Abstract
Antibiotic resistance is a persistent health care problem worldwide. Evidence for the negative consequences of subtherapeutic feeding in livestock production has been mounting while the antibiotic pipeline is drying up. In recent years, there has been a paradigm shift in our perception of antibiotics. Apart from its roles in self-defense, antibiotics also serve as inter-microbial signaling molecules, regulators of gene expression, microbial food sources, and as mediators of host immune response.
“The time may come when penicillin can be bought by anyone in the shops. Then there is the danger that the ignorant man may easily under-dose himself and by exposing his microbes to nonlethal quantities of the drug make them resistant.”
~Alexander Fleming
The Growing Threat of Antibiotic Resistance
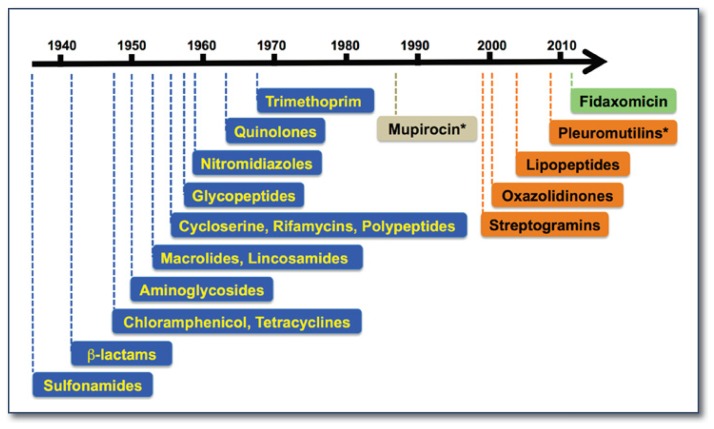
In 1945, the Scottish bacteriologist Alexander Fleming, who received the Nobel Prize for the discovery of penicillin, delivered the above cautionary statement in his Nobel lecture. His prophecy has since been fulfilled. Following the development of penicillin and sulfonamide (by the 1939 Nobel Laureate Gerhard Domagk), the serendipitous discoveries of antibiotics in the 1940s had inspired intense research into new human therapeutics. Fourteen classes of antibiotics representing six distinct mechanisms of action were introduced to clinical and veterinary uses before 1970 (See Figure 1).1 They became the miracle drugs of the twentieth century and enabled common life-threatening infections to be treated for the first time. Yet after decades of extensive use (misuse, overuse, and habitual use), the medical community is facing an unprecedented challenge in combating antibiotic resistance. At the same time, bottlenecks have slowed drug discovery and development.
Figure 1.
The depleting antibiotic pipeline. Antibiotic development suffered a long draught between 1970–2001. *: topical application.
Every year at least two million people fall ill to hospital-acquired infections and approximately 100,000 of the cases are fatal, the majority due to resistant bacterial and fungal strains. The empty pipeline for antibiotics and the rapid erosion of drug efficacy pose serious health risks in modern medicine. As antibiotics become ineffective, cases of hospital-acquired pneumonia will rise, life-threatening septicemia will become a greater concern in abdominal surgery, and the risks of opportunistic infections will be the limiting factor for patients undergoing cancer treatments and transplant surgery. According to World Health Organization estimates, new antibiotic-resistant strains have claimed more lives than HIV, influenza, and traffic accidents combined in the past decade. The infections are caused by members of gram-positive bacteria, including methicillin- and vancomycin-resistant Staphylococcus aurues (MRSA and VRSA), multidrug-resistant and extremely drug-resistant Mycobacterium tuberculosis (MDR-TB and XDR-TB), and Clostridium difficile.1 Most recently, the emergence of the gram-negative enterobacteria and vibrio species carrying the New Delhi metallo-β-lactamase-1 (NDM-1) gene not only left infected patients untreatable by the last line of drug, carbapenem, but also raised worldwide concern over the spread of the resistant gene through the transmissible plasmid.2
Falling Behind the Bugs
Antibiotics are generally prescribed in restricted quantity and for a short period, so the profits associated with new antibiotic discovery are considered much less lucrative than those associated with long-term treatments for chronic diseases such as cardiovascular diseases and diabetes. However, that is not to say that the pharmaceutical industry lacks incentive solely due to financial burden and lengthy regulatory hurdles. In reality, the development of novel antimicrobial drugs is scientifically daunting. Almost all new drugs approved after 1990 were either complete synthetic (oxazolidinones) or semi-synthetic derivatives of existing scaffolds originated from soil-dwelling actinomycetes (See Figure 1).1,3 The discovery of new classes of antibiotics with distinct scaffolds and novel modes of action has been constrained by the lack of chemical diversity in current combinatorial libraries, the narrow scope of structural- and target-based screening approaches, off-target signals in whole-cell screening, and poor success in developing effective screens to validate potential targets that were identified through comparative genomic analysis.1,4
Mechanisms of Antibiotic Resistance and Tolerance
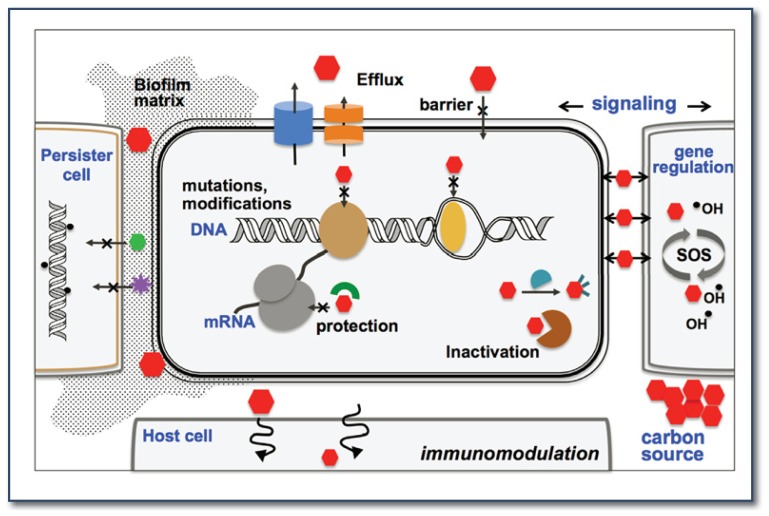
There are hundreds of antibiotics on the market, but they are very similar in that they each interfere with microbial cell growth by inhibiting only a handful of drug targets. These sites are usually macromolecules of essential biosynthesis pathways, such as the ribosome (protein synthesis), topoisomerase, DNA and RNA polymerase (nucleic acid synthesis), cell wall and membrane (peptidoglycan and lipid synthesis), and dihydrofolate reductase (folate metabolism) (See Figure 2).3 Not surprisingly, antibiotic resistance often evolves by reprogramming and camouflaging these targets through at least four general mechanisms: immunity, efflux, target modification, and drug inactivation. Some bacteria are immune to a drug simply because they have an impermeable barrier or produce protective proteins (TetM and FusB) that mask the drug binding sites. Active efflux involves multidrug resistance transporters that eject antibiotics directly to the outside of the cells or secrete them into the periplasm. Degradation and covalent modification can disarm the antibiotics. Furthermore, modifications of the target entail mutations in the target proteins or enzymatically modifying the targets that disable drug binding, for example, by methylation of the ribosomal RNA (See Figure 2).3,5,6
Figure 2.
Multiple roles of antibiotics in natural and clinical environments and the mechanisms of resistance.
Research in our lab focuses on studying the molecular mechanism of resistance mediated by rRNA methylation that is induced by subinhibitory concentrations of antibiotic. In staphylococci and streptococci, the erm genes encode for the methyltransferase enzymes that modify the drug target in 23S rRNA of the bacterial ribosome and reduce antibiotic binding. The expression of erm is regulated at the translational level by at least one upstream leader peptide via an unusual ribosome stalling mechanism. A threshold concentration of antibiotic, such as erythromycin, binds to a ribosome translating the leader peptide and causes the ribosome to stall. The stalling denatures an inhibitory mRNA hairpin in the intergenic region that masks the translation initiation site of erm and thereby facilitates the synthesis of Erm enzyme.7 Clinical studies have shown that Erm leader peptides are highly diverse and the antibiotic inducibility is drug type-specific, but the underlying mechanism is poorly understood. Our major goal is to understand the selective induction of antibiotic resistance in different erm homologous systems, specifically to compare the molecular interactions of the regulatory leader peptide with the ribosome in the presence and absence of antibiotic ligands. The work could potentially lead to rational drug design for non-inducer antibiotics.
In some cases, antibiotic tolerance arises when the microbes are able to withstand the drug killing effect by an altered metabolic pathway, such as an attenuation of hydroxyl radical production. This observation has provided an explanation on how seemingly unrelated genetic mutations away from a primary target can result in increased drug tolerance.8,9 Because most antibiotics are derived from bacterial natural products, the producers possess self-defense mechanisms that contribute to resistance. Recent studies have shown that horizontal gene transfer is extensive between environmental, clinical, and commensal bacteria by means of plasmids, transposons, integrons, and bacteriophages. Widespread antibiotic use is another complication, which exerts strong selective pressure in bacterial population and consequently induces mutagenesis in hypermutator strains. This evolution encourages the formation of persister cells that usually survive in biofilms (bacterial aggregates) (See Figure 2)6,10 and has hampered the complete eradication of pathogens during chronic infections.
Reservoirs of Antibiotic Resistance
According to a recent survey from the FDA, approximately 13.1 million kg (~80%) of antimicrobial drugs in the United States are sold for animal husbandry and fisheries annually. In contrast, only 3.3 million kg are administered for human use.11 Nontherapeutic and subtherapeutic uses of antibiotics in agribusiness began in the 1950s for inclusion in feed and water for poultry and livestock to prevent disease and to promote growth. The application fattens the animal faster and helps to mitigate the impact of disease spread because animals are regularly raised in crowded, unsanitary, and stressful conditions. The result is reduced cost of time and feed. Recognized scientific studies have repeatedly shown that low-dose antibiotics not only propagate resistant bacteria, but also disseminate these strains into nearby communities as well as the food supply.6,12–14 During the last few years, the new MRSA strain CC398 and a multidrug resistant strain of Salmonella enterica serovar Heidelberg have been found in high abundance in US retail turkey products, farm animals, and even humans who have had no direct contact with the animals.14 Resistant determinants are also widespread in soil, sewage, and water systems, at high prevalence compared with background resistance in nature.12,13,15,16 The example of bidirectional zoonotic exchange is an alarming sign of inadvertent creation of resistant strains that are too powerful for current medicine, given that the seven classes of antibiotics used in livestock production are also commonly used in treating human infections. To help preserve long-term effectiveness of medically important antimicrobials, FDA officials in April 2012 called for a voluntary halt to the routine use of several performance-enhancing antibiotics except for circumstances where prescription of such drugs is needed to treat and control animal illness. While farmers and stakeholders have argued tenaciously that eliminating antibiotics would result in larger economic loss and have little or no impact on reducing human infections, alternatives to antibiotics in agriculture should be explored in the United States. Approaches such as improving livestock hygienic practices and reducing overcrowding have had substantial success in Europe in the decades following the bans of antibiotics in animal production.
Multiple Roles of Antibiotics in Non-Clinical Settings
The popular press tends to paint microbes as disease-causing agents that should be eliminated using antibiotics, and that the role of antibiotics is to inhibit the cohabitating competitors. In contrast to these conventional views, a growing body of evidence has demonstrated that antibiotics serve a role far beyond that of a direct weapon. It has been shown that all antibiotics, regardless of their target and mode of action, exhibit the phenomenon of “hormesis” that is manifested by biphasic response of high-dose inhibition and low-dose stimulation.17
At subinhibitory doses, the concentration likely to be found in the environment, antibiotics act as signaling molecules to modulate gene expression and intercellular signaling (See Figure 2).8,17–19 For instance, aminoglycoside (AG) antibiotics such as tobramycin could interact with the AG response regulator and trigger secondary messenger cyclic diguanylate, which in turn leads to biofilm formation.20 In another example, subtherapeutic antibiotic treatment of young mice increased the metabolic activity of their gut microbiome by altering the expression of genes involved in the conversion of carbohydrates to short-chain fatty acids.21 Furthermore, bacterial cells exposed to subinhibitory concentrations of different antibiotics displayed distinct transcription profiles that are associated with pleiotropic effects in metabolic processes.17,22 Of note, it was found that the production of hydroxyl radicals induced by low antibiotic treatment could generate subpopulations of mutant strains that are resistant to other antibiotics but not the one applied.8 The isolation of naturally occurring bacteria that can subsist on both natural and synthetic antibiotics as a sole carbon source (See Figure 2)23 further expands the unrecognized role of antibiotics within bacterial communities. These discoveries have ignited the idea of removing antibiotic “pollution” from the environment by using the bacteria’s ability to devour antibiotics. Finally, quorum-sensing molecules that are important for mediating intra- and interspecies communication could exhibit antimicrobial activity at high concentrations,24 which further illustrates the common dual role of naturally occurring small molecules and suggests a potential resource for antimicrobial discovery.
Immunomodulatory Benefits of Macrolide Antibiotics
Relatively little is known about the beneficial effects of antibiotics on eukaryotic cell responses, except for the adverse effects usually associated with drug toxicity or compromised innate immunity. Interestingly, long-term macrolide prophylaxes are common pulmonary practices for the treatment of patients with chronic airway diseases, such as chronic obstructive pulmonary disease (COPD) and diffuse panbronchiolitis (DPB). Macrolides belong to the polyketide class of antibiotics that exert their bacteriostatic efficacy by inhibiting protein synthesis. Although the reduction of airway inflammation can in part be attributed to the antimicrobial activity of macrolide, the effects are believed to be negligible because the treatment dosage is too low to fight infection. The biological effects of macrolides are multifaceted but the exact mechanism is not well understood. They have been found to modulate (attenuate) the production of proinflammatory cytokines, to reduce epithelial expression of adhesion molecule, and to enhance phagocytosis and inhibition.25,26 Development of a macrolide-based anti-inflammatory drug devoid of the antimicrobial properties is currently of potential clinical interest.
The Future is an Arms Race: Bugging Out
Preventing and controlling infectious diseases relies on widespread collaboration in drug discover y, the judicious use of available antibiotics, improved hygiene, and well-developed methodologies for disease diagnosis. At the top of the list is drug discover y, which is essential to the long-term success of combating disease and requires a reinvigorated search in underexplored microbial niches. Robust screens designed to avoid rediscovering known scaffolds, leveraging existing libraries of synthetic molecules, and exploring the metagenomics approach to access the secondary metabolites from unculturable bacteria are all equally important components in drug discovery in need of further refinement.
In the short-term, antibiotic combination therapies that generate synergistic effects may slow the rise of resistance. To conserve drug effectiveness, tighter regulation on misuse and abuse of antibiotics must be imposed. More efficient and precise diagnostic tools tailored for individual patients will help guide more appropriate drug therapies based on the identified pathogen. Ultimately, it is through research and understanding of the microbes and the biological effects of each molecule at the metabolic level within the cell and within the microbial communities that will lead to new insights into antibiotic design. With an ever-greater range of new genomic and chemical biology tools, the race against antibiotic resistance is just the beginning of a new era for modern medicine.
Acknowledgments
M.N.F. Yap is a Pew scholar in the biomedical sciences, supported by the The Pew Charitable Trusts, and is supported by grant R00GM094212 from the National Institutes of Health and Saint Louis University startup and President’s Research Fund.
Biography
Mee-Ngan F. Yap, PhD, is an Assistant Professor in the Edward A. Doisy Department of Biochemistry and Molecular Biology Saint Louis University School of Medicine.
Contact: myap1@slu.edu

Footnotes
Disclosure
None reported.
References
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009 Aug 28;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011 Dec;19(12):588–595. doi: 10.1016/j.tim.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 3.von Nussbaum F, Brands M, Hinzen B, Weigand S, Habich D. Antibacterial natural products in medicinal chemistry--exodus or revival? Angew Chem Int Ed Engl. 2006 Aug 4;45(31):5072–5129. doi: 10.1002/anie.200600350. [DOI] [PubMed] [Google Scholar]
- 4.Chopra I. The 2012 Garrod Lecture: Discovery of antibacterial drugs in the 21st century. J Antimicrob Chemother Epub. 2012 doi: 10.1093/jac/dks436. [DOI] [PubMed] [Google Scholar]
- 5.Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007 Mar 23;128(6):1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010 Apr;8(4):251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 7.Ramu H, Mankin A, Vazquez-Laslop N. Programmed drug-dependent ribosome stalling. Mol Microbiol. 2009 Feb;71(4):811–824. doi: 10.1111/j.1365-2958.2008.06576.x. [DOI] [PubMed] [Google Scholar]
- 8.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010 Feb 12;37(3):311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee HH, Collins JJ. Microbial environments confound antibiotic efficacy. Nat Chem Biol. 2012 Jan;8(1):6–9. doi: 10.1038/nchembio.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sommer MO, Dantas G. Antibiotics and the resistant microbiome. Curr Opin Microbiol. 2011 Oct;14(5):556–563. doi: 10.1016/j.mib.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 11.Food and Drug Administration DoHaHS. “Antimicrobials sold or distributed for use in food-producing animals” and “An estimate of human antibacterial drug sales”. 2009–2011. http://www.fda.gov/downloads/ForIndustry/UserFees/AnimalDrugUserFeeActADUFA/UCM231851.pdf; http://www.fda.gov/downloads/Drugs/DrugSafety/InformationbyDrugClass/UCM319435.pdf.
- 12.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012 Aug 31;337(6098):1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heuer H, Schmitt H, Smalla K. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr Opin Microbiol. 2011 Jun;14(3):236–243. doi: 10.1016/j.mib.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Oppliger A, Moreillon P, Charriere N, Giddey M, Morisset D, Sakwinska O. Antimicrobial Resistance of Staphylococcus aureus Strains Acquired by Pig Farmers from Pigs. Appl Environ Microbiol. 2012 Nov;78(22):8010–8014. doi: 10.1128/AEM.01902-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Looft T, Johnson TA, Allen HK, et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruden A, Arabi M, Storteboom HN. Correlation between upstream human activities and riverine antibiotic resistance genes. Environ Sci Technol. 2012 Nov 6;46(21):11541–11549. doi: 10.1021/es302657r. [DOI] [PubMed] [Google Scholar]
- 17.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006 Oct;9(5):445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Romero D, Traxler MF, Lopez D, Kolter R. Antibiotics as signal molecules. Chem Rev. 2011 Sep 14;111(9):5492–5505. doi: 10.1021/cr2000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008 Apr;11(2):161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005 Aug 25;436(7054):1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- 21.Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012 Aug 30;488(7413):621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002 Dec 24;99(26):17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dantas G, Sommer MO, Oluwasegun RD, Church GM. Bacteria subsisting on antibiotics. Science. 2008 Apr 4;320(5872):100–103. doi: 10.1126/science.1155157. [DOI] [PubMed] [Google Scholar]
- 24.LoVetri K, Madhyastha S. Antimicrobial and antibiofilm activity of quorum sensing peptides and Peptide analogues against oral biofilm bacteria. Methods Mol Biol. 2010;618:383–392. doi: 10.1007/978-1-60761-594-1_24. [DOI] [PubMed] [Google Scholar]
- 25.Blasi F, Mantero M, Aliberti S. Antibiotics as immunomodulant agents in COPD. Curr Opin Pharmacol. 2012 Jun;12(3):293–299. doi: 10.1016/j.coph.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 26.Zarogoulidis P, Papanas N, Kioumis I, Chatzaki E, Maltezos E, Zarogoulidis K. Macrolides: from in vitro anti-inflammatory and immunomodulatory properties to clinical practice in respiratory diseases. Eur J Clin Pharmacol. 2012 May;68(5):479–503. doi: 10.1007/s00228-011-1161-x. [DOI] [PubMed] [Google Scholar]