantially in response to ABA exposure. In the presence of 100 μM ABA, H2O2 production in individually silenced SpMPK1 and SpMPK2 plants increased by 6.6% and 5.7%, respectively, which was significantly more than the increase seen in the control plants (p < 0.05) (shown in Figure 3B). The individually silenced SpMPK3 plants and both groups (SpMPK1/SpMPK2 and SpMPK1/SpMPK2/SpMPK3) of co-silenced plants displayed a hypersensitivity to ABA, evidenced by increases of 9%, 8.5% and 12.8%, respectively, in H2O2 production, which were significantly greater than those of plants with individually silenced SpMPK1 and SpMPK2 genes (p < 0.01). The data presented here demonstrate that SpMPK1, SpMPK2, and SpMPK3 may be involved in ABA-H2O2 signaling related to stress tolerance by regulating the production of H2O2. Future studies are needed, because it is not yet known whether H2O2 acts upstream or downstream of the SpMPKs.
Figure 3.
H2O2 production and CATs transcription levels of gene-silenced plants. (A) Detection of ABA-induced H2O2 production by DAB staining. Leaves of control, individual gene-silenced (SpMPK1, SpMPK2, or SpMPK3) and combined gene-silenced (SpMPK1/SpMPK2 or SpMPK1/SpMPK2/SpMPK3) plants were treated with 100 μM ABA for 2 h and transferred to 100 mg·mL−1 DAB solution for 8 h. The presence of H2O2 in the leaves was visualized as a dark brown color; (B) The amount of H2O2 in leaves of gene-silenced plants. The data represent means ± SD of two leaves from each of six gene-silenced plants, respectively. Asterisk, p < 0.05; double asterisks, p < 0.01; (C) The transcription levels of H2O2 scavenging genes (CAT1 and CAT2) in VIGS plants treated with 100 μM ABA. The experiments are representative of 10 gene-silenced plants, respectively.
Catalase, produced by the expression of the CAT genes, is an important enzyme that catalyzes the decomposition of H2O2. However, in the ABA signaling pathway, the transcription levels of CAT1 were reduced in individually silenced SpMPK3 plants and in SpMPK1/SpMPK2/SpMPK3 co-silenced plants, but not in the other gene-silenced plants. Furthermore, we did not observe a significant decrease of CAT2 transcription levels in any of the gene-silenced plants (Figure 3C). The lack of correlation between CAT transcriptional levels and the H2O2 levels of gene-silenced plants suggests that ABA-mediated H2O2 production may be governed by multiple genes that function in different pathways. These results suggest that CAT1 may be involved in the feedback regulation of H2O2 signaling and that SpMPK3 may regulate the H2O2 signaling by mediating the expression of CAT1.
3. Discussion
Drought is one of the greatest threats to agriculture production worldwide. Previous findings have shown that MAPKs are involved in the tolerance-related signaling networks associated with various stressors, including drought stress. In this study, we found that inhibiting the function of SpMPK1, SpMPK2, and SpMPK3 genes in tomato plants reduced their tolerance to drought.
As is commonly known, the movement of stomata is tightly linked to water loss. In drought conditions, plants produce ABA, which then triggers a signaling cascade that causes stomata to close, thus preventing water loss [31]. Therefore, stomatal behavior may reflect a plant’s tolerance for drought stress. In this study, the functions of SpMPK1, SpMPK2, and SpMPK3 as they relate to stomatal behavior were investigated. Although inhibiting SpMPK1, SpMPK2, and SpMPK3 resulted in the formation of clustered stomata, we did not observe remarkable differences between the control plants and the gene-silenced plants (both the individually silenced and co-silenced plants) (shown in Figure S1). These results suggest that additional MPKs besides SpMPK1, SpMPK2, and SpMPK3, may exist in the tomato that redundantly regulates stomatal development. However, all the SpMPKs investigated here participated in ABA- and H2O2-induced stomatal closure, and the function of SpMPK1 may be redundant with that of SpMPK2. It has been reported that a given MAPKKK or MAPKK can activate more than one target kinase, which means that MAPKKKs and MAPKKs function as divergence points in signal transduction [32]. Thus, plants can produce thousands of different combinations of MAPK cascades to regulate stress responses, such that many of the cascades overlap [33]. In tomato plants, MAPKs are generally grouped into four subfamilies [34]. SpMPK1, SpMPK2, and SpMPK3 belong to the same subfamily (subfamily A) because their sequences are highly homologous. Therefore, functional redundancies between SpMPK1, SpMPK2, and SpMPK3 are unsurprising.
Previous studies have shown that ABA can activate AtMPK3 and AtMPK6 in Arabidopsis species [15–17,35]. In agreement with these results, we found that SpMPK3 and SpMPK1/2 (homologous to AtMPK3 and AtMPK6) were also up-regulated in the presence of ABA (Figure S2). Additionally, silencing SpMPK1, SpMPK2, and SpMPK3 impaired ABA-induced stomatal closure. Thus, it is speculated that these genes act downstream of ABA by regulating ABA signaling to mediate drought tolerance in tomato plants. In addition, H2O2 is known to be an intracellular messenger that can evoke specific cellular responses under different stimuli [36–39]. Previous studies have shown that ABA triggers the production of H2O2, and H2O2 mediates ABA-induced stomatal closure; these processes are thought to be regulated by MAPK signaling [26,40]. In this study, we demonstrated that silencing SpMPK1, SpMPK2, and SpMPK3 worked to impair stomatal closure, indicating that SpMPK genes may act downstream of H2O2. However, we also observed increased H2O2 levels in gene-silenced plants exposed to ABA, which alternatively suggests that SpMPK1, SpMPK2, and SpMPK3 may be involved in ABA-induced H2O2 production. In addition, silencing the SpMPK3 gene attenuated the transcription level of CAT1, the translated product of which scavenges H2O2. These results demonstrate that SpMPK3 is involved in the feedback regulation of H2O2 production and CAT1 expression. For SpMPK1 and SpMPK2, increased levels of H2O2 were observed in the individually silenced and co-silenced plants, but CAT transcription levels were not affected, which suggests that some unknown H2O2-scavening genes may exist in SpMPK1/SpMPK2-mediated H2O2 signaling.
MAPK cascades represent the primary way in which plant cellular functions are controlled in response to a number of external signals. As shown in our results, the complementary and commutable functions of SpMPK1, SpMPK2, and SpMPK3 in drought tolerance reveal the complexity and crosstalk associated with MAPK cascades. Because MAPKs are regulated post-translationally by phosphorylation and they act on various substrates, the loss of a functional gene product might not pinpoint the function of a MAPK cascade. Therefore, a complementary gain-of-function assay on SpMPKs would help us understand the complex roles of SpMPKs in the tolerance responses of tomatoes. Although 16 MAPK family genes have been identified in tomatoes to date, no systematic investigations on MAPKK and MAPKKK family genes have yet been performed. Thus, identifying different combinations of MAPKKK-MAPKK-MAPK cascades under stimulus-specific stressors would be a highly complex undertaking.
4. Experimental Section
4.1. Plant Material and Growth Conditions
Seeds of S. pimpinellifolium L03708, kindly supplied by AVRDC—The World Vegetable Center, were grown in cell trays filled with matrix in artificial climate incubator at 21 ± 2 °C with 50% relative humidity and a 16 h photoperiod with light intensity ranging from 300 to 400 μmol·m−2·s−1. Nine- to 10-day-old seedlings (cotyledons fully expanding and true leaves just emerging) were subjected to VIGS tests by inoculation with Agrobacterium suspensions (as shown below).
4.2. VIGS Constructs
The pTRV1 and pTRV2 VIGS vectors described in Liu et al. [41] were obtained from Dr. Dinesh-Kumar (Yale University, New Haven, CT, USA). Fragments for individually and combined silencing SpMPK1, SpMPK2, SpMPK3 were obtained by polymerase chain reaction (PCR) using cDNA synthesized with PrimeScript RT reagent Kit (Takara, Dalian, China) from total RNA extracted from leaf tissues of L03708 using Trizol reagent (Invitrogen, Carlsbad, CA, USA) as a template and the primer pairs listed in Table S1. The PCR products were cloned into the pMD-18T vector, and then recombined into pTRV2 plasmid digested by Xho I/Sac I. The recombinant constructs were electroporated into cells of Agrobacterium tumefaciens GV3101. The resultant Agrobacterium strains were used in the indicated VIGS experiments.
4.3. Infiltration of pTRV-Containing A. tumefaciens Cultures into Cotyledons
A. tumefaciens GV3101 strains carrying each TRV derivative were grown at 28 °C in LB medium containing antibiotics (50 μg/mL of kanamycin, 10 μg/mL of rifampicin and 50 μg/mL of gentamycin), and bacterial suspensions (OD600 = 2) were prepared for infiltration on cotyledons following a similar procedure as described in Velásquez et al. [42]. The cotyledons of young seedlings were infiltrated with Agrobacteria by using a 1-mL syringe. Agroinfiltrated plants were maintained at 18–22 °C in a growth chamber with a 16 h light period for the indicated periods of time.
4.4. RNA Isolation and Quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and then treated with DNase I (Fermentas, Glen Burnie, MD, USA) to clean out DNA. cDNA synthesized from 1 μg of total RNA using the PrimeScript RT reagent Kit (Takara, Dalian, China) was utilized for quantitative RT-PCR. Quantitative RT-PCR was performed using SYBR Premix Ex Taq II (TaKaRa, Dalian, China) on iQ5 Real-Time PCR Detection System (BIO-RAD Corp., Hercules, CA, USA). The PCR cycling conditions were as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 10 s, 55 °C for 10 s and 72 °C for 20 s. The melting curve was routinely performed after 40 cycles to verify primer specificity. The 2−ΔΔCt method was applied to calculate the change of expression of each gene [43]. S. pimpinellifolium elongation factor 1-α (EF1α) mRNA level was used as internal control for normalization [44]. All primer sequences are given in Table S1.
4.5. Measurement of Stomatal Aperture
Leaves of control, individual gene-silenced (SpMPK1, SpMPK2, or SpMPK3) and combined gene-silenced (SpMPK1/SpMPK2 or SpMPK1/SpMPK2/SpMPK3) plants at 25–30 dpi were used for the observation of matured stomata. Detached leaves were incubated in stomata opening solution (10 mM MES, pH 6.1, in a 50 mM KCl solution), and illuminated with 250 to 350 μmol m−2 s−1 light for 2 h and then transferred to the stomatal opening solution supplemented with 50 μM ABA or 50 mM H2O2 respectively [45]. Subsequently, epidermal strips were carefully prepared from the adaxial surface of the leaf, mounted on glass slides and observed using an Olympus PD72 microscope (Olympus Corp., Tokyo, Japan). The ratio of width to length of the stomata was measured using Image Pro Plus software (Media Cybernetics Inc., Rockville, MD, USA). Over 20 guard cells from each sample of ten gene-silenced plants were used to measure stomatal aperture.
4.6. DAB Staining
DAB staining was used to observe the accumulation of H2O2[32,46]. Leaves of individual and combined gene-silenced plants at 25–30 dpi were treated with 100 μM ABA for 2 h, then incubated with 1 mg·mL−1 DAB solution for 8 h. Following the incubation, DAB-stained samples were submerged in bleaching solution (ethanol:acetic acid:glycerol, 3:1:1) and boiled in a water bath for 15–20 min to remove chlorophyll. The presence of H2O2 in leaves was visualized as a dark brown color.
4.7. Drought Tolerance Assay
Gene-silenced plants at 25–30 dpi were subjected to drought stress by ceasing watering for 15 days. After re-watering for the following three days, surviving plants were counted. For the experiment of water loss, detached leaves were placed in petri dishes at room temperature with approximately 60% humidity under dim light. The weights of leaves were measured over time (0–6 h). Water loss of leaf was expressed as a percentage of initial fresh weight.
4.8. Tissue Sampling for Assays
Leaf samples for RNA analysis, water loss of leaves, and stomatal aperture or DAB staining assays were collected at 25–30 dpi. The development and extent of gene silencing were assessed by monitoring the photo-bleaching pattern of PHYTOENE DESATURASE-silenced plants according to Liu et al. [41]. For each plant, the fifth to ninth true leaves were used for their high silencing efficiency (shown in Figure S3). Samples for RNA analysis, five leaflets from the true leaves (fifth-ninth), were collected and frozen immediately, whereas the other leaflets (total of 15–25) were utilized for the other assays.
4.9. Statistical Analysis
R 2.15.2, an open-source software, is used for the statistical analysis. The data were subjected to univariate ANOVA analysis of variance followed by a post hoc test. Values were computed as the means ± SD of three or more independent experiments.
5. Conclusions
In conclusion, plants with individually silenced SpMPK1, SpMPK2, and SpMPK3 genes, as well as co-silenced SpMPK1/SpMPK2 and SpMPK1/SpMPK2/SpMPK3 genes, had lower survival rates than control plants when they were exposed to drought-like conditions. In addition, silencing these SpMPKs resulted in impaired stomatal closure and increased H2O2 production when plants were exposed to ABA. In conclusion, this study demonstrates that SpMPK1, SpMPK2, and SpMPK3 may play important roles in the drought tolerance of tomato plants. In drought tolerance, the functions of SpMPK1 and SpMPK2 may be redundant, and they may overlap with that of SpMPK3. Future studies will employ genetic transformation experiments to further explore how SpMPKs function in the drought tolerance of tomato plants.
Supplementary Information
Acknowledgments
We owe gratitude to AVRDC-The World Vegetable Center for supplying the experimental material. This work is funded by the “13115” Technology Innovation Project of Shaanxi Province (#2009ZDKG-14), and the Science and Technology Coordination and Innovation Project of Shaanxi Province (#2011KTCL02-03).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ichimura K., Shinozaki K., Tena G., Sheen J., Henry Y., Champion A., Kreis M., Zhang S., Hirt H., Wilson C. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002;7:301–308. doi: 10.1016/s1360-1385(02)02302-6. [DOI] [PubMed] [Google Scholar]
- 2.Tena G., Asai T., Chiu W.-L., Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 2001;4:392–400. doi: 10.1016/s1369-5266(00)00191-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S., Klessig D.F. MAPK cascades in plant defense signaling. Trends Plant Sci. 2001;6:520–527. doi: 10.1016/s1360-1385(01)02103-3. [DOI] [PubMed] [Google Scholar]
- 4.Pitzschke A., Schikora A., Hirt H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009;12:421–426. doi: 10.1016/j.pbi.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Komis G., Illés P., Beck M., Šamaj J. Microtubules and mitogen-activated protein kinase signalling. Curr. Opin. Plant Biol. 2011;14:650–657. doi: 10.1016/j.pbi.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Jonak C., Ökrész L., Bögre L., Hirt H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002;5:415–424. doi: 10.1016/s1369-5266(02)00285-6. [DOI] [PubMed] [Google Scholar]
- 7.Lampard G.R., Lukowitz W., Ellis B.E., Bergmann D.C. Novel and expanded roles for MAPK signaling in Arabidopsis stomatal cell fate revealed by cell type-specific manipulations. Plant Cell Online. 2009;21:3506–3517. doi: 10.1105/tpc.109.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Ngwenyama N., Liu Y., Walker J.C., Zhang S. Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell Online. 2007;19:63–73. doi: 10.1105/tpc.106.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q., Xie Q.-G., Smith-Becker J., Navarre D.A., Kaloshian I. Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signaling cascades. Mol. Plant Microbe Interact. 2006;19:655–664. doi: 10.1094/MPMI-19-0655. [DOI] [PubMed] [Google Scholar]
- 10.Kandoth P.K., Ranf S., Pancholi S.S., Jayanty S., Walla M.D., Miller W., Howe G.A., Lincoln D.E., Stratmann J.W. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA. 2007;104:12205–12210. doi: 10.1073/pnas.0700344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melech-Bonfil S., Sessa G. The SlMKK2 and SlMPK2 genes play a role in tomato disease resistance to Xanthomonas campestris pv. vesicatoria. Plant Signal. Behav. 2011;6:154–156. doi: 10.4161/psb.6.1.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirayama T., Shinozaki K. Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–351. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 13.Iuchi S., Kobayashi M., Taji T., Naramoto M., Seki M., Kato T., Tabata S., Kakubari Y., Yamaguchi-Shinozaki K., Shinozaki K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 14.Alzwiy I.A., Morris P.C. A mutation in the Arabidopsis MAP kinase kinase 9 gene results in enhanced seedling stress tolerance. Plant Sci. 2007;173:302–308. [Google Scholar]
- 15.Xing Y., Jia W., Zhang J. AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 2007;58:2969–2981. doi: 10.1093/jxb/erm144. [DOI] [PubMed] [Google Scholar]
- 16.Hwa C.-M., Yang X.-C. The AtMKK3 pathway mediates ABA and salt signaling in Arabidopsis. Acta Physiol. Plant. 2008;30:277–286. [Google Scholar]
- 17.Xing Y., Jia W., Zhang J. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 2008;54:440–451. doi: 10.1111/j.1365-313X.2008.03433.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang X.-J., Zhu S.-Y., Lu Y.-F., Zhao R., Xin Q., Wang X.-F., Zhang D.-P. Two coupled components of the mitogen-activated protein kinase cascade MdMPK1 and MdMKK1 from apple function in ABA signal transduction. Plant Cell Physiol. 2010;51:754–766. doi: 10.1093/pcp/pcq037. [DOI] [PubMed] [Google Scholar]
- 19.Xing Y., Jia W., Zhang J. AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol. Biol. 2009;70:725–736. doi: 10.1007/s11103-009-9503-0. [DOI] [PubMed] [Google Scholar]
- 20.Petersen M., Brodersen P., Naested H., Andreasson E., Lindhart U., Johansen B., Nielsen H.B., Lacy M., Austin M.J., Parker J.E. Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell. 2000;103:1111–1120. doi: 10.1016/s0092-8674(00)00213-0. [DOI] [PubMed] [Google Scholar]
- 21.Nakagami H., Soukupová H., Schikora A., Zárský V., Hirt H. A mitogen-activated protein kinase kinase kinase mediates reactive oxygen species homeostasis in Arabidopsis. J. Biol. Chem. 2006;281:38697–38704. doi: 10.1074/jbc.M605293200. [DOI] [PubMed] [Google Scholar]
- 22.Gao M., Liu J., Bi D., Zhang Z., Cheng F., Chen S., Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18:1190–1198. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 23.Pitzschke A., Djamei A., Bitton F., Hirt H. A major role of the MEKK1–MKK1/2–MPK4 pathway in ROS signalling. Mol. Plant. 2009;2:120–137. doi: 10.1093/mp/ssn079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudesblat G.E., Iusem N.D., Morris P.C. Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 2007;173:713–721. doi: 10.1111/j.1469-8137.2006.01953.x. [DOI] [PubMed] [Google Scholar]
- 25.Liu P.F., Chang W.C., Wang Y.K., Chang H.Y., Pan R.L. Signaling pathways mediating the suppression of Arabidopsis thaliana Ku gene expression by abscisic acid. Biochim. Biophys. Acta-Gene Regul. Mech. 2008;1779:164–174. doi: 10.1016/j.bbagrm.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Jammes F., Song C., Shin D., Munemasa S., Takeda K., Gu D., Cho D., Lee S., Giordo R., Sritubtim S. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. USA. 2009;106:20520–20525. doi: 10.1073/pnas.0907205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desikan R., Cheung M.K., Bright J., Henson D., Hancock J.T., Neill S.J. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. J. Exp. Bot. 2004;55:205–212. doi: 10.1093/jxb/erh033. [DOI] [PubMed] [Google Scholar]
- 28.Kim T.-H., Böhmer M., Hu H., Nishimura N., Schroeder J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang P., Song C.P. Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 2008;178:703–718. doi: 10.1111/j.1469-8137.2008.02431.x. [DOI] [PubMed] [Google Scholar]
- 30.Cho D., Shin D., Jeon B.W., Kwak J.M. ROS-mediated ABA signaling. J. Plant Biol. 2009;52:102–113. [Google Scholar]
- 31.MacRobbie E. Signal transduction and ion channels in guard cells. Philos. Trans. R. Soc. Lond. Ser. B. 1998;353:1475–1488. doi: 10.1098/rstb.1998.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thordal-Christensen H., Zhang Z., Wei Y., Collinge D.B. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley—Powdery mildew interaction. Plant J. 1997;11:1187–1194. [Google Scholar]
- 33.Ichimura K., Casais C., Peck S.C., Shinozaki K., Shirasu K. MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 2006;281:36969–36976. doi: 10.1074/jbc.M605319200. [DOI] [PubMed] [Google Scholar]
- 34.Kong F., Wang J., Cheng L., Liu S., Wu J., Peng Z., Lu G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene. 2012;499:108–120. doi: 10.1016/j.gene.2012.01.048. [DOI] [PubMed] [Google Scholar]
- 35.Brock A.K., Willmann R., Kolb D., Grefen L., Lajunen H.M., Bethke G., Lee J., Nürnberger T., Gust A.A. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 2010;153:1098–1111. doi: 10.1104/pp.110.156109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levine A., Tenhaken R., Dixon R., Lamb C. H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell. 1994;79:583–593. doi: 10.1016/0092-8674(94)90544-4. [DOI] [PubMed] [Google Scholar]
- 37.Zhu D., Scandalios J.G. Differential accumulation of manganese-superoxide dismutase transcripts in maize in response to abscisic acid and high osmoticum. Plant Physiol. 1994;106:173–178. doi: 10.1104/pp.106.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Desikan R., Reynolds A., Hancock J., Neill S. Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 1998;330:115–120. doi: 10.1042/bj3300115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Ye N., Liu R., Chen M., Zhang J. H2O2 mediates the regulation of ABA catabolism and GA biosynthesis in Arabidopsis seed dormancy and germination. J. Exp. Bot. 2010;61:2979–2990. doi: 10.1093/jxb/erq125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pei Z.-M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G.J., Grill E., Schroeder J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature. 2000;406:731–734. doi: 10.1038/35021067. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Schiff M., Dinesh-Kumar S. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313x.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 42.Velásquez A.C., Chakravarthy S., Martin G.B. Virus-induced gene silencing (VIGS) in Nicotiana benthamian a and tomato. J. Vis. Exp. 2009 doi: 10.3791/1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Gong P., Zhang J., Li H., Yang C., Zhang C., Zhang X., Khurram Z., Zhang Y., Wang T., Fei Z. Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 2010;61:3563–3575. doi: 10.1093/jxb/erq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S., Choi H., Suh S., Doo I.-S., Oh K.-Y., Choi E.J., Taylor A.T.S., Low P.S., Lee Y. Oligogalacturonic acid and chitosan reduce stomatal aperture by inducing the evolution of reactive oxygen species from guard cells of tomato and Commelina communis. Plant Physiol. 1999;121:147–152. doi: 10.1104/pp.121.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daudi A., Cheng Z., O’Brien J.A., Mammarella N., Khan S., Ausubel F.M., Bolwell G.P. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell Online. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.