Abstract
Molluscs are a diverse animal phylum with a formidable fossil record. Although there is little doubt about the monophyly of the eight extant classes, relationships between these groups are controversial. We analysed a comprehensive multilocus molecular data set for molluscs, the first to include multiple species from all classes, including five monoplacophorans in both extant families. Our analyses of five markers resolve two major clades: the first includes gastropods and bivalves sister to Serialia (monoplacophorans and chitons), and the second comprises scaphopods sister to aplacophorans and cephalopods. Traditional groupings such as Testaria, Aculifera, and Conchifera are rejected by our data with significant Approximately Unbiased (AU) test values. A new molecular clock indicates that molluscs had a terminal Precambrian origin with rapid divergence of all eight extant classes in the Cambrian. The recovery of Serialia as a derived, Late Cambrian clade is potentially in line with the stratigraphic chronology of morphologically heterogeneous early mollusc fossils. Serialia is in conflict with traditional molluscan classifications and recent phylogenomic data. Yet our hypothesis, as others from molecular data, implies frequent molluscan shell and body transformations by heterochronic shifts in development and multiple convergent adaptations, leading to the variable shells and body plans in extant lineages.
1. Introduction
Molluscs are a morphologically megadiverse group of animals with expansive body plan modifications. There is no doubt about the monophyly of Mollusca as a whole or of any of the eight extant molluscan classes, based on strong morphoanatomical evidence and the consensus of molecular studies [1]. Despite a number of important recent studies, resolving ingroup molluscan topology remains contentious (Figure 1(a)) and a major challenge of invertebrate evolution [2].
Figure 1.
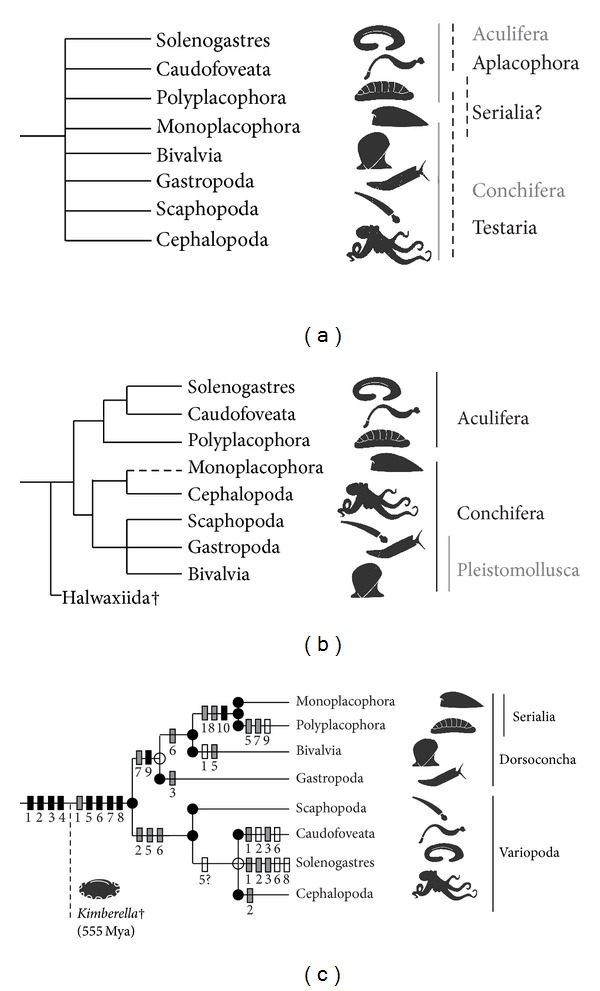
Schematic trees of molluscan relationships. (a) showing traditional proposed subdivisions. (b) consensus tree of two recent molluscan phylogenies inferred from large-scale genomic data by Kocot et al. [5] and Smith et al. [8]. The traditional concepts of Aculifera and Conchifera are supported but with differing positions of scaphopods. Monoplacophora is missing in the data set of Kocot et al. [5] (dotted line reflects the position of Monoplacophora in Smith et al. [8]). (c) the preferred multilocus tree with morphological features indicated numerically on branches. Unfilled dots indicate maximum Bayesian node support, filled dots additional high (>75%) bootstrap support in ML analyses. The Ediacaran fossil genus Kimberella corresponds to the description of molluscan stem-group features (1–4, below); crown group taxa originating in the Cambrian and later are united by additional features. Black boxes indicate first appearance of features; grey boxes indicate significant adaptive change; unfilled boxes indicate trait reversals: (1) radula: bipartite in stem molluscs and paedomorphic aplacophorans; broadened, on cartilages and specialised in crown molluscs, stereoglossate-like in Serialia; lost in Bivalvia (and several gastropods); (2), foot with broad gliding sole: transformed into digging foot in variopods (and derived bivalves), narrowed and reduced in aplacophorans, and forming the funnel in cephalopods; (3) circumpedal mantle cavity, miniaturised and anteriorly dislocated in torted gastropods while placed posteriorly in vermiform molluscs; (4) separate mantle covered with cuticula (with calcareous spicules in chitons, aplacophorans, and probably Kimberella); (5) dorsal shell: duplicated/fragmented in bivalves and chitons, lost in aplacophorans (and members of most other classes); (6) head with paired appendages: multiplied into feeding tentacles in variopods; trait for head reduction in bivalves plus Serialia and aplacophorans; (7) pericardium: heart fused around intestine in Dorsoconcha; (8) paired ctenidia: expanded to serially repeating gills in Serialia (and nautiloid cephalopods) and reduced in Solenogastres and some gastropod lineages; (9) complex stomach with style (reduced in carnivorous subgroups and chitons; convergently (?) present in a caudofoveate family); (10) paired eightfold dorsoventral muscles; (11) (not shown) statocysts (lost convergently in chitons and aplacophorans); (12) (not shown) suprarectal visceral commissure (subrectal convergently in chitons and aplacophorans).
Other studies have not had access to suitable material for broad taxon sampling, in particular for monoplacophorans, a class of small deep-sea molluscs that still remain rare and largely inaccessible [3, 4]. Among several recent studies on molluscan phylogeny, most use a subset of classes [5–7]; only one phylogenomic study so far has included all eight classes [8].
Multigene studies on ribosomal proteins [6] and housekeeping genes [7] and two broad phylogenomic (EST-based) data sets [5, 8] supported a monophyletic clade Aculifera. This clade comprises those molluscs with a partial or entire body covered by a cuticle with calcareous spicules or scales and is composed of shell-less vermiform molluscs (aplacophoran) and shell-plate bearing Polyplacophora (chitons). The opposing clade Conchifera (incorporating the five classes with a “true” shell) remains controversial; phylogenomic studies recovered a monophyletic clade Conchifera [5, 8], but ribosomal protein multigene and housekeeping gene analyses showed paraphyletic Conchifera [6, 7].
A contradictory alternative hypothesis was proposed by earlier ribosomal RNA-dominated multilocus studies that included Monoplacophora and recovered this class as the sister to Polyplacophora [4, 9, 10]. This clade “Serialia” combines conchiferan and aculiferan members and is thus incompatible with results of recent molecular studies or the morphological Testaria (i.e., Conchifera + Polyplacophora) hypothesis (Figure 1). This result was widely criticised in the literature (e.g., [11]). Yet initial deficiencies [12] of the study by Giribet et al. [9] were addressed by Wilson et al. [10] and Serialia recovered again in a partially overlapping data set by Meyer et al. [13] and independently by Kano et al. [4].
The single phylogenomic data set with a monoplacophoran species also indicated some signal for Serialia, though weaker than that supporting a relationship of cephalopods and monoplacophorans within Conchifera [8]. Phylogenomic data sets cannot yet cover the same density of taxon sampling relative to targeted gene approaches, and while systematic errors of phylogenomic analyses have been explored recently (e.g., [14–16]), there is already a suite of tools available for addressing well-known pitfalls of ribosomal RNA-based sequences (e.g., [17–20]). All data sets may still contribute to ongoing investigations of phylogeny if used and interpreted with care.
Where published topologies differ radically from concepts born from morphoanatomical hypotheses, these results have often been dismissed as artefacts even by the studies' own authors. In addition to the “Serialia” concept, several studies over the last decade have repeatedly recovered Caudofoveata sister to Cephalopoda (e.g., [6, 9, 10, 21–23]). But this pattern has low support values [6, 12]. The position of scaphopods is also highly variable, sometimes in a clade with gastropods and bivalves [5, 7, 8] or sister to aplacophorans and cephalopods [9, 10, 21]. With only eight major clades to rearrange, it could be a serious handicap that many studies exploring molluscan topology have had to exclude one (e.g., [5, 7, 21]) to three (e.g., [4, 6]) classes, and all but one previous study [10] used single-taxon exemplars for at least one [9] to as many as three [7, 8] of those clades. More and better quality data from the monoplacophorans are necessary to resolve molluscan relationships and particularly the two mutually exclusive hypotheses Serialia and Aculifera. We assembled a large multilocus data set for molluscs, including novel sequences of three monoplacophoran species (added to previously published data for only two species, Veleropilina seisuimaruae and Laevipilina hyalina). To determine the plausibility of this new topology, we applied several tests for phylogenetic informativity, saturation of sites, and compositional heterogeneity within the molecular data sets and have also considered our results against other molecular, morphological, and fossil evidence. Finally we calculated a new time tree via a relaxed molecular clock approach, using multiple sets of fossil calibration points.
Applying carefully calibrated molecular clocks on broad extant taxon sets and reconstructing characters on dated ancient lineages are indispensable for interpretation of enigmatic key fossils such as Halkieria or Nectocaris that may form part of the early evolutionary history of the group (e.g., [24–27]). We present an alternative view on molluscan evolution that supports the Serialia hypothesis and demonstrates that the debate on pan-molluscan relationships is still in progress.
2. Material & Methods
2.1. DNA Extraction, PCR, and Sequencing
DNA from 12 molluscan taxa, including 3 previously unsampled monoplacophoran species, was extracted using the Qiagen Blood and Tissue Kit (Qiagen, Hilden) by following the manufacturer's instructions. Amplifications of the four standard marker fragments, partial 16S, partial 18S, partial 28S, and complete H3, were carried out under PCR conditions and with primer pairs shown (see Supplementary Material available online at http://dx.doi.org/10.1155/2013/407072). Sequencing reactions were operated on an ABI 3730 48 capillary sequencer of the sequencing service of the Department of Biology of the LMU Munich by using the amplification primers. Newly generated sequences were edited in Sequencher version 4.7 (Gene Codes Inc., Ann Arbor, MI, USA).
2.2. Taxon and Gene Sampling
To compile a comprehensive and dense taxon sampling for resolving deep molluscan relationships, we expanded earlier published data sets [9, 10] by our own and archived (Genbank) data, including a broad selection of outgroups and initially including any molluscs with substantial sequence information available for five standard marker fragments (partial 16S rRNA, partial or complete 18S rRNA, partial 28S rRNA, complete H3, and partial COI). In some poorly sampled but significant ingroup clades we also included species with fragmentary sequence data. Previously unpublished, partial 16S, complete 18S and 28S, complete H3, and partial COI sequences of Veleropilina seisuimaruae were provided separately by one of the authors (YK). The total initial data set comprised 158 taxa (141 molluscan and 17 outgroup taxa; Suppl. Table 2).
2.3. Data Cleaning and Alignment
All the downloaded and new single sequences, including all 28S sequences, and all individual amplicons for 18S sequences in Solenogastres, were cross-checked against the nucleotide database of BLAST [29] by using the blastn algorithm. Potentially aberrant or problematic fragments were removed from the data sets (Suppl. Table 3A).
In some bivalve 28S sequences a dubious part of ca. 500 bp was detected in an otherwise homogeneous molluscan alignment. This portion differed substantially in most bivalve taxa but not in all and was highly heterogeneous also in closely related species. No pattern could be observed, so we removed the dubious region (Suppl. Table 3B).
The 18S sequences of Solenogastres were partially excluded due to contamination. Retained sequences of Epimenia species (E. sp., E. australis, and E. babai) were aligned separately with the first uncontaminated sequences of Meyer et al. [13], and resulting large gaps were cut by hand according to the template sequences of Micromenia fodiens, Simrothiella margaritacea and Wirenia argentea (Meyer sequences in [13]).
Patellogastropoda has aberrant 18S and 28S sequences with many indels causing highly incongruent alignments (own observations), leading to long branches and attraction artefacts in previous [13] and our own analyses. Patellogastropoda clustered with long branched Cephalopoda and Solenogastres under different regimes (Table 1). To verify the correct position of Patellogastropoda within or outside other Gastropoda a more focused data set was generated comprising only gastropod taxa plus some selected, short-branched outgroup taxa, that is, two bivalves, two polyplacophorans, one annelid, and one kamptozoan. This alignment is more homogeneous, and patellogastropods appear as a moderately long branch in a rather derived position within the Gastropoda (Suppl. Figure 2). So we confirm that patellogastropods show aberrant evolution leading to long branch attraction artefacts in broader data sets [13]; therefore we excluded this clade from the main analyses.
Table 1.
Preanalyses comparing different taxon sampling and masking strategies; Mafft [30] and RNAsalsa [18] are alignment methods; Aliscore [17, 31] and Gblocks [35] are masking methods.
| Dataset | Alignment treatment | Alignment length (bp) | Major changes in tree topology, compared to main topology (Figure 2, Supplementary Figure 1) |
|---|---|---|---|
| Total set (158 taxa) | Mafft-cut and paste inconsistent blocks in 18S and 28S fragments-Aliscore | 10318 | Annelida s.l. sister to Mollusca; Aplacophora monophyletic (Caudofoveata sister to Solenogastres); Patellogastropoda clusters with Cephalopoda |
| Total set (158 taxa) | Mafft-RNAsalsa-Aliscore | 7597 | Mollusca non-monophyletic; Caudofoveata, Solenogastres, Cephalopoda, and Scaphopoda cluster with Annelida s.l.; Neritimorpha basal sister to remaining Gastropoda; Patellogastropoda sister to partial Vetigastropoda (Lepetelloida + Vetigastropoda s.s.) |
| Total set (158 taxa) | Mafft-RNAsalsa-Gblocks | 4083 | Nemertea + Entoprocta + Cycliophora is sister to Mollusca; Heterobranchia is sister to remaining Mollusca; Patellogastropoda clusters with Solenogastres and Cephalopoda |
| Large set (142 taxa, excluding Patellogastropoda) | Mafft-Gblocks | 5550 | Annelida s.l. + Entoprocta + Cycliophora is sister to Mollusca |
| Large set (142 taxa, excluding Patellogastropoda) | Mafft-Aliscore | 8721 | Main analyses (Figure 2, Supplementary Figure 1) |
Single alignments (per fragment) were created with Mafft version 6.847b [30] with the implemented E-INS-i algorithm. Alignments of 16S, 18S, and 28S rRNA were masked with Aliscore version 5.1 [17, 31] by running 10,000,000,000 replicates. All ambiguous positions were automatically cut with Alicut version 2.0 [17, 31] to remove highly variable positions that could lead to aberrant phylogenetic signals. The alignments of protein coding genes H3 and COI were manually checked for stop codons using MEGA5 [32]. The single data sets were concatenated automatically using FASconCAT version 1.0 [33]. This procedure resulted in a total alignment of 142 taxa with 8721 bp in length and a proportion of 60% gaps (Suppl. Table 5). Where taxon sampling had to be modified, for example, removing taxa or dubious gene fragments, this was done in the initial single data sets and the complete procedure of alignment, masking and concatenation was carried out again.
Final analyses were computed with the large data set excluding Patellogastropoda (142-taxon set), a targeted taxon subset (81-taxon set, alignment length 8367 bp, proportion of gaps 57%) after pruning fast-evolving species or derived members of densely sampled undisputed clades, and the gastropod data set (all gastropods including Patellogastropoda plus selected slowly evolving outgroups). Moreover, we generated and analysed diverse data sets for control reasons to test interclass topologies: the 142- and 81-taxon sets without Aplacophora, the 142-taxon set without long-branched Cephalopoda and Solenogastres, the 142-taxon set with COI and H3 coded as amino acids (142-taxon set amino acid), and one data set that comprises only 18S, 28S, and H3 fragments of the 142-taxon set (Suppl. Table 5). The concatenated sequence matrices of the two main analyses (142-taxon set and 81-taxon set) were deposited at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S14594). New sequences generated herein were deposited at Genbank (Suppl. Table 2).
2.4. Preanalyses of the Data
Since saturated sequences have minimal or no phylogenetic signal and could even lead to anomalous results, we measured substitution saturation of the protein coding genes, namely, H3 and COI, with Xia's method implemented in DAMBE version 5.2.31 [37]. We used default parameters, and the proportion of invariable sites was specified. The method was executed for all three codon positions together, for combined first and second codon positions, and for third codon position separately. In both cases, H3 and COI, the index of substitution saturation (Iss) values of all three codon positions in combination were significantly smaller than critical index of substitution saturation (Iss.c) values. This was also true for the alignments of first and second codon positions. This assumes that those positions conserve phylogenetic signal and are useful for further analyses. In the case of third codon positions only, substantial saturation could be observed (Iss significantly higher than Iss.c). All results are shown in Supplementary Table 6. Although substitution saturation was observed in third codon positions of H3 and COI, we ran additional analyses with the complete sequence information (1st, 2nd, and 3rd codon positions) to implement potential phylogenetic signal for lower taxonomic levels.
To crosscheck the phylogenetic results of the data sets with and without excluded third codon positions of protein coding genes we conducted the same analyses with all three codon positions included, using distinct models of evolution for the three different codon positions and without third codon positions of H3 and COI.
Testing the evolutionary models for all genes and in case of COI and H3 for every single codon position and for codon positions one and two versus position three was carried out with the programs Modeltest version 3.7 [38] (for complete alignments) and MrModeltest version 2.3 [39] (for codon positions) by the help of PAUP* version 4b10 for Windows [40]. With the amino acid alignments of H3 and COI we additionally tested for the best fitting amino acid model of evolution using ProtTest version 2.4 [41]. As RAxML provides only a part of the models that can potentially be tested by ProtTest we only selected those models in our ProtTest analysis (DAYHOFF, DCMUT, JTT, MTREV, WAG, RTREV, CPREV, VT, BLOSUM62, and MTMAM). The resulting best models for all genes (16S, 18S, 28S, H3, and COI), distinct codon positions of H3 and COI, and amino acid alignments of H3 and COI as well as the corresponding proportions of invariant sites and the gamma distribution shape parameters are shown in Supplementary Table 4.
2.5. Phylogenetic Analyses
Maximum Likelihood (ML) analyses for all data sets were executed using RAxML-HPC for Windows [28] and RAxML version 7.2.6 [28] on the Linux cluster of the Leibniz Computer Centre. Parameters for the initial rearrangement settings and the rate categories were optimised under the GTRCAT model of evolution and a partition by genes (16S, 18S, and 28S) and codon positions (COI, H3) by conducting the hardway analysis described by Stamatakis [42].
First, a set of 10 randomised Maximum Parsimony (MP) starting trees was generated. Second, based on this set of starting trees, the ML trees with a specified setting of initial rearrangements (−i 10) and with an automatically determined initial rearrangement setting had to be inferred. Third, the number of rate categories was adjusted. Initial setting –c 10 was augmented by increments of 10 up to –c 50 for all MP starting trees. The fourth step was to execute 200 inferences on the original alignments. Finally, values of 1000 bootstrap topologies were mapped on the best-scoring ML tree.
Bayesian analyses for selected data sets were conducted with MrBayes v. 3.1.2 [43]. Partitioning with corresponding models of evolution, substitution rates and nucleotide frequencies were applied according to the results of Modeltest [38], MrModeltest [39], and ProtTest [41]. One tree was sampled every 1000 generations. If the average standard deviation of split frequencies declined 0.01 after 5 million generations the analysis was stopped. If not, analysis was continued with another 5 million generations. If the average standard deviation of split frequencies still did not decrease, the log likelihood values were examined with Tracer version 1.5 [44]. If the run reached stationarity, the analysis was stopped. Burn-in was set to 2500 after 5 million generations and to 5000 after 10 million generations.
2.6. Molecular Clock Analyses
Time estimations were performed with the software package BEAST version 1.6.1 [34]. The program is based on the Bayesian Markov Chain Monte Carlo (MCMC) method and therefore can take into account prior knowledge of the data. That is used when nodes in the topology are calibrated and the rate of molecular evolution along the branches is estimated.
We used nine fossil calibration points (Suppl. Table 7) with their corresponding prior distributions and assumed a relaxed clock with a lognormal distribution [45] of the rates for each branch (Suppl. Table 7). This setting is recommended because it additionally gives an indication of how clock-like the data are [46]. Calibration points were set with a minimum bound according to Jörger et al. [47]. To reduce computing time we used the targeted (81-taxa) data set for time estimations. The topology was constrained according to the resulting tree of the phylogenetic analyses.
An Xml-file with all information on data, calibration points, priors and the settings for the MCMC options was created with BEAUti version 1.4.7 [34]. Gamma-shaped priors for all nine calibration points were used (Suppl. Table 7). We assumed that the lower bound of each calibration point is not more than 10% of its maximum age. In case that the next older fossil is within these 10% boundary we used the maximum age of that fossil as lower bound for the younger fossil [48].
Detailed partitioning of genes (16S, 18S, and 28S) and codon positions of COI and H3 and the constraint tree topology were added by hand to the Xml-file. The analysis was executed for 30 million generations, sampling one tree every 1000 generations on the Linux cluster of the Leibniz Computer Centre. The implemented program Tracer version 1.5 [44] was used to confirm that posterior probabilities had reached stationarity. Burn-in was set to 25% (7500), so 22,500 trees were effectively analysed with TreeAnnotator version 1.6.1 [34] to form the summary tree. Further, to check the reliability of our fossils, we repeated the same analysis several times and always omitted one calibration point (Table 2; Suppl. Table 7).
Table 2.
Sensitivity tests of individual calibration nodes used for relaxed molecular clock time estimates of major molluscan groups. Table shows influence of single calibration points on node ages of all other calibration points.
| Excluded calibration point | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| None | Diversification of Mollusca | Split Serialia/Bivalvia | Origin of Cephalopoda | Split Polyplacophora/Monoplacophora | Origin of Pteriomorpha | Origin of Caenogastropoda | Diversification of Scaphopoda | Split Astarte/Cardita | Diversification of Polyplacophora | ||
| Calibrated nodes | Diversification of Mollusca | 551.02 | 683.50* | 550.58 | 549.76 | 551.52 | 551.55 | 551.59 | 551.68 | 552.10 | 551.72 |
| Split Serialia/Bivalvia | 530.93 | 533.98 | 523.58* | 530.80 | 530.88 | 530.36 | 530.82 | 531.01 | 530.82 | 531.01 | |
| Origin of Cephalopoda | 504.92 | 511.40 | 504.40 | 431.05* | 504.26 | 504.85 | 504.75 | 503.96 | 504.16 | 504.35 | |
| Split Polyplacophora/Monoplacophora | 493.06 | 493.44 | 491.79 | 493.68 | 431.68* | 493.47 | 493.13 | 493.47 | 493.79 | 493.14 | |
| Origin of Pteriomorpha | 475.06 | 474.81 | 474.20 | 474.43 | 474.97 | 376.65* | 474.56 | 475.20 | 474.16 | 474.82 | |
| Origin of Caenogastropoda | 421.49 | 422.82 | 421.77 | 421.91 | 422.61 | 421.54 | 326.50* | 421.95 | 420.85 | 421.61 | |
| Diversification of Scaphopoda | 359.95 | 360.02 | 359.97 | 359.55 | 360.12 | 359.59 | 359.37 | 382.50* | 359.19 | 359.60 | |
| Split Astarte/Cardita | 325.43 | 325.40 | 325.20 | 324.98 | 325.45 | 324.55 | 325.37 | 325.09 | 44.34* | 325.63 | |
| Diversification of Polyplacophora | 233.44 | 233.49 | 233.37 | 233.18 | 233.75 | 233.42 | 233.63 | 233.57 | 232.71 | 243.67* | |
Bold ages marked with an asterisk (∗) indicate time estimations without calibration of this node.
2.7. Testing Hypotheses
Several existing hypotheses about the molluscan interrelationships (Table 3) were tested by executing Approximately Unbiased tests (AU tests) implemented in Treefinder version of October 2008 [36]. Therefore the input constraint trees were computed with RAxML-HPC [28] by using the –g-option and the associated partition by genes and codon positions. Those input tree topologies were tested in Treefinder with maximum number of replicates under the GTR model.
Table 3.
Testing alternative topologies against various data sets. Results of Approximately Unbiased (AU) tests with Treefinder [36], various schemes. P-values of AU Test executed on selected taxon and data sets. Tested tree topologies were constrained in RAxML [28]. Only meaningful tests have been executed. P-value > 0.05: constrained topology is not rejected; P-value < 0.05: constrained topology is rejected significantly; P-value = 0: constrained topology is rejected with high significance.
| Constrained topology | 142-taxon set. all markers | 81-taxon set. all markers | 142-taxon set. 18S + 28S + H3 | Aplacophora removed from 142-taxon set. all markers |
|---|---|---|---|---|
| Sinusoida | 0.4244 | Not tested | 0.2652 | 0.0383 |
| Mollusca + Kamptozoa | 0.0 | Not tested | 0.0 | 0.0 |
| Mollusca + Annelida | 0.7421 | 0.7097 | 0.4090 | 0.3876 |
| Testaria | 0.0 | 0.0 | 0.0 | Not tested |
| Aculifera | 0.0 | 0.0 | 0.0 | Not tested |
| Aplacophora | 0.6908 | 0.3651 | 0.7730 | Not tested |
| Conchifera | 0.0 | 0.0 | 0.0 | 0.0333 |
| Pleistomollusca | 0.6665 | 0.0 | 0.0863 | 0.1927 |
| Monoplacophora + Cephalopoda | 0.1389 | 0.0632 | 0.0 | 0.2779 |
| Scaphopoda + Gastropoda + Bivalvia | 0.0154 | 0.0 | 0.0 | 0.1065 |
| Scaphopoda + Cephalopoda | 0.1913 | 0.0 | 0.2527 | 0.6914 |
| Scaphopoda + Cephalopoda + Gastropoda | 0.0 | 0.0 | 0.0 | 0.7232 |
| Scaphopoda + Gastropoda | 0.8850 | 0.9452 | 0.0573 | 0.8271 |
| Diasoma (Scaphopoda + Bivalvia) | 0.0 | 0.0 | 0.0 | 0.0 |
| Monophyletic Protobranchia | 0.0219 | 0.0 | 0.1085 | 0.0188 |
| Dorsoconcha | 0.6830 | 0.1097 | 0.3503 | 0.4048 |
| Variopoda | 0.3170 | 0.8903 | 0.6497 | 0.5952 |
3. Results and Discussion
3.1. Analyses
Analysing traditional multilocus markers for several large taxon sets with Maximum Likelihood and Bayesian methods under different alignment and masking regimes (Table 1, Suppl. Table 5), we recovered consistent phylogenetic trees (Figure 1(c)) with monophyletic Mollusca in contrast to other studies with similar markers [9, 10, 19, 21] and strong support for the monophyly of all molluscan classes, including Bivalvia (also in contrast to some earlier studies [9, 19, 21]).
Our approach included rigorously testing of all amplicons before and after alignment, which led to the exclusion of aberrant or problematic, previously published sequences from the data set (Suppl. Table 3). Criticism of previous accounts using the same set of markers has included the incomplete representation of taxa and the varying extant of missing data [12, 49]. Missing data is a common burden of multilocus studies and will be more severe for phylogenomic approaches [14, 15]. Our preanalyses showed that dubious sequences or ambiguous parts of alignments had much greater effect on the outcome than selecting taxa with the highest amount of data available. Rather than maximizing sequences per species, we concentrated on increasing taxon sampling to minimise potential branch lengths. Our quality controlled 158-taxon set includes 17 lophotrochozoan outgroups. Analytical trials on different subsets of nonmolluscan outgroups altered outgroup topology and support values of some basal ingroup nodes but did not change the ingroup topology (Figure 1).
Alignment issues involved in ribosomal RNA data were addressed by an array of measures proven to be beneficial ([20]; see Section 2). Potential homoplasy in protein coding genes (especially the third codon positions) in our preferred multilocus analysis was addressed by additionally running the analysis with those fragments (COI and H3) encoded as amino acids. This had little effect on the topology but supported monophyletic Aplacophora. We applied a variety of alignment tools, including masking (Aliscore [17]) and refinement algorithms based on secondary structures (RNAsalsa [18]) and applied compartmentalised analyses of taxon clusters causing obvious alignment problems. Excluding patellogastropods (142-taxon set, Suppl. Figure 1; see Section 2) did not change our molluscan backbone topology (Figure 1(c)) but improved alignments. Separately analysing gastropods plus some slowly evolving outgroup taxa shows patellogastropods cluster with vetigastropods (Suppl. Figure 2). Our main aim was to elucidate molluscan relationships at the class level; thus we further pruned outgroups and fast-evolving members from more densely sampled ingroups (such as heterobranch gastropods) and used an 81-taxon set presented here in our main analysis (Figure 2).
Figure 2.
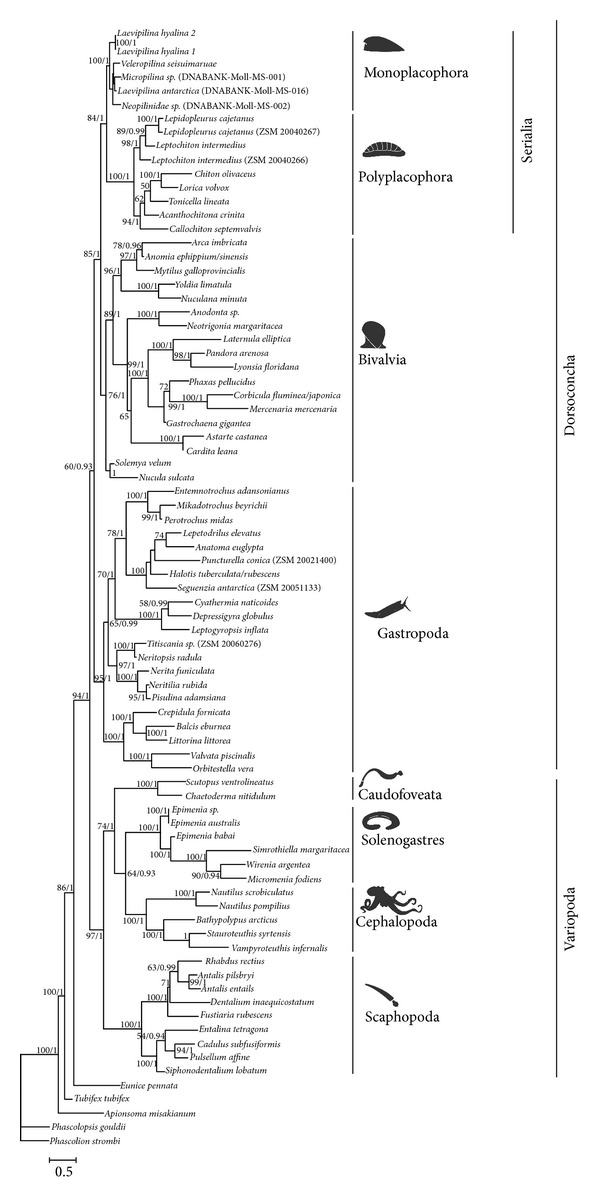
Preferred molluscan tree. Maximum Likelihood analysis (RAxML [28], hardway) of pruned 81-taxon set; values at nodes refer to bootstrap support (1000 pseudoreplicates, first value) and posterior probabilities obtained from the Bayesian analysis (second value).
3.2. The Basal Molluscan Dichotomy
In our new tree, the phylum Mollusca is divided into two clades (Figure 1(c), Figure 2, Suppl. Figure 1). The first clade is composed of Gastropoda sister to a clade of Bivalvia and Serialia (Monoplacophora + Polyplacophora). For convenience we will refer to this clade as “Dorsoconcha”; the name refers to the (plesiomorphic) presence of a dorsal shell for members of this clade, though modified to two lateral valves in bivalves and to (7-)8 dorsal plates in chitons, and the shell internalised or lost multiple times especially among gastropods.
Gastropods, bivalves, and monoplacophorans are commonly considered to be united by their single shell (secondarily split in bivalves) built by a shell gland at the mantle border (and by the entire mantle roof secreting organic matrix and calcareous layers letting the shell grow thicker, or repair damage). Chitons are traditionally excluded from the hypothetical clade “Conchifera” on the basis of their eight shell plates. The chiton girdle is also covered by a cuticle with embedded calcareous and organic sclerites, similar to the body cuticle of the shell-less aplacophorans, but according to our results, this is convergent and may reflect the different, single versus multicellular spicule formation in these taxa [50]. That chitons cluster with monoplacophorans rather than aplacophorans is congruent to previous molecular approaches that included monoplacophoran exemplars [4, 9, 10, 13]. The exception is the phylogenomic study by Smith et al. [8], in which a single monoplacophoran, Laevipilina hyalina, robustly clustered with cephalopods in the main analyses, though parts of the genes used also showed signal supporting an association with chitons.
In the second major molluscan clade, Scaphopoda are sister to a clade of vermiform Caudofoveata and Solenogastres, plus Cephalopoda. Herein we will call this clade “Variopoda,” referring to the various derived foot attributes of its members: the digging foot in Scaphopoda, reduced narrow gliding sole or completely lost in (adult) aplacophorans, and transformed in cephalopods possibly building parts of tentacles and funnel. Dorsoconcha appears as a monophyletic group although bootstrap support is low (60%), and the Variopoda is strongly supported in all Maximum Likelihood analyses; Bayesian posterior probabilities are high for both nodes (Figure 2, Suppl. Figure 1).
The placement of aplacophorans within Variopoda is unconventional, but a sister relationship between Scaphopoda and Cephalopoda has been previously put forward [51, 52]. Previous multilocus approaches with broad taxon sampling (i.e., more than one exemplar of each aplacophoran class) are actually not in general disagreement with Variopoda, since contaminated aplacophoran sequences may account for occasionally aberrant topologies [9, 10, 13]. Inner scaphopod topology resolves the two currently recognised groups Dentaliida and Gadilida, as does Cephalopoda splitting into modern Nautilida and Coleoidea, and is congruent with previous classifications [53].
We calculated time trees with a Bayesian molecular clock approach (Figure 3) using a mix of younger and older calibration points (Suppl. Table 7). We also tested sets of calibrations successively excluding each single calibration point used (Table 2) to minimise circularity involved by calculating individual node times [54]. All our time trees confirm a Precambrian origin of Mollusca (Table 2, Suppl. Table 8) in agreement with previous studies [7], and 95% confidence time bars of all our time trees allow for a Cambrian origin of those classes with a reliable fossil record (Figure 3). As a further sensitivity test we also calculated a time tree from a data set excluding aplacophorans; the topologies are congruent and node ages almost identical, confirming general time estimates (not shown).
Figure 3.
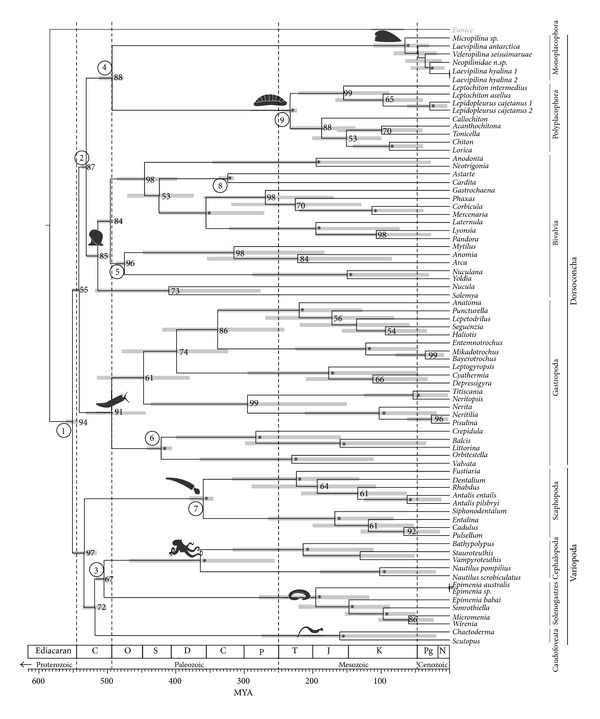
Chronogram of molluscan evolution. Divergence times (million years before present, Ma) estimated from BEAST version 1.6.1 [34] under an uncorrelated lognormal relaxed clock model; bars refer to the 95% highest posterior density. All nodes show maximum posterior probabilities (1.0, not indicated) from a run with 108 generations (25% burn-in). Numbers at nodes refer to bootstrap support values (>50%; asterisks are 100%) obtained from separate Maximum Likelihood analysis (RAxML [28], hardway, 1000 pseudoreplicates) of the same data set. Circled digits indicate calibrated nodes. Details of calibration can be found in Supplementary Table 7. Omitting Cambrian calibrations shifts molluscan diversification deeper into the Precambrian (for sensitivity analyses see Table 2).
Molluscan diversification occurred at an extremely rapid pace after the initial origination of the shell (Figure 3). Short branches at the base of the ingroup can be artefacts of signal erosion in deep nodes [55], but as we discuss below, the rapid early evolution of Mollusca is also supported by the fossil record. Our molecular clock indicates a potential time frame of only around 20–40 million years from the first shelled molluscs (ca. 560–540 Ma) to the presence of differentiated variopod, dorsoconch, gastropod, bivalve, and serialian stem lineages (ca. 520 Ma). The shell was central for rapid evolutionary success of molluscs, and shell modification and divergence are correlated with adaptive radiations during this early period.
3.3. Evaluating Molecular Data Sets
All recent multigene and phylogenomic studies [5–8] have tested the effects of gene sampling, analytical methods, and inference programs; like our results, their topologies were more or less robust, also against varying outgroup selection. Sensitivity analyses do not attribute the major split into Variopoda (or parts thereof) and Dorsoconcha or the recovery of Serialia to LBA effects. Yet our multilocus study uses fewer markers and nucleotides than “next-generation sequencing” studies [5–8], so it may be more prone to inadequate signal of certain markers or stochastic errors.
Split decomposition analyses of an earlier multilocus set [9] usually recovered the single monoplacophoran species among bivalves [12], consistent with a Dorsoconcha clade. Splitstree analyses (not shown) of our improved data set still show overall polytomy and some individual taxa are clearly misplaced in the network (e.g., the gastropod Crepidula clusters with cephalopods). Overall, most dorsoconch terminals are separated from variopods. Within Dorsoconcha, monoplacophorans cluster with chitons and bivalves. A lack of tree-like structure and a priori split support, especially in a large and heterogeneous taxon set, may not necessarily mean that there is too little signal for phylogenetic analyses; it just means that there is conflict that may or may not be resolved applying current models of sequence evolution.
Nuclear ribosomal RNA genes were shown to be informative even on deeper levels than basal molluscs, if treated adequately [20]. Other, supposedly faster-evolving mitochondrial markers (partial COI, 16S) were stringently masked herein, partitioned when necessary or excluded when saturated (Suppl. Tables 4–6). Combined analysis incorporates multiple tempos of evolution experienced by the different loci and is therefore more representative of deep evolutionary patterns. Our backbone topology is robust against varying the taxon and marker sets, masking and partitioning regimes, models of evolution, and methods of analyses (Table 1, Suppl. Tables 4 and 5).
3.4. Evaluating Alternative Morphological and Molecular Concepts
We directly evaluated the statistical fit of major competing morphology- or molecular-based concepts constraining our topologies and calculating their likeliness according to our data set. Using our preferred 81-taxon set with all markers, but also under most other schemes, the AU test rejects all the higher molluscan textbook concepts [1]: the Testaria, Aculifera, Conchifera, Cyrtosoma, and Diasoma hypotheses, with the highest possible statistical support; the same AU tests do not reject Dorsoconcha nor Variopoda (Table 3). We also tested our data against three new molecular concepts (Figure 1(b)): Pleistomollusca (Bivalvia + Gastropoda) established by Kocot et al. [5] and the clades of Monoplacophora and Cephalopoda [8] versus other conchiferans (Scaphopoda, Gastropoda, and Bivalvia) [5, 8]. Only the clade of Monoplacophora and Cephalopoda was not rejected with significant support in any of the main analyses, but all these groups received much lower AU values than our unconstrained topology.
While several recent phylogenomic studies recover Aculifera [5, 7, 8], the Serialia concept has been tested only by Smith et al. [8], by inclusion of a single monoplacophoran species. Though association with cephalopods is preferred, there is a weaker signal also for Serialia [8]. Kano et al. [4] recovered Serialia but did not include any aplacophoran taxa in their data set. The Serialia as a concept cannot be dismissed yet, and our dense taxon sampling herein, though based on far fewer sequences than recent phylogenomic approaches [5, 8], still may allow for a more differentiated and perhaps more correct view on molluscan interclass relationships.
The association of cephalopods and aplacophorans has been recovered previously but dismissed as an artefact of high substitution rates in rRNA genes [6, 13, 21]. But our results cannot easily be explained by long branch attraction (LBA) effects (contra [13]). Branch lengths of scaphopods and caudofoveates are moderate, and the variopod node is stable against removal of putative long branched taxa showing accelerated evolutionary rates or biased base compositions [13], such as the branches of Solenogastres or Cephalopoda or both (trees not shown).
Molluscan evolution, whatever the underlying tree, is known to be laden with convergence at all taxon levels, including morphological features previously suspected to be informative for deep phylogeny (e.g., [56–58]). Conclusions derived from single organ systems, or the shell alone, are not able to exclude alternative interpretations. Coding hypothetical bauplans rather than existing representatives has been criticised [59, 60] and may lead to erroneous assumptions especially in groups with uncertain internal topology such as gastropods or aplacophorans. Morphocladistic approaches to date (e.g., [61–63]) all recovered Testaria, but this hypothesis is not supported by any molecular approaches.
Our proposed topology and any other nontestarian hypothesis imply that ancestral molluscs were complex rather than simple. This means that many anatomical characters inherited by descendants may be plesiomorphic and thus not informative, or could have been reduced or lost repeatedly, implying a high level of homoplasy. In fact, early molluscan phylogeny may have been shaped by habitat-induced selective pressure combined with heterochronic processes (e.g., [64]). This combination may lead to concerted morphological parallelisms powerful enough to obfuscate any phylogenetic signal, which has been found to be the case in heterobranch gastropods (e.g., [47, 65, 66]). It is possible to disentangle even highly homoplastic and heterochronic groups (e.g., [67–69]) if detailed and reliable microanatomical data are available on a dense ingroup taxon sampling, which is, however, not yet available for most molluscs. Unfortunately none of the many competing morphology-based hypotheses on molluscan class interrelationships available at present appears to represent a reliable benchmark for evaluating molecular topologies.
3.5. Topologies Tested against the Fossil Record
Molluscan diversification has been widely assumed to originate from a basal “monoplacophoran” bauplan [59], although early single shelled molluscs cannot be reliable separated from gastropods or any nonmonoplacophoran univalve [70]. The earliest calcareous molluscan-like shells, including undisputed molluscs, appear in the uppermost Precambrian, in the late Nemakit-Daldynian ca. 543 Ma [70]. Polyplacophoran shell plates first appear in the Late Cambrian, almost 50 My later [7, 71]. This does not support the Testaria hypothesis that would suggest that chitons evolved before the invention of a true “conchiferan” shell. There are dubious disarticulated microscopic chiton-like plates [72] from the early Meishuchunian (likely Early Tommotian) of China, but these still appeared later rather than earlier than the very first undisputed conchiferan shells. The Aculifera concept with monoplacophorans sister to other members of Conchifera or our molecular basal dichotomy are both fully compatible with the origin of molluscan shells latest at the Precambrian/Cambrian boundary.
The earliest tryblidian monoplacophorans are recorded from the Late Cambrian [73]. Older, nontryblidian “monoplacophorans” do not show serialised muscle scars and thus cannot be considered part of the crown-group. Yet the earliest reliable bivalves with elaborated hinge and ligament (Fordilla, Pojetaia) appear earlier, in the Early Tommotian ([74]; ca. 535 Ma). Both Aculifera and our basal dichotomy are not contradicted by the early appearance of bivalves. Under an Aculifera topology, chiton-like stem members could appear soon after a terminal Precambrian split separating Aculifera and Conchifera. Interpreting Early to Middle Cambrian sachitids (halwaxiids) as stem aculiferans would help fill this gap [7], but these taxa show a chronological sequence of shell plate loss rather than acquisition, which may be contrary to a progressive transition to chitons. The mosaic taxon Phthipodochiton, which has been proposed as a stem aplacophoran, does not appear until the Ordovician [75, 76]; other fossils from the Silurian, combining aplacophoran and polyplacophoran features with some soft tissue preservation, have also been used to support the Aculifera hypothesis [77]. These could also simply represent further disparity in extinct Polyplacophora. Regardless, there is compelling evidence from molecular systematics as well as fossil evidence that aplacophorans lost their ancestral shell (or shell plates) secondarily, and many other groups show repeated shell-loss or evolution to a vermiform body plan.
The topologies recovered by Vinther et al. [7] and Kocot et al. [5] support Aculifera but also imply that cephalopods are sister to Aculifera [7] or represent the earliest-diverging conchiferans [5] (excluding monoplacophorans from the analysis). However, there is no evidence for cephalopod-like fossils appearing earlier than, for example, bivalves. Similarly, bivalves are derived within Conchifera in the topology of Smith et al. [8], which is contradicted by the early fossil record of bivalves. In contrast, our basal dichotomy could fit with the many univalve small shelly fossils occurring earlier in the fossil record than bivalves, and both monoplacophorans and polyplacophorans appear later, actually at a similar time in the Latest Cambrian, and as predicted by a split of Serialia into Monoplacophora and Polyplacophora.
3.6. The Timing of Early Molluscan Evolution
The molluscan stem is Precambrian according to all our molecular time trees. The Vendian (555 Ma) body fossil Kimberella was discussed as a mollusc [78], but not widely accepted as such, and rather treated as lophotrochozoan stem member or “no more specifically than as a bilaterian” [79]. According to previous constrained (e.g., [7]) and our less constrained time trees (Table 2, Suppl. Table 8), however, Kimberella appears late enough in the fossil record to be considered as a potential stem mollusc. The other recent molecular clock for Mollusca puts the stem Mollusca even deeper [4], but Kimberella is within the 95% HPD interval for the split of the basal dichotomy also recovered herein. Having confirmed the conceptual basis of our proposed topology is not rejected by evidence in the fossil record, we further consider the timing of the radiation of specific clades proposed by our molecular clock analyses (Figure 3).
Cap-shaped Helcionellidae from the terminal Precambrian (e.g., Latouchella) are putative monoplacophorans according to the seminal study by Runnegar and Pojeta [80] or a separate molluscan class [81] or, based on nonserial muscle scars, gastropods [70]. Our time tree suggests that Nemakit-Daldynian and Earliest Tommotian molluscs with symmetrical cap-shaped shells with large openings are stem molluscs (or in the stem of one part of the basal dichotomy). In contrast, helicoid shells from the same period such as Aldanellidae (e.g., [82]) could well be gastropods, whether or not the animal was torted [70, 82].
Early Tommotian Watsonella, formerly known as Heraultipegma (the putatively earliest rostroconch), is a laterally compressed, bivalve-like univalve [70], possibly with dorsomedially decalcified or even bivalved shell [83]. This and other laterally compressed Watsonellidae may pre-date the first reliable Bivalvia (Early to Middle Tommotian Fordilla; [74] versus [70]) by some million years and thus could well be stem bivalves (or offshoots of the dorsoconch stem) according to our time tree (Figure 3).
It is important to note that neither reliable Monoplacophora (sensu Tryblidia) nor reliable Polyplacophora (i.e., Paleoloricata) are known before Late Cambrian, and this is confirmed in our chronograms (Figure 3). Yu [84] interpreted the Early Cambrian Merismoconchia as having eight pairs of muscles on a pseudometameric shell, linking 8-plated chitons with single shelled monoplacophorans in a transitional row of shell fusion. The similarity of merismoconchs with both serialian classes is curious, and their early occurrence in the pretrilobite Meishucun Stage suggests they could be early stem Serialia. The microscopic merismoconchs with their ventrally still connected shell segments and seven observed pairs of muscle scars may have been a transitional stage in how to make a foot efficient for sucking and a shell more flexible to adapt to uneven hard substrates. According to our time tree (Figure 3), chiton-like shell “fragmentation” into fully separated plates occurred much later, after splitting from single-shelled monoplacophoran-like ancestors.
The Cambrian (Atdabanian) Halkieria and related Middle Cambrian halwaxiids could also be interpreted as stem Serialia (Figure 3). A role as ancestral lophotrochozoans for halwaxiids as suggested by Edgecombe et al. [79] is not supported by our analysis.
According to our time tree (Figure 3), Yochelcionellidae, conspicuous Tommotian to Middle Cambrian shells that have a “snorkel,” could be part of the gastropod radiation as suggested by Parkhaev [70], or members of the dorsoconch stem lineage, or variopod stem members. The latter possibility is especially intriguing, since Yochelcionellidae evolved a “flow-through” water system with two shell openings; a dorsal shell elongates laterally and fuses ventrally, and the body axis shifts towards anterior growth extending head and foot out of a now tube-like shell. This condition is displayed by living and fossil variopods (i.e., scaphopods, cephalopods, and nonwatsonellid Rostroconchia).
Our results show that scaphopods could have split off from the variopod stem earlier, that is, in the Early Cambrian, but the oldest potential scaphopods in the familiar modern tusk-like shape are from the Ordovician [85] or even post-Devonian [86]. There is a vast record of Middle Cambrian tube-like shells that may be unrecognised parts of the early scaphopod diversification that started much earlier and morphologically less constrained than previously expected [87].
Knightoconus, a Middle to Late Cambrian large “monoplacophoran” conical shell with internal septa but no siphuncle [88], was described as a stem cephalopod [80] but subsequently questioned (e.g., [89]) and ultimately suspected to be a brachiopod [90]. Knightoconus could fit stratigraphically with stem cephalopods based on our evidence (Figure 3), but its morphological interpretation remains in doubt. The earliest reliable cephalopod fossils are the small bodied, septate, and siphuncle-bearing Plectronoceras from the Late Cambrian. Some versions of our analysis used Plectronoceras as a soft bound calibration point; by not using Plectronoceras, the origin of cephalopods shifts considerably towards the Silurian (Table 2).
Recently, shell-less and coleoid-shaped Lower Cambrian Nectocaris pteryx was regarded as a cephalopod [24], but this was immediately rejected on several lines of argument [91, 92]. Other putative Early Cambrian nectocaridids such as Vetustovermis [93] are superficially similar to Nectocaris in having a pair of long cephalic tentacles and stalked eyes but show a ventral foot separated from the supposedly wing-like mantle. Interpreting Nectocaris as having an axial cavity with gills and a funnel would provide synapomorphies for interpreting Nectocarididae as stem cephalopods [24, 94]. Molecular clock estimates can provide further insight to such contentious interpretations; according to our time estimates (which excluded nectocaridids as potential calibration points), Nectocaris is too ancient to be a cephalopod (Figure 3). If Nectocaris could be accepted as molluscan based on its contentious morphological interpretation, our time trees would be compatible with the idea that nectocaridids are stem variopods or within the stem of an aplacophoran/cephalopod or aplacophoran clade. Nectocaridid features with superficial similarities to coleoid cephalopods [24, 94] instead could be ancestral attributes of variopods: an anteriorly elongated body with head, long and flexible head tentacles, putative preoral hood, and a more or less reduced foot.
The fossil record offers shells and body fossils which, by their occurrence and morphology, at least hypothetically fill our time tree with life. The topology and timing of our hypothesis of early molluscan evolution is not rejected by fossil evidence.
3.7. Dorsoconcha
Molecular, morphological, and palaeontological evidence support (or fail to reject) our basal molluscan dichotomy. The clade Dorsoconcha includes most shelled molluscs and 98% of living species in four classes: Gastropoda, Bivalvia, Polyplacophora, and Monoplacophora.
We note two inferred potential morphological synapomorphies of Dorsoconcha, both relating to the digestive system and both somewhat ambiguous: the intestine is surrounded by the pericardium in basal lineages of gastropods, bivalves, and in monoplacophorans and may be positionally homologous in chitons (Figure 1(c) character 7) and a rotating enzymatic crystalline style (or protostyle; Figure 1(c) character 9). Many basal, noncarnivorous molluscs have a more or less well-developed stomach separated into sorting zones, but only dorsoconchs (and a family of caudofoveates [61]) have the complex style; this was secondarily lost in chitons, which have a derived position in our proposed topology.
Most previous studies on the phylogeny of molluscs have been driven by the Conchifera concept [1, 95] and emphasised the opinion that Serialia violates putative conchiferan synapomorphies [12]. Such features all are plesiomorphic for dorsoconchs in our topology (Figure 1(c)). We note several potential apomorphies for Serialia (Figure 1(c)): the serial (seven or) eightfold (octoserial) dorsoventral pairs of muscle bundles, with two pairs of intertwined muscle bundles in chitons and also partly present in large Neopilina [95, 96]; serial gills in a circumpedal mantle cavity; a highly similar cerebral nerve cord; and a longitudinal elongation of the dorsoventrally flattened body, to mention just some (Figure 1(c)). The most prominent feature of Serialia, serial paired foot retractors, is also present in bivalves, but octoserial retractors appear in Ordovician Babinka and not in the earliest known bivalves in the Cambrian [97] (Figure 1(c) character 10). While head and buccal apparatus are reduced almost completely in bivalves, Serialia elaborated the buccal mass evolving highly similar radulae and the radula bolster. Similar foot and radula structures in patellogastropod limpets [61] could be either plesiomorphic or convergent, because Patellogastropoda are either an isolated early-diverging gastropod group or relatively recently derived within Vetigastropoda [98, 99].
From this topological result and the available fossil evidence, we propose that the last common ancestor of monoplacophorans and chitons was cap-shelled and adapted to epibenthic life in shallow waters, rasping algae or other microorganisms from rocky substrates (Figure 1). In this scenario, chitons are not primitive molluscs but rather a derived group, potentially adapted to high-energy marine shores. Monoplacophorans initially also were shallow water dwellers [73] but could have colonised deeper waters during the Palaeozoic, where modern monoplacophorans still occur [100]. The Cenozoic or Late Cretaceous molecular dating of the diversification of living monoplacophorans and their short inner branches ([4], Figure 3) are compatible with earlier assumptions of pronounced anagenetic changes in the long stem line of these so-called “living fossils” [4, 100].
3.8. Variopoda
The clade Variopoda (Figure 1(c)) groups the scaphopods, aplacophorans, and cephalopods together in all our analyses, and it is very well supported. We infer several features of variopods, including an apparent propensity for habitat-induced transformations (noted in the taxon epithet; Figure 1(c) character 2). Some other roughly hypothesised apomorphies may refer to a clade of scaphopods and cephalopods only, that is, to variopods only under the assumption that aplacophorans represent highly paedomorphic and thus aberrant offshoots (see below): lateral extension of a primitively dorsal cap-like shell forming a tube; twisting the growth axis during ontogeny from initial dorsoventral to an anterior body extension, translocating head foot and mantle cavity with anal opening anteriorly; formation of a ring-like dorsoventral muscle insertion; multiplication of cephalic tentacles into prey-capturing feeding tentacles; and at least partly using muscle antagonist rather than merely hydrostatic systems in these tentacles (convergently in gastropod cephalic sensory tentacles); a hood is formed anterior to the mouth; and muscular retraction of the foot is used to pump water, waste, and gametes through/out the mantle cavity.
A clade of scaphopods and cephalopods repeatedly has been proposed based on morphological data, sometimes with one or the other or both together allied with gastropods [1], and was recovered by molecular data [52] and broadly within some pan-molluscan molecular phylogenies [10, 21]. In contrast, morphocladistic neontological [101] and palaeontological studies (e.g., [80, 102]) advocated the Diasoma concept suggesting scaphopods as sister to bivalves with a rostroconch ancestor. Developmental data showing different ontogeny of shells have not supported the latter opinion [103]. Diasoma has been equivocally recovered within one mitogenomic analysis ([104], but see [105] for limitations of protein coding mitochondrial genes), and in one supplementary analysis of transcriptome data [8]. Similar features such as a digging foot could be interpreted as convergent adaptations to infaunal life.
The two aplacophoran classes Caudofoveata and Solenogastres have never been associated with either scaphopods or cephalopods in morphological studies. In our analyses aplacophorans are usually paraphyletic, but some permutations, in particular when excluding (the faster-evolving, but stringently masked) COI and 16S markers, recover a clade Aplacophora sister to Cephalopoda. Aplacophora as a clade is not rejected by AU analyses of the combined 5-marker set either (Table 3). A single origin of vermiform body plans in the cephalopod stem lineage could arguably be more parsimonious than arising twice independently. Monophyly of Aplacophora is indicated by all recent studies using multiple nuclear protein coding genes and phylogenomic data sets ([5, 7, 8]; Figure 1(b)) but not neuroanatomy [106].
Aplacophorans may share an inferred tendency of modifying the ancestral foot, they have an elongated body with a foot (or head) shield with strong retractor muscle in caudofoveates, and the atrial cavity especially in Solenogastres could be interpreted as a modified preoral hood, as remnants of a hypothesised variopod body plan. Yet there is no morphological indication for a specifically aplacophoran-cephalopod clade. Interpretation of the vermiform molluscan morphology as progenetically derived rather than reflecting a basal molluscan condition (also assumed under the Aculifera concept) actually allows for hypotheses that resolve them at any position in the molluscan tree (or makes their position impossible to recover using currently available anatomical data). Assuming that aplacophorans (once or twice independently) initially evolved into interstitial secondary worms could be correlated with precerebral ganglia present in caudofoveates [106]; these transformations have evolved many times independently in interstitial worm-like gastropod groups, which are likely progenetic [47]. Calcareous spicules also evolved many times convergently in different interstitial shell-less gastropod lineages [47] and a protective dorsal cuticle covering the body evolved within progenetic corambid sea slugs [67, 68]. “Regressive” [sensu [107]] traits in aplacophorans such as miniaturisation, losses of shell, tentacles, and cephalisation have been attributed to progenesis [64]. The serial dorsoventral muscle grid of aplacophorans resembles early ontogenetic stages observed in other molluscs [108] and could be paedomorphic, but it is still an adaptive innovation for nonlarval stages. The narrow bipartite radulae of aplacophorans are specialised tools for microcarnivory but also resemble some stem molluscan radula types [56]; evidence from Cambrian fossils is more congruent with an ancestral unipartite radula [109].
Our topology places aplacophorans in an unconvential position; however, there is consensus among all recent molecular studies that aplacophorans represent derived rather than plesiomorphic members of Mollusca (Figure 1). These notes on the specific feature of aplacophorans therefore are of general interest to resolving the pattern and tempo of molluscan evolution, regardless of differences between our new topology and other studies.
3.9. Molluscan Ancestors
The origin of molluscs is a long-standing question, and speculations on the “hypothetical ancestral mollusk” depend on character-polarity and even topological assumptions [1, 59]. Broad genomic analyses (e.g., [14–16, 22]) recovered molluscs as an early-derived offshoot of Lophotrochozoa (Spiralia), as had been proposed on morphological grounds [110]. Modern morphological studies suggest entoprocts as sister to molluscs [1], a view supported by mitochondrial genomics [105]. MicroRNA data [111] suggest Annelida is the sister to Mollusca, as recovered (but never robustly supported) by most of our analyses with a large outgroup taxon set (Suppl. Figure 1A). Our analyses did not resolve a consistent sister group to Mollusca. Yet permutations and pruning of our outgroup sampling did not affect ingroup topologies. We regard the molluscan sister group as an unanswered question, but not necessarily problematic to the question of internal molluscan phylogeny (if ingroup taxon sampling is sufficiently dense).
Our initial morphological character mapping (Figure 1(c)) suggests that the last common ancestor of living molluscs (“LAM”) was a single-shelled conchiferan with a complex body, single (or few) paired shell retractors, single paired gills in a circumpedal mantle cavity, and an elaborated (cephalised) anterior body portion. There is little reason to assume that this hypothetical LAM resembled a chiton or monoplacophoran (e.g., [25]) or to suspect a segmented body organisation (e.g., [112]). Instead, the LAM may have resembled an untorted gastropod with a cap-like shell, perhaps similar to Latouchella, as assumed by morphologists before the discovery of the supposedly “living fossil” Neopilina and still advocated by some palaeontologists [70].
Our assessment of potential morphological apomorphies (Figure 1(c)) and the molecular clock results (Figure 3) would suggest that the Vendian (555 Ma) Kimberella [78] is a candidate stem-group mollusc appearing before the evolution of a dorsal shell field. The interpretation of Kimberella is controversial [113], but the true stem molluscs probably did have a large, bilaterally symmetrical body with subapical mouth on a snout with a likely bipartite radula [114], a broad ventral foot, many dorsoventral muscle bundles, and a dorsal mantle covered with a resistant dorsal cuticle with mineralised spicules, which are all molluscan features, but lacking a shell [115, 116]. During the latest Precambrian rise of predators and successive development of sediment bottoms [25], molluscan larvae or early juveniles may have calcified their plesiomorphic cap-shaped mantle cuticle for protective reasons. Answering Yochelson [117], the mollusc made a shell, but then the shell made the molluscs.
4. Conclusions
Only one (if any) of the dozens of proposed hypotheses on molluscan phylogeny reflects the true tree. Both the traditional palaeontological concept, with monoplacophorans giving rise to all other molluscan lineages, and the widely accepted morphocladistic Testaria hypothesis, with progressive evolution from vermiform molluscs to chitons and conchiferans [62, 118], are not supported by molecular evidence and are apparently incompatible with the chronological appearance of reliable fossils representing major molluscan lineages.
The Aculifera concept has been supported by phylogenomic results [2, 119], whose dichotomy is not inherently contradicted by the available fossil record if the last common molluscan ancestor was small and complex and had a shell (i.e., was conchiferan rather than chiton-like). Yet the branching patterns of living clades in available phylogenomic topologies appear to be incongruent with stratigraphic evidence. The debate on molluscan phylogeny can only be progressed using all available evidence, integrating morphological, fossil, and molecular data. To provide meaningful insights, molecular approaches must include all eight molluscan classes and cover the well-known diversity of living taxa.
Our results, despite using traditional markers that cover arguably less data than next-generation approaches, are based on a comprehensive taxon set with data quality checked exhaustively at all levels. Topologies recovered still may suffer from poor sampling especially of aplacophoran lineages and from heterogeneous evolution of ingroup clades such as cephalopods or patellogastropods. The data available, while extensive and of high quality, are small in comparison to the total genetic diversity of the phylum under study.
Nevertheless, our data sets, regimes, and analyses support and refine the Serialia hypothesis [9]. The topological results inferred herein cannot be refuted by recent research on shell building gene expression and mollusc palaeontology. In many well-studied molluscan taxa, shells are reduced or duplicated, bodies adapted to different environments and life styles such as benthic, interstitial, or pelagic realms, and features such as mantle cavities and radulae repeatedly were transformed, often drastically and rapidly. Heterochronic processes could already have occurred in the Palaeozoic, which would be consistent with the disparity known in living molluscs but which could also obscure deeper phylogenetic signal in morphological analyses. Ultimately, such complex diversification could have led to the fossil and extant molluscs that stand apart from other (noninsect) animals in terms of species diversity, body disparity, and variation of life traits. The true reconstruction of the early radiation of molluscs still is one of the major unresolved issues in evolutionary biology. Independent molecular evidence, such as microRNAs or phylogenomic data on a similarly comprehensive and dense taxon sampling as used herein, will be needed to further test these hypotheses.
Supplementary Material
Figure S1. Maximum Likelihood tree of 142-taxon set; A. Tree with collapsed Gastropoda; B. Expanded gastropod clade.
Figure S2. Maximum Likelihood tree of total gastropod taxon set.
Table S1. Primer sequences and PCR conditions.
Table S2. Taxa and markers used in the initial total analyses (158-taxon data set).
Table S3. Sequences that were not considered in the analyses. A. Ambiguous BLAST results. B. Bivalve taxa with 2nd amplicon of 28S showing aberrant features.
Table S4. Models of evolution used for main analyses.
Table S5. Data and taxon partitions used in different iterations of the core analyses, including gene regions, total dataset length (bp) and gaps.
Table S6. Substitution saturation tested with DAMBE.
Table S7. Fossil calibration points and settings for prior distribution of calibrated nodes.
Table S8. Time estimates of calibration points and major molluscan groups.
Authors' Contributions
I. Stöger carried out the molecular genetic studies, performed the sequence alignments and the phylogenetic analyses, and drafted the paper. J. D. Sigwart participated in the design of the study and contributed to writing the paper; Y. Kano participated in original fieldwork and in the molecular genetic studies; T. Knebelsberger participated in the molecular genetic studies; B. A. Marshall carried out original fieldwork and helped draft the paper; E. Schwabe participated in original fieldwork and helped draft the paper; M. Schrödl conceived and designed the study, participated in original fieldwork, contributed to molecular genetic studies, and contributed to writing the paper.
Acknowledgments
Monoplacophoran material was collected during the expedition ANDEEP SYSTCO, from commercial fisheries bycatch, and by Taeko and Shoichi Kimura. Dirk Metzler (Munich) helped with molecular clocks, Patrick Kück (Bonn) performed some of the analyses with removed putative long branched molluscan classes by the help of a bioinformatic pipeline, Korbinian Trumpp provided computational support, and Gerhard Haszprunar (Munich) is thanked for critical discussions. Anonymous referees on a previous, short version of this paper greatly contributed to improve analyses and interpretations. The authors are grateful for additional comments from David Lindberg (Berkeley) and an anonymous reviewer. This study was funded by the Deep Metazoan Priority Programme of the German Research Foundation (DFG667/9-1 to MS). Further support came from the GeoBioCenter LMU and the Leibniz Computer Centre (both Munich).
References
- 1.Haszprunar G, Schander C, Halanych KM. Relationships of higher taxa. In: Ponder WF, Lindberg DR, editors. Phylogeny and Evolution of the Mollusca. Berkeley, Calif, USA: University of California Press; 2008. pp. 19–32. [Google Scholar]
- 2.Telford MJ, Budd GE. Invertebrate evolution: bringing order to the molluscan chaos. Current Biology. 2011;21(23):R964–R966. doi: 10.1016/j.cub.2011.10.029. [DOI] [PubMed] [Google Scholar]
- 3.Wilson NG, Huang D, Goldstein MC, Cha H, Giribet G, Rouse GW. Field collection of Laevipilina hyalina McLean, 1979 from southern California, the most accessible living monoplacophoran. Journal of Molluscan Studies. 2009;75(2):195–197. [Google Scholar]
- 4.Kano Y, Kimura S, Kimura Y, Warén A. Living Monoplacophora: morphological conservatism or recent diversification? Zoologica Scripta. 2012;41(5):471–488. [Google Scholar]
- 5.Kocot KM, Cannon JT, Todt C, et al. Phylogenomics reveals deep molluscan relationships. Nature. 2011;477(7365):452–456. doi: 10.1038/nature10382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer A, Witek A, Lieb B. Selecting ribosomal protein genes for invertebrate phylogenetic inferences: how many genes to resolve the Mollusca? Methods in Ecology and Evolution. 2011;2(1):34–42. [Google Scholar]
- 7.Vinther J, Sperling EA, Briggs DEG, Peterson KJ. A molecular palaeobiological hypothesis for the origin of aplacophoran molluscs and their derivation from chiton-like ancestors. Proceedings of the Royal Society B. 2012;279(1732):1259–1268. doi: 10.1098/rspb.2011.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SA, Wilson NG, Goetz FE, et al. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature. 2011;480(7377):364–367. doi: 10.1038/nature10526. [DOI] [PubMed] [Google Scholar]
- 9.Giribet G, Okusu A, Lindgren AR, Huff SW, Schrödl M, Nishiguchi MK. Evidence for a clade composed of molluscs with serially repeated structures: monoplacophorans are related to chitons. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(20):7723–7728. doi: 10.1073/pnas.0602578103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson NG, Rouse GW, Giribet G. Assessing the molluscan hypothesis Serialia (Monoplacophora + Polyplacophora) using novel molecular data. Molecular Phylogenetics and Evolution. 2010;54(1):187–193. doi: 10.1016/j.ympev.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen C, Haszprunar G, Ruthensteiner B, Wanninger A. Early development of the aplacophoran mollusc Chaetoderma . Acta Zoologica. 2007;88(3):231–247. [Google Scholar]
- 12.Wägele JW, Letsch H, Klussmann-Kolb A, Mayer C, Misof B, Wägele H. Phylogenetic support values are not necessarily informative: the case of the Serialia hypothesis (a mollusk phylogeny) Frontiers in Zoology. 2009;6(1):p. 12. doi: 10.1186/1742-9994-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer A, Todt C, Mikkelsen NT, Lieb B. Fast evolving 18S rRNA sequences from Solenogastres (Mollusca) resist standard PCR amplification and give new insights into mollusk substitution rate heterogeneity. BMC Evolutionary Biology. 2010;10(1):p. 70. doi: 10.1186/1471-2148-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology. 2011;9(3) doi: 10.1371/journal.pbio.1000602.e1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippe H, Brinkmann H, Copley RR, et al. Acoelomorph flatworms are deuterostomes related to Xenoturbella . Nature. 2011;470(7333):255–260. doi: 10.1038/nature09676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pick KS, Philippe H, Schreiber F, et al. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Molecular Biology and Evolution. 2010;27(9):1983–1987. doi: 10.1093/molbev/msq089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misof B, Misof K. A Monte Carlo approach successfully identifies randomness in multiple sequence alignments: a more objective means of data exclusion. Systematic Biology. 2009;58(1):21–34. doi: 10.1093/sysbio/syp006. [DOI] [PubMed] [Google Scholar]
- 18.Stocsits RR, Letsch H, Hertel J, Misof B, Stadler PF. Accurate and efficient reconstruction of deep phylogenies from structured RNAs. Nucleic Acids Research. 2009;37(18):6184–6193. doi: 10.1093/nar/gkp600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallatt J, Craig CW, Yoder MJ. Nearly complete rRNA genes assembled from across the metazoan animals: effects of more taxa, a structure-based alignment, and paired-sites evolutionary models on phylogeny reconstruction. Molecular Phylogenetics and Evolution. 2010;55(1):1–17. doi: 10.1016/j.ympev.2009.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Paps J, Baguñà J, Riutort M. Lophotrochozoa internal phylogeny: new insights from an up-to-date analysis of nuclear ribosomal genes. Proceedings of the Royal Society B. 2009;276(1660):1245–1254. doi: 10.1098/rspb.2008.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Passamaneck YJ, Schander C, Halanych KM. Investigation of molluscan phylogeny using large-subunit and small-subunit nuclear rRNA sequences. Molecular Phylogenetics and Evolution. 2004;32(1):25–38. doi: 10.1016/j.ympev.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 22.Dunn CW, Hejnol A, Matus DQ, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 23.Hejnol A, Obst M, Stamatakis A, et al. Assessing the root of bilaterian animals with scalable phylogenomic methods. Proceedings of the Royal Society B. 2009;276(1677):4261–4270. doi: 10.1098/rspb.2009.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith MR, Caron J-B. Primitive soft-bodied cephalopods from the Cambrian. Nature. 2010;465(7297):469–472. doi: 10.1038/nature09068. [DOI] [PubMed] [Google Scholar]
- 25.Caron J-B, Scheltema AH, Schander C, Rudkin D. A soft-bodied mollusc with radula from the Middle Cambrian Burgess Shale. Nature. 2006;442(7099):159–163. doi: 10.1038/nature04894. [DOI] [PubMed] [Google Scholar]
- 26.Vinther J, Nielsen C. The early Cambrian Halkieria is a mollusc. Zoologica Scripta. 2005;34(1):81–89. [Google Scholar]
- 27.Morris SC, Caron J-B. Halwaxiids and the early evolution of the lophotrochozoans. Science. 2007;315(5816):1255–1258. doi: 10.1126/science.1137187. [DOI] [PubMed] [Google Scholar]
- 28.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods in Molecular Biology. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 31.Kück P, Meusemann K, Dambach J, et al. Parametric and non-parametric masking of randomness in sequence alignments can be improved and leads to better resolved trees. Frontiers in Zoology. 2010;7:p. 10. doi: 10.1186/1742-9994-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kück P, Meusemann K. FASconCAT, Version 1. 0. Zool. Bonn, Germany: Forschungsmuseum A. Koenig; 2010. [Google Scholar]
- 34.Drummond AJ, Rambaut A. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7(1):754–755. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 36.Jobb G, Von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evolutionary Biology. 2004;4:p. 18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity. 2001;92(4):371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 38.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 39.Nylander JAA. MrModeltest V2. 3. Program Distributed by the Author. Uppsala, Sweden: Evolutionary Biology Centre, Uppsala University; 2008. [Google Scholar]
- 40.Swofford DL. PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods). Version 4. Sunderland, Mass, USA: Sinauer Associates; 2003. [Google Scholar]
- 41.Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21(9):2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- 42.Stamatakis A. The RAxML 7.0.4 Manual. Department of Computer Science, Ludwig-Maximilian-Universität München, Germany.
- 43.Huelsenbeck JP, Ronquist F. MRBAYES: bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 44.Rambaut A, Drummond AJ. Tracer v1. 4. 2007, http://tree.bio.ed.ac.uk/software/tracer/
- 45.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biology. 2006;4(5):p. e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Drummond AJ, Ho SYW, Rawlence N, Rambaut A. A Rough Guide to BEAST 1.4. 2007. [Google Scholar]
- 47.Jörger KM, Stöger I, Kano Y, Fukuda H, Knebelsberger T, Schrödl M. On the origin of Acochlidia and other enigmatic euthyneuran gastropods, with implications for the systematics of Heterobranchia. BMC Evolutionary Biology. 2010;10(1):p. 323. doi: 10.1186/1471-2148-10-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hipsley CA, Himmelmann L, Metzler D, Müller J. Integration of bayesian molecular clock methods and fossil-based soft bounds reveals early cenozoic origin of African lacertid lizards. BMC Evolutionary Biology. 2009;9(1):p. 151. doi: 10.1186/1471-2148-9-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wägele JW, Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evolutionary Biology. 2007;7:p. 147. doi: 10.1186/1471-2148-7-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuhashi T, Schwarzinger C, Miksik I, Smrz M, Beran A. Molluscan shell evolution with review of shell calcification hypothesis. Comparative Biochemistry and Physiology B. 2009;154(3):351–371. doi: 10.1016/j.cbpb.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 51.Waller TR. Origin of the molluscan class Bivalvia and a phylogeny of major groups. In: Johnston PA, Haggard JW, editors. Bivalves: An Eon of Evolution. Calgary, Canada: University of Calgary Press; 1998. pp. 1–45. [Google Scholar]
- 52.Steiner G, Dreyer H. Molecular phylogeny of Scaphopoda (Mollusca) inferred from 18s rDNA sequences: support for a Scaphopoda-Cephalopoda clade. Zoologica Scripta. 2003;32(4):343–356. [Google Scholar]
- 53.Ponder WF, Lindberg DR. Phylogeny and Evolution of the Mollusca. Berkeley, Calif, USA: University of California Press; 2008. [Google Scholar]
- 54.Donoghue PCE, Benton MJ. Rocks and clocks: calibrating the Tree of Life using fossils and molecules. Trends in Ecology and Evolution. 2007;22(8):424–431. doi: 10.1016/j.tree.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 55.Mossel E, Steel M. How much can evolved characters tell us about the tree that generated them? In: Gascuel O, editor. Mathematics of Evolution and Phylogeny. Oxford, UK: Oxford University Press; 2005. pp. 384–412. [Google Scholar]
- 56.Scheltema AH, Kerth K, Kuzirian AM. Original molluscan radula: comparisons among Aplacophora, Polyplacophora, Gastropoda, and the Cambrian fossil Wiwaxia corrugata . Journal of Morphology. 2003;257(2):219–244. doi: 10.1002/jmor.10121. [DOI] [PubMed] [Google Scholar]
- 57.Shigeno S, Sasaki T, Haszprunar G. Central nervous system of Chaetoderma japonicum (Caudofoveata, Aplacophora): implications for diversified ganglionic plans in early molluscan evolution. Biological Bulletin. 2007;213(2):122–134. doi: 10.2307/25066628. [DOI] [PubMed] [Google Scholar]
- 58.Lundin K, Schander C, Todt C. Ultrastructure of epidermal cilia and ciliary rootlets in Scaphopoda. Journal of Molluscan Studies. 2009;75(1):69–73. [Google Scholar]
- 59.Lindberg DR, Ghiselin : MT. Fact, theory and tradition in the study of molluscan origins. Proceedings of the California Academy of Sciences. 2003;54:663–686. [Google Scholar]
- 60.Prendini L. Species or supraspecific taxa as terminals in Cladistic analysis? groundplans versus exemplars revisited. Systematic Biology. 2001;50(2):290–300. doi: 10.1080/10635150118650. [DOI] [PubMed] [Google Scholar]
- 61.Haszprunar G. Is the Aplacophora monophyletic? A cladistic point of view. American Malacological Bulletin. 2000;15(2):115–130. [Google Scholar]
- 62.Haszprunar G, Wanninger A. Molluscs. Current Biology. 2012;22(13):R510–R514. doi: 10.1016/j.cub.2012.05.039. [DOI] [PubMed] [Google Scholar]
- 63.Salvini-Plawen L, Steiner G. Synapomorphies and plesiomorphies in higher classification of Mollusca. In: Taylor J, editor. Origin and Evolutionary Radiation of the Mollusca. Oxford, UK: Oxford University Press; 1996. pp. 29–51. [Google Scholar]
- 64.Scheltema AH. Phylogenetic position of Sipuncula, Mollusca and the progenetic Aplacophora. In: Taylor J, editor. Origin and Evolutionary Radiation of the Mollusca. Oxford, UK: Oxford University Press; 1996. pp. 53–58. [Google Scholar]
- 65.Schrödl M, Jörger KM, Klussmann-Kolb A, Wilson NG. Bye bye “Opisthobranchia”! a review on the contribution of mesopsammic sea slugs to euthyneuran systematics. Thalassas. 2011;27(2):101–112. [Google Scholar]
- 66.Schrödl M, Jörger KM, Wilson NG. A reply to Medina et al. (2011): crawling through time: transition of snails to slugs dating back to the Paleozoic based on mitochondrial phylogenomics. Marine Genomics. 2011;4(4):301–303. doi: 10.1016/j.margen.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 67.Martynov A, Brenzinger B, Hooker Y, Schrödl M. 3D-anatomy of a new tropical Peruvian nudibranch gastropod species, Corambe mancorensis, and novel hypotheses on dorid gill ontogeny and evolution. Journal of Molluscan Studies. 2011;77(2):129–141. [Google Scholar]
- 68.Martynov A, Schrödl M. Phylogeny and evolution of corambid nudibranchs (Mollusca: Gastropoda) Zoological Journal of the Linnean Society. 2011;163(2):585–604. [Google Scholar]
- 69.Schrödl M, Neusser TP. Towards a phylogeny and evolution of Acochlidia (Mollusca: Gastropoda: Opisthobranchia) Zoological Journal of the Linnean Society. 2010;158(1):124–154. [Google Scholar]
- 70.Parkhaev PY. The early molluscan radiation. In: Ponder WF, Lindberg DR, editors. Phylogeny and Evolution of the Mollusca. Berkeley, Calif, USA: University of California Press; 2008. pp. 33–69. [Google Scholar]
- 71.Smith AG. Amphineura. In: Moore RC, editor. Treatise on Invertebrate Palaeontology, Part 1: Mollusca 1. New York, NY, USA: Geological Society of America; 1960. pp. 141–176. [Google Scholar]
- 72.Yu W. Stratigraphy and Palaeontology Boundary in China Precambrian-Cambrian Boundary. Vol. 1. Jiangsu, China: Nanjing Institute of Geology and Palaeontology Academia Sinica, Nanjing University; 1987. Yangtze micromolluscan fauna in Yangtze region of China with notes on Precambrian-Cambrian boundary; pp. 19–275. [Google Scholar]
- 73.Lindberg DR. Monoplacophorans and the origin and relationships of mollusks. Evolution. 2009;2(2):191–203. [Google Scholar]
- 74.Pojeta J., Jr. Cambrian Pelecypoda (Mollusca) American Malacological Bulletin. 2000;15(2):157–166. [Google Scholar]
- 75.Sigwart JD, Sutton MD. Deep molluscan phylogeny: synthesis of palaeontological and neontological data. Proceedings of the Royal Society B. 2007;274(1624):2413–2419. doi: 10.1098/rspb.2007.0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sutton MD, Sigwart JD. A chiton without a foot. Palaeontology. 2012;55(2):401–411. [Google Scholar]
- 77.Sutton MD, Briggs DEG, Siveter DJ, Siveter DJ, Sigwart JD. A Silurian armoured aplacophoran and implications for molluscan phylogeny. Nature. 2012;490:94–97. doi: 10.1038/nature11328. [DOI] [PubMed] [Google Scholar]
- 78.Fedonkin MA, Waggoner BM. The late Precambrian fossil Kimberella is a mollusc-like bilaterian organism. Nature. 1997;388(6645):868–871. [Google Scholar]
- 79.Edgecombe GD, Giribet G, Dunn CW, et al. Higher-level metazoan relationships: recent progress and remaining questions. Organisms Diversity and Evolution. 2011;11(2):151–172. [Google Scholar]
- 80.Runnegar B, Pojeta J., Jr. Molluscan phylogeny: the paleontological viewpoint. Science. 1974;186(4161):311–317. doi: 10.1126/science.186.4161.311. [DOI] [PubMed] [Google Scholar]
- 81.Yochelson EL. An alternative approach to the interpretation of the phylogeny of ancient molluscs. Malacologia. 1978;17:185–191. [Google Scholar]
- 82.Harper JA, Rollins HB. The bellerophont controversy revisited. American Malacological Bulletin. 2000;15(2):147–156. [Google Scholar]
- 83.Dzik J. Evolution of “small shelly fossils” assemblages of the early Paleozoic. Acta Palaeontologica Polonica. 1994;39(3):247–313. [Google Scholar]
- 84.Yu W. On merismoconchids. Acta Oceanologica Sinica. 1983;23:432–446. [Google Scholar]
- 85.Gubanov AP, Peel JS. Cambrian monoplacophoran molluscs (Class Helcionelloida) American Malacological Bulletin. 2000;15(2):139–145. [Google Scholar]
- 86.Yochelson EL, Holland CH. Dentalium saturni Goldfuss, 1841 (Eifelian: Mollusca): complex issues from a simple fossil. Paläontologische Zeitschrift. 2004;78(1):97–102. [Google Scholar]
- 87.Peel JS. Pinnocaris and the origin of scaphopods. Acta Palaeontologica Polonica. 2004;49(4):543–550. [Google Scholar]
- 88.Yochelson E, Flower RH, Webers GF. The bearing of new Late Cambrian monoplacophoran genus Knightoconus upon the origin of Cephalopoda. Lethaia. 1973;6(3):275–309. [Google Scholar]
- 89.Webers GF, Yochelson EL. Carboniferous Scaphopoda (Mollusca) and non-scaphopods from Scotland. Scottish Journal of Geology. 2011;47(1):67–79. [Google Scholar]
- 90.Dzik J. Brachiopod identity of the alleged monoplacophoran ancestors of cephalopods. Malacologia. 2010;52(1):97–113. [Google Scholar]
- 91.Kröger B, Vinther J, Fuchs D. Cephalopod origin and evolution: a congruent picture emerging from fossils, development and molecules: extant cephalopods are younger than previously realised and were under major selection to become agile, shell-less predators. BioEssays. 2011;33(8):602–613. doi: 10.1002/bies.201100001. [DOI] [PubMed] [Google Scholar]
- 92.Mazurek D, Zatoń M. Is Nectocaris pteryx a cephalopod? Lethaia. 2011;44(1):2–4. [Google Scholar]
- 93.Chen J-Y, Huang D-Y, Bottjer DJ. An Early Cambrian problematic fossil: Vetustovermis and its possible affinities. Proceedings of the Royal Society B. 2005;272(1576):2003–2007. doi: 10.1098/rspb.2005.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smith MR, Caron J-B. Nectocaris and early cephalopod evolution: reply to Mazurek & Zatoń. Lethaia. 2011;44(4):369–372. [Google Scholar]
- 95.Salvini-Plawen L. The Significance of the Placophora for Molluscan Phylogeny. Vol. 65. Elko, Nev, USA: Venus; 2006. [Google Scholar]
- 96.Wingstrand KG. On the anatomy and relationships of recent Monoplacophora. Galathea Report. 1985;16:7–94. [Google Scholar]
- 97.Giribet G. Bivalvia. In: Ponder WF, Lindberg DR, editors. Phylogeny and Evolution of the Mollusca. Berkeley, Calif, USA: University of California Press; 2008. pp. 105–141. [Google Scholar]
- 98.Aktipis SW, Giribet G. A phylogeny of Vetigastropoda and other “archaeogastropods”: re-organizing old gastropod clades. Invertebrate Biology. 2010;129(3):220–240. [Google Scholar]
- 99.Aktipis SW, Giribet G. Testing relationships among the vetigastropod taxa: a molecular approach. Journal of Molluscan Studies. 2012;78(1):12–27. [Google Scholar]
- 100.Haszprunar G. Monoplacophora (Tryblidia) In: Ponder WF, Lindberg DR, editors. Phylogeny and Evolution of the Mollusca. Berkeley, Calif, USA: University of California Press; 2008. pp. 97–104. [Google Scholar]
- 101.Simone LRL. Comparative morphology among representatives of main taxa of Scaphopoda and basal protobranch Bivalvia (Mollusca) Papeis Avulsos de Zoologia. 2009;49(32):405–457. [Google Scholar]
- 102.Pojeta J, Runnegar B. The paleontology of rostroconch mollusks and the early history of the phylum Mollusca. U.S. Geological Survey Professional Paper. 1976;986:1–88. [Google Scholar]
- 103.Wanninger A, Haszprunar G. The expression of an engrailed protein during embryonic shell formation of the tusk-shell, Antalis entalis (Mollusca, Scaphopoda) Evolution and Development. 2001;3(5):312–321. doi: 10.1046/j.1525-142x.2001.01034.x. [DOI] [PubMed] [Google Scholar]
- 104.Yokobori S-I, Iseto T, Asakawa S, et al. Complete nucleotide sequences of mitochondrial genomes of two solitary entoprocts, Loxocorone allax and Loxosomella aloxiata: implications for lophotrochozoan phylogeny. Molecular Phylogenetics and Evolution. 2008;47(2):612–628. doi: 10.1016/j.ympev.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 105.Stöger I, Schrödl M. Mitogenomics does not resolve deep molluscan relationships (yet?) Molecular Phylogenetics and Evolution. 2013;69(2):376–392. doi: 10.1016/j.ympev.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 106.Faller S, Rothe BH, Todt C, Schmidt-Rhaesa A, Loesel R. Comparative neuroanatomy of Caudofoveata, Solenogastres, Polyplacophora, and Scaphopoda (Mollusca) and its phylogenetic implications. Zoomorphology. 2012:1–22. [Google Scholar]
- 107.Pelseneer MP. Sur le quatrième orifice palléal des Pélécypodes. Comptes Rendus Hebdonnaires des Séances de l’Académie des Sciences. 1890;110:154–156. [Google Scholar]
- 108.Wanninger A, Haszprunar G. Chiton myogenesis: perspectives for the development and evolution of larval and adult muscle systems in molluscs. Journal of Morphology. 2002;251(2):103–113. doi: 10.1002/jmor.1077. [DOI] [PubMed] [Google Scholar]
- 109.Smith MR. Mouthparts of the Burgess Shale fossils Odontogriphus and Wiwaxia: implications for the ancestral molluscan radula. Proceedings of the Royal Society B. 2012;279(1745):4287–4295. doi: 10.1098/rspb.2012.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Salvini-Plawen L. Origin, Phylogeny and Classification of the Phylum Mollusca. Vol. 9. Madrid, Spain: Iberus; 1990. [Google Scholar]
- 111.Wheeler BM, Heimberg AM, Moy VN, et al. The deep evolution of metazoan microRNAs. Evolution and Development. 2009;11(1):50–68. doi: 10.1111/j.1525-142X.2008.00302.x. [DOI] [PubMed] [Google Scholar]
- 112.Hoare RD. Considerations on Paleozoic Polyplacophora including the description of Plasiochiton curiosus n. gen. and sp. American Malacological Bulletin. 2000;15(2):131–137. [Google Scholar]
- 113.Ivantsov AY. Paleontological evidence for the supposed Precambrian occurrence of mollusks. Paleontological Journal. 2010;44(12):1552–1559. [Google Scholar]
- 114.Seilacher A, Hagadorn JW. Early molluscan evolution: evidence from the trace fossil record. Palaios. 2010;25(9):565–575. [Google Scholar]
- 115.Fedonkin MA, Simonetta A, Ivantsov AY. New data on Kimberella, the Vendian mollusc-like organism (White Sea region, Russia): palaeoecological and evolutionary implications. Geological Society Special Publication. 2007;(286):157–179. [Google Scholar]
- 116.Ivantsov AY. New reconstruction of Kimberella, problematic Vendian metazoan. Paleontological Journal. 2009;43(6):601–611. [Google Scholar]
- 117.Yochelson EL. Concerning the concept of extinct classes of Mollusca: or what may/may not be a class of mollusks. American Malacological Bulletin. 2000;15(2):195–202. [Google Scholar]
- 118.Haszprunar G, Ruthensteiner B. Monoplacophora (Tryblidia)-some unanswered questions. American Malacological Bulletin. 2013;31(1):189–194. [Google Scholar]
- 119.Kocot K. Recent advances and unanswered questions in deep molluscan phylogenetics. American Malacological Bulletin. 2013;31(1):195–208. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Maximum Likelihood tree of 142-taxon set; A. Tree with collapsed Gastropoda; B. Expanded gastropod clade.
Figure S2. Maximum Likelihood tree of total gastropod taxon set.
Table S1. Primer sequences and PCR conditions.
Table S2. Taxa and markers used in the initial total analyses (158-taxon data set).
Table S3. Sequences that were not considered in the analyses. A. Ambiguous BLAST results. B. Bivalve taxa with 2nd amplicon of 28S showing aberrant features.
Table S4. Models of evolution used for main analyses.
Table S5. Data and taxon partitions used in different iterations of the core analyses, including gene regions, total dataset length (bp) and gaps.
Table S6. Substitution saturation tested with DAMBE.
Table S7. Fossil calibration points and settings for prior distribution of calibrated nodes.
Table S8. Time estimates of calibration points and major molluscan groups.


