Abstract
Chromatin is a supramolecular assembly of DNA and histone proteins, organized into nucleosome repeat units. The dynamics of chromatin organization regulates DNA accessibility to eukaryotic transcription and DNA repair complexes. However, the structural and dynamic properties of chromatin at high concentrations characteristic of the cellular environment (> ~200 mg/ml) are largely unexplored at the molecular level. Here, we apply magic angle spinning nuclear magnetic resonance to directly probe the dynamic histone protein regions in 13C,15N-enriched recombinant nucleosome arrays at cellular chromatin concentrations and conditions designed to emulate distinct states of DNA condensation, with focus on the flexible H3 and H4 N-terminal tails which mediate chromatin compaction. 2D 1H-13C and 1H-15N spectra reveal numerous correlations for H3 and H4 backbone and side-chain atoms, enabling identification of specific residues making up the dynamically disordered N-terminal tail domains. Remarkably, we find that both the H3 and H4 N-terminal tails are overall dynamic even in a highly condensed state. This significant conformational flexibility of the histone tails suggests that they remain available for protein binding in compact chromatin states to enable regulation of heterochromatin. Furthermore, our study provides a foundation for quantitative structural and dynamic investigations of chromatin at physiological concentrations.
Eukaryotic DNA is dynamically organized into chromatin fibers, which regulate essential functions of the genome including transcription and DNA repair.1,2 The basic building block of chromatin is the nucleosome core particle, which contains ~146 base pairs (bp) of DNA wrapped 1.65 times around a histone protein octamer containing two copies each of histones H2A, H2B, H3 and H4.3 The nucleosome X-ray structure has been determined to near atomic-resolution4 and reveals a compact helical core with ~15–30% of the histone sequences protruding from the core as largely unstructured, and presumably flexible, N-terminal tail domains (Figure 1A). The crystal structure of a tetranucleosome has also been solved,5 but the low (9 Å) resolution of this structure precludes the definition of N-terminal histone tail conformations.
Figure 1.
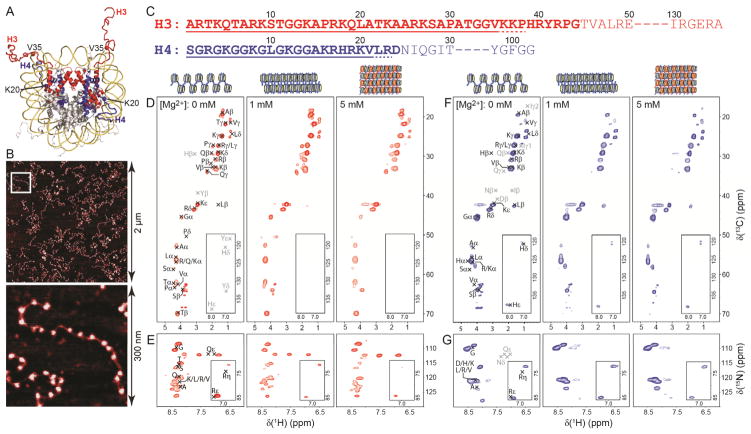
(A) Nucleosome crystal structure (PDB entry 1KX5).4 Histones H3 and H4 are colored in red and blue, respectively, with selected residues located near the N-terminal tail boundaries highlighted. (B) Representative atomic force microscopy images of 17-mer nucleosome arrays used for the NMR studies. (C) Amino acid sequences of histones H3 and H4. N-terminal residues that are relatively unstructured in nucleosome core particle crystals are highlighted in bold. Residues determined in the present study to comprise the flexible N-terminal H3 and H4 tails in highly concentrated 17-mer nucleosome arrays irrespective of the degree of array compaction are underlined with solid lines. Additional residues that possibly belong to the dynamically disordered histone tail domains but cannot be unambiguously identified from the NMR data are underlined with dashed lines. (D,E) 2D 1H-13C (D) and 1H-15N (E) correlation spectra of 17-mer nucleosome arrays containing 13C,15N-enriched H3 and prepared with 0, 1 and 5 mM Mg2+, corresponding to extended, folded and aggregated chromatin conformation, respectively, as indicated. The spectra were recorded at 500 MHz 1H frequency, 11.111 kHz MAS rate and 30 °C using a 2D refocused INEPT pulse scheme described in detail previously.26 Each spectrum was recorded with acquisition times of 18 ms and 30 ms in the 1H and 13C/15N dimensions, respectively, and a total measurement time of ~48 h. Resonance assignments based on the average chemical shift values in the BioMagResBank database corresponding to residues located in the flexible N-terminal histone tail are labeled in black font. Indicated in gray font are the approximate locations of signals from unique residues bordering the flexible tail that would be observed if those residues were sufficiently mobile. Note that several narrow signals visible at 13C frequency of ~65–70 ppm, particularly for the 0 mM Mg2+ sample, arise from residual sucrose from the sucrose gradient purification procedure. (F,G) Same as panels (D,E) but for 17-mer nucleosome arrays containing 13C,15N-labeled H4.
In human cells, single chromatin fibers are chains of ~100,000 nucleosomes located in the nucleus at extremely high concentrations of over 200 mg/ml,6 with each fiber organized into distinct chromosome territories.7 In vitro, longer (> ~10–12-mer) nucleosome arrays form a variety of higher order structures in presence of Mg2+, ranging from an extended beads-on-a-string type “10-nm” fiber in the absence of Mg2+ to a folded “30-nm” fiber at intermediate (~1 mM) Mg2+ concentration to highly condensed aggregates at high Mg2+ concentrations.8,9 The 30-nm chromatin fiber has been observed in a few distinct cell types.10,11 Interestingly, however, recent small-angle X-ray scattering studies indicate that the 30-nm fiber is not the dominant structural form of chromatin in mitotic chromosomes,12 suggesting that the high cellular concentrations of chromatin may impact its higher order structure.
It is well-established that the positively charged N-terminal tails of histones H3 and H4 mediate the compaction of chromatin into 30 nm diameter fibers and interfiber condensation in vitro, apparently through interactions with DNA and/or acidic regions on the histone octamer surface of neighboring nucleosomes.13–15 Remarkably, peptides with sequences corresponding to the H4 N-terminal tail are also able to mediate self-association of H4 tail-less nucleosome arrays,16 suggesting that charge neutralization plays an important role in chromatin compaction. The mobile N-terminal histone tails have been previously probed by solution nuclear magnetic resonance (NMR) in ~20 mg/ml samples of single nucleosomes17–19 and oligonucleosomes20 in the absence of Mg2+. Of particular note is the recent elegant study of mononucleosomes containing 13C,15N-enriched histones by Bai and co-workers,19 which found that flexible H3 and H4 domains encompass residues 1–36 and 1–15, respectively, based on sets of backbone amide signals detected in 15N-1H heteronuclear single quantum coherence spectra. On the other hand, the structure and dynamics of H3 and H4 tails in compacted chromatin fibers remain largely unknown, with the analysis of these domains to date being limited to modeling21–23 and relatively indirect biochemical and biophysical approaches such as mutagenesis combined with analytical centrifugation or chemical cross-linking14,24 and hydrogen-deuterium exchange coupled with solution NMR.25 Interestingly, the recent hydrogen-deuterium exchange NMR study of 12-mer nucleosome arrays25 concluded that the H3 tail forms stable folded structures in highly condensed chromatin fibers. This is in contrast with observation of a dynamically disordered H3 N-terminus in soluble single nucleosomes19 and suggests possible differences in the conformational flexibilities of histone N-terminal domains between uncompacted and compacted chromatin.
Here, we apply one- and two-dimensional (1D and 2D) magic-angle spinning (MAS) NMR to probe histones H3 and H4 within isotopically 13C,15N-enriched recombinant 17-mer nucleosome arrays at very high densities of ~200–400 mg/ml typical of the cellular environment. The arrays were incubated with 0, 1 or 5 mM Mg2+, conditions that respectively yield extended chromatin, compact 30 nm chromatin fibers, and a highly condensed chromatin state with self-association between multiple fibers.9 The major advantage of the MAS NMR methodology in the present context is that it enables the amino acid residues comprising the rigid and flexible histone protein segments to be monitored directly under physiologically-relevant conditions, in a manner similar to that previously demonstrated for a variety of large macromolecular complexes including membrane and fibrillar protein assemblies.26–28
The nucleosome arrays used in this study were reconstituted from a 3,046 bp DNA template containing 17 tandem repeats of a Widom 601 nucleosome positioning sequence variant with 30 bp of linker DNA29 (Figure S1) and histone octamer containing 13C,15N-labeled H3 or H4 (see Supporting Information (SI) for details). Sucrose gradient purified 17-mer arrays were analyzed by composite gel electrophoresis, BamHI and AvaI restriction enzyme digestions and atomic force microscopy (AFM) (Figure 1B) to assess their level of saturation with histone octamer. These analyses (Figures S2–S3 and accompanying SI text) show that the array samples are highly homogeneous, effectively (> 95%) saturated with 17 histone octamers, and stable during the course of the MAS NMR experiments. As noted above, different levels of chromatin condensation were induced by increasing amounts of Mg2+.9 The extent of self-association of the different array samples was readily evident in their physical appearance and was quantitatively confirmed by using a UV absorbance based sedimentation assay30 (Figure S4). For the NMR measurements the array samples were concentrated to ~200–400 mg/ml by ultracentrifugation, which is typical of chromatin densities found in cells.
The presence of flexible and rigid segments of histones H3 and H4 in the context of the highly concentrated 17-mer nucleosome arrays was first assessed using a set of 1D 13C refocused INEPT and cross-polarization MAS NMR spectra recorded as a function of temperature between −20 and 30 °C and described in the SI (Figures S5–S7). The key result from this set of experiments is that at temperatures of ca. −10 °C and above both H3 and H4 contain highly mobile domains spanning ~25% of the protein sequences, irrespective of the degree of array compaction. Next, we performed a more detailed analysis of the flexible H3 and H4 domains by 2D MAS NMR methods. Figure 1 shows a series of 1H-13C and 1H-15N correlation spectra recorded at 30 °C using a 2D refocused INEPT pulse scheme26,27 as a function of increasing Mg2+ concentration for arrays containing 13C,15N-H3 (panels D and E) or H4 (panels F and G). As expected based on the 1D INEPT 13C spectra, the 2D 1H-13C and 1H-15N data sets display multiple intense resonances for both histones H3 and H4. A qualitative inspection of the 1H-15N spectra reveals that all detectable backbone amide protons resonate in a narrow range between 8.0–8.5 ppm, characteristic of unstructured proteins.31 Indeed, by assuming that the flexible H3 and H4 tails are located at the N-termini and behave as ensembles of random-coil-like states we have been able to account for the vast majority of the observed protein signals by mapping onto the spectra the average residue- and site-specific chemical shifts obtained from the BioMagResBank database. While the spectral sensitivity and resolution achievable for these array samples were not sufficient to establish sequence specific resonance assignments de novo, analysis of the 2D NMR spectra in conjunction with the H3 and H4 amino acid sequences nevertheless permitted a relatively precise identification of the residues encompassing the flexible H3 and H4 N-terminal domains in the 17-mer arrays as follows. For both histones resonances characteristic of the various amino acids were accounted for starting from the N-terminus, until signals from one or more amino acids in the sequence that should be clearly discernible in the spectra if sufficiently mobile were no longer detectable. It is important to note that while identification of amino acid types was fairly straightforward, the number of residues contributing to the individual resonances could not be readily determined due to the complicating effect of local protein dynamics on signal intensities.
For H3 a number of unique resonances were observed corresponding to residues in the range 1–38, including for example Gα, Aβ, Sβ, Tβ, Vγ, Lδ and Kε signals in the 1H-13C spectra as well as Qε, Rε and Rη signals in the 1H-15N spectra. Particularly remarkable is the observation of the Vγ signals. Combined with the fact that V35 is the first valine encountered in the sequence, this finding indicates that the flexible H3 tail extends to at least position 35. Equally crucial is the fact that no side-chain 1H-13C signals characteristic of His or Tyr were observed, which indicates that H39 and Y41 are relatively immobile and defines the H3 tail boundary. In the absence of sequential resonance assignments it is impossible to unambiguously establish from these data whether residues K36-P38 are sufficiently mobile to be detectable, because multiple other residues of the same type that yield observable signals are located between positions 1 and 35. It is also unlikely that any of the H3 signals originate from protein regions other than the flexible N-terminal tail—in particular, it is improbable that the Vγ signal could be attributed to any of the other five valines located in the core domain of H3. The reason is that, in addition to the absence of His and Tyr resonances, we also do not observe any characteristic Ile side-chain signals (there are 7 isoleucines in H3 located between positions 51 and 130). Altogether, we conclude that flexible N-terminal H3 tails in concentrated 17-mer nucleosome arrays span amino acids 1–35 and possibly several additional residues in the 36–38 regime (Figure 1C), while the remaining H3 residues (certainly 39–130) are largely immobile.
Analogous analysis of the H4 spectra identified unique signals arising from multiple residues in the range 1–21, including for example Gα, Aβ, Vγ, Lδ, Hδ, Hε and Kε signals in the 1H-13C spectra as well as Rε and Rη signals in the 1H-15N spectra. Key is the observation of the Aβ, Vγ, Hδ and Hε signals because A15, H18 and V21 are the first Ala, His and Val residues in the H4 sequence. Also critical is the finding that characteristic side-chain signals from Asp, Asn, Ile, Gln and Thr, located in the range 24–30, are missing, which clearly defines the flexible tail boundary. Given that no Tyr and Phe resonances are detected either—there are two Phe and four Tyr residues in H4, with Y98 and F100 located near the C-terminus—we conclude that flexible N-terminal H4 tails in the 17-mer arrays span residues 1–21 while most of the remaining residues are relatively rigid (Figure 1C) (note that it is possible that H4 residues L22 and R23 are also somewhat flexible, but this cannot be unambiguously established due to the presence of other leucines and arginines in the 1–21 segment).
In addition to the identification of specific amino acids making up the flexible N-terminal domains of histones H3 and H4 in concentrated chromatin, a central finding of our study is that by and large the same residues remain dynamic in uncompacted, folded and highly condensed nucleosome arrays as judged by the similarity of the 2D 1H-13C and 1H-15N NMR spectra in Figure 1. In order to compare the extent of protein dynamics between the different array samples we performed 15N INEPT based NMR experiments to monitor the transverse relaxation rates of the amide protons exclusively for residues encompassing the flexible histone tails (Figure S8). Although rather qualitative, these studies reveal that collectively the amide 1H coherences relax somewhat more rapidly for arrays incubated with 1 mM and 5 mM Mg2+ relative to those prepared in the absence of Mg2+, particularly for H3. This finding is suggestive of a subtle overall reduction in the flexibility of the histone tail domains in the most highly condensed and concentrated arrays, which would lead to less efficient motional averaging of the 1H-1H dipolar couplings and consequently faster 1H coherence decay. We also recorded complementary electron paramagnetic resonance (EPR) spectra of the 17-mer arrays containing nitroxide spin labeled analogs of histone H3 at two locations including the N-terminus and residue 35 near the flexible tail boundary. The EPR spectra (Figure S9) display nearly identical, relatively narrow lineshapes for all Mg2+ concentrations indicating significant flexibility of the H3 N-terminal tail irrespective of the degree of compaction, and, importantly, no detectable broad components characteristic of immobilized protein segments. The moderate line broadening observed for position 35 relative to the N-terminus is consistent with the expected reduction in mobility of the nitroxide spin probe in the vicinity of the nucleosomal DNA.
In summary, we have demonstrated that MAS NMR spectroscopy permits the direct and unequivocal analysis of flexible histone tails in large nucleosome arrays at cellular concentrations. A strong correlation exists between the most dynamic histone residues identified in the current study, amino acids 1–35 in H3 and 1–21 in H4, and the crystal structure of the nucleosome core particle which reveals the same residues as being largely unstructured. Our data are also generally in agreement with the NMR study of single nucleosomes in solution,19 which reports that flexible H3 and H4 tails encompass residues 1–36 and 1–15, respectively. The finding that H4 residues 16–21 display significant mobility in the array samples could be indicative of somewhat different properties of these domains in the context of single nucleosomes at relatively low concentration versus highly concentrated arrays. Nevertheless, we also cannot discount the possibility that this difference stems from the fact that the solution NMR study focused solely on the protein backbone, while MAS NMR techniques monitor both the backbone and side-chain signals. Conversely, our results do not support the main conclusion of a recent hydrogen-deuterium exchange NMR study that the H3 N-terminal tail forms stable folded structures in highly condensed nucleosome arrays.25 The origin of the discrepancy between the hydrogen-deuterium exchange study and the MAS NMR data is unclear. However, given that the former method does not visualize mobile residues directly but rather infers information about protein dynamics from amide proton occupancies, it is plausible that the histone tails in the highly condensed arrays could remain flexible yet concurrently somewhat protected from solvent exchange on the relatively short exchange time scales investigated. It is also noteworthy that the significant flexibility of histone tails in condensed nucleosome arrays revealed by MAS NMR is compatible with the EPR data for H3 presented in this study as well as the previous observation of mobile histone protein domains in oligonucleosomes by natural abundance 13C solution NMR at low and moderate ionic strengths,20 with the caveat that those experiments were done on relatively dilute nucleosome array samples.
The finding that H3 and H4 N-terminal tail domains are flexible even in highly condensed nucleosome arrays strongly suggests that chromatin compaction does not involve specific, high-affinity protein-protein or protein-DNA contacts that would lead to their immobilization. However, given the well-established importance of H3 and H4 histone tails in the folding of chromatin fibers,13–15 it is feasible that these flexible domains participate in the chromatin condensation process via multiple weak, transient interactions that shield the electrostatic repulsion between DNA moieties associated with different nucleosome units. The overall conformational flexibility of the H3 and H4 tails indicates that they are accessible to the numerous regulatory proteins that bind histone tails to regulate transcription even in compact heterochromatin at extremely high cellular densities. This provides a mechanism by which transcription activating complexes could directly gain access to histone tails to initiate conversions from heterochromatin to euchromatin. Factors in addition to histone tails that compact chromatin are also dynamic. Heterochromatin Protein 1 (HP1), which binds trimethylated lysine 9 within H3, and linker histone H1, which binds near the nucleosome DNA entry-exit region, facilitate heterochromatin formation.32,33 Both HP134 and H135 rapidly exchange in vivo, suggesting that multiple factors, which compact chromatin, function through dynamic mechanisms. Further studies of histone tails and the proteins that bind them are required to determine the role of histone tail dynamics in the conversion between heterochromatin and euchromatin.
Supplementary Material
Acknowledgments
This work was supported by grants from the Center for Emergent Materials at Ohio State (NSF MRSEC, DMR-0820414), the National Institutes of Health (R01GM094357 to C.P.J. and R01GM083055 to M.G.P.) and the Camille & Henry Dreyfus Foundation (Camille Dreyfus Teacher-Scholar Award to C.P.J.), and an OSU-CCC Pelotonia Predoctoral Fellowship (J.A.N.). The authors thank Dr. Swagata Chakraborty for assistance with the preparation of 13C,15N-labeled histone protein samples.
Footnotes
Notes
The authors declare no competing financial interests.
Supporting Information. Materials and methods, detailed descriptions of gel electrophoresis, AFM, UV-based sedimentation and 1D variable temperature NMR studies, and supplementary figures with gel, AFM, NMR, and EPR data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Woodcock CL, Ghosh RP. Cold Spring Harb Perspect Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Reinberg D. Curr Opin Genet Dev. 2011;21:175–186. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 4.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 5.Schalch T, Duda S, Sargent DF, Richmond TJ. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 6.Dehghani H, Dellaire G, Bazett-Jones DP. Micron. 2005;36:95–108. doi: 10.1016/j.micron.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Cremer T, Cremer M. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen JC, Ausio J, Stanik VH, van Holde KE. Biochemistry. 1989;28:9129–9136. doi: 10.1021/bi00449a026. [DOI] [PubMed] [Google Scholar]
- 9.Hansen JC. Annu Rev Biophys Biomol Struct. 2002;31:361–392. doi: 10.1146/annurev.biophys.31.101101.140858. [DOI] [PubMed] [Google Scholar]
- 10.Belmont AS, Bruce K. J Cell Biol. 1994;127:287–302. doi: 10.1083/jcb.127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bystricky K, Heun P, Gehlen L, Langowski J, Gasser SM. Proc Natl Acad Sci USA. 2004;101:16495–16500. doi: 10.1073/pnas.0402766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joti Y, Hikima T, Nishino Y, Kamada F, Hihara S, Takata H, Ishikawa T, Maeshima K. Nucleus. 2012;3:404–410. doi: 10.4161/nucl.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luger K, Richmond TJ. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 14.Dorigo B, Schalch T, Bystricky K, Richmond TJ. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 15.Zheng C, Lu X, Hansen JC, Hayes JJ. J Biol Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 16.Chodaparambil JV, Barbera AJ, Lu X, Kaye KM, Hansen JC, Luger K. Nat Struct Mol Biol. 2007;14:1105–1107. doi: 10.1038/nsmb1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cary PD, Moss T, Bradbury EM. Eur J Biochem. 1978;89:475–482. doi: 10.1111/j.1432-1033.1978.tb12551.x. [DOI] [PubMed] [Google Scholar]
- 18.Hilliard PR, Jr, Smith RM, Rill RL. J Biol Chem. 1986;261:5992–5998. [PubMed] [Google Scholar]
- 19.Zhou BR, Feng H, Ghirlando R, Kato H, Gruschus J, Bai Y. J Mol Biol. 2012;421:30–37. doi: 10.1016/j.jmb.2012.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith RM, Rill RL. J Biol Chem. 1989;264:10574–10581. [PubMed] [Google Scholar]
- 21.Arya G, Schlick T. J Phys Chem A. 2009;113:4045–4059. doi: 10.1021/jp810375d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potoyan DA, Papoian GA. J Am Chem Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 23.Korolev N, Lyubartsev AP, Nordenskiold L. Biophys J. 2006;90:4305–4316. doi: 10.1529/biophysj.105.080226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan PY, Lu X, Hansen JC, Hayes JJ. Mol Cell Biol. 2007;27:2084–2091. doi: 10.1128/MCB.02181-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato H, Gruschus J, Ghirlando R, Tjandra N, Bai Y. J Am Chem Soc. 2009;131:15104–15105. doi: 10.1021/ja9070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helmus JJ, Surewicz K, Surewicz WK, Jaroniec CP. J Am Chem Soc. 2010;132:2393–2403. doi: 10.1021/ja909827v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M. J Am Chem Soc. 2005;127:12965–12974. doi: 10.1021/ja0530164. [DOI] [PubMed] [Google Scholar]
- 28.Siemer AB, Arnold AA, Ritter C, Westfeld T, Ernst M, Riek R, Meier BH. J Am Chem Soc. 2006;128:13224–13228. doi: 10.1021/ja063639x. [DOI] [PubMed] [Google Scholar]
- 29.Poirier MG, Bussiek M, Langowski J, Widom J. J Mol Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu X, Simon MD, Chodaparambil JV, Hansen JC, Shokat KM, Luger K. Nat Struct Mol Biol. 2008;15:1122–1124. doi: 10.1038/nsmb.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wüthrich K. NMR of Proteins and Nucleic Acids. Wiley; New York, NY: 1986. [Google Scholar]
- 32.Grewal SI, Jia S. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 33.McBryant SJ, Hansen JC. Adv Exp Med Biol. 2012;725:15–26. doi: 10.1007/978-1-4614-0659-4_2. [DOI] [PubMed] [Google Scholar]
- 34.Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 35.Lever MA, Th’ng JP, Sun X, Hendzel MJ. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.