Abstract
A major problem in the discovery of new biologically active compounds from natural products is the re-isolation of known compounds. Such re-isolations waste time and resources, distracting chemists from more promising leads. To address this problem, dereplication strategies are needed that enable crude extracts to be screened for the presence of known compounds before isolation efforts are initiated. In a project to identify anticancer drug leads from filamentous fungi, a significant dereplication challenge arises, as the taxonomy of the source materials are rarely known, and, thus, the literature cannot be probed to identify likely known compounds. An ultra-performance liquid chromatography-photodiode array-high-resolution tandem mass spectrometric (UPLC-PDA-HRMS-MS/MS) method was developed for dereplication of fungal secondary metabolites in crude culture extracts. A database was constructed by recording HRMS and MS/MS spectra of fungal metabolites, utilizing both positive- and negative-ionization modes. Additional details, such as UV-absorption maxima and retention times, were also recorded. Small-scale cultures that showed cytotoxic activities were dereplicated before engaging in the scale-up or purification processes. Using these methods, approximately 50% of the cytotoxic extracts could be eliminated from further study after the confident identification of known compounds. The specific attributes of this dereplication methodology include a focus on bioactive secondary metabolites from fungi, the use of a 10-min chromatographic method, and the inclusion of both HRMS and MS/MS data.
Many natural product drug discovery programs utilize bioactivity-directed fractionation methodologies for the isolation of lead compounds from crude extracts, in which the bioassay results guide the purification processes.1,2 While countless studies demonstrate the utility of this approach in identifying new drug leads, with taxol and camptothecin possibly the most well-known examples,3–5 it is often criticized for resulting in the re-isolation and re-characterization of previously known compounds. This criticism of natural products research grows in significance annually, as over 246,000 compounds have been described from Nature, with approximately 4,000 new ones added each year.6 Thus, to expedite the discovery of new leads, and to avoid the re-isolation of previously known compounds, it is crucial to discriminate between known vs. new compounds as early as possible. This process, which is termed “dereplication”,7 enables the efficient use of human and financial resources,8,9 so that efforts can be focused on the discovery of structurally novel compounds.8,10
Early dereplication strategies focused largely on botanical extracts. The ability to characterize a plant’s taxonomy (at least to the genus level) based on morphology can simplify the dereplication process, as it enables botanical extracts to be screened for known chemical compounds specific to the taxa under investigation. Some examples include colchicinoids in Colchicum spp.,11 acetogenins in plants of the Annonaceae,12 phenolics in the genus Lippia,13 steroidal alkaloids in Buxus spp.,14 and triterpenoids in the genus Actaea.15
In contrast, dereplication can be more complicated for organisms of unknown taxonomy.16,17 This is the case for our ongoing research aimed at identifying anticancer drug leads from nonsporulating filamentous fungi.18,19 Fungi are known to produce structurally diverse secondary metabolites that display a wide range of biological activities. Although fungi are relatively under-investigated,20,21 a major challenge with these organisms as a source of bioactive lead compounds is the production of mycotoxins, which can be observed across different fungal species. Examples of these include aflatoxins, ochratoxins, trichothecenes, citreoviridin, fumonisins, and various indole-derived tremorgenics.22 Indeed, a promising side application for the presently described dereplication procedure is the rapid identification of mycotoxins in foods, as mycotoxins continue to present safety challenges for the food supply.23,24 While molecular methods have accelerated the process,25 assignment of fungal taxonomy can be a tedious procedure, and regardless, only about 100,000 of the estimated 1.5 to 5.1 million species of fungi have been ascribed a scientific name.20,21 Hence, the taxonomy of fungi being studied in natural products programs is often not known at all, or only determined after compounds have been isolated. Thus, there is a critical need for better strategies to dereplicate crude fungal extracts for the presence of known chemical entities, including mycotoxins and other biologically relevant compounds. In doing so, resources for drug discovery can be devoted into those samples most likely to yield new chemical entities.19
Several dereplication methods have been reported in the literature for fungal secondary metabolites; however, all of these are hampered by at least one, if not several, limitations, including long run times (30 min or more), low-resolution mass measurements, and/or lack of confirmatory MS-MS data.16,26–29 A recent LC-UV/vis-MS-based dereplication strategy utilized UV spectra (acquired with a photodiode array detector) and ESI+/ESI− time-of-flight MS for assignment of 719 microbial natural product and mycotoxin reference standards.27 While effective, only 17% (29 compounds) of our database of cytotoxic fungal compounds overlapped with the standards used, as this earlier procedure did not have a focus on compounds with biological activity.27 Moreover, a 30-min chromatographic method was utilized,27 which may be too long for routine processing of scores of samples simultaneously, particularly for shared instruments. Finally, CID MS/MS data were not reported,27 which serve to fingerprint and confirm the identity of dereplicated compounds. Thus, the goal of the present study was to harness the powerful resolution and short analysis time afforded by ultra-performance liquid chromatography (UPLC) coupled to the outstanding mass accuracy of an Orbitrap mass spectrometer to develop a rapid and effective method to dereplicate biologically active fungal extracts. In addition, MS/MS and UV (photodiode array) spectra were employed as an integral part of the strategy, so as to differentiate isobaric compounds often present in natural product extracts. Finally, given the likelihood of the co-elution of compounds in complex mixtures separated over short (<10 min) chromatographic run times, a data analysis strategy was incorporated (ACD/IntelliXtract), aimed at rapidly deconvoluting complex LC/MS chromatograms.
RESULTS AND DISCUSSION
The foundation of the present UPLC-PDA-HRMS-MS/MS dereplication procedure was the construction of a database for the identification of more than 170 fungal secondary metabolites via recording chromatographic retention times, UV data, and full-scan (high-resolution) mass spectra and MS/MS spectra in both positive and negative electrospray ionization (ESI) modes. The chromatographic and tune methods were developed using a mixture of 36 compounds representing diverse structural classes that ranged from terpenenoids to polyketides to depsipeptides. To test the developed method, a mixture of ten structurally diverse compounds was prepared and analyzed (Figures S1 and S2, Supporting Information). The resulting 10-min chromatographic method and associated high-resolution mass spectra could be used to discriminate between all the compounds in the database. Of these, only two resorcylic acid lactones, 7-epi-zeaenol and 15-O-desmethyl-5Z-7-oxozeaenol,30 did not ionize in the ESI+ mode, whereas 17 compounds, mostly trichothecenes,31 did not ionize in the ESI− mode (Table S1, Supporting Information). Also, the molecular ion peaks of aphidicolin and viridicatumtoxin were the only ones not detected in the positive-ion mode, due to the facile loss of H2O; however, the molecular ion peak for the latter was observed in the negative-ion mode. All MS/MS fragmentation data were collected using collisionally induced dissociation (CID) with 30% collision energy, and the ten most intense fragments for each compound were utilized in the database (Table S1, Supporting Information).
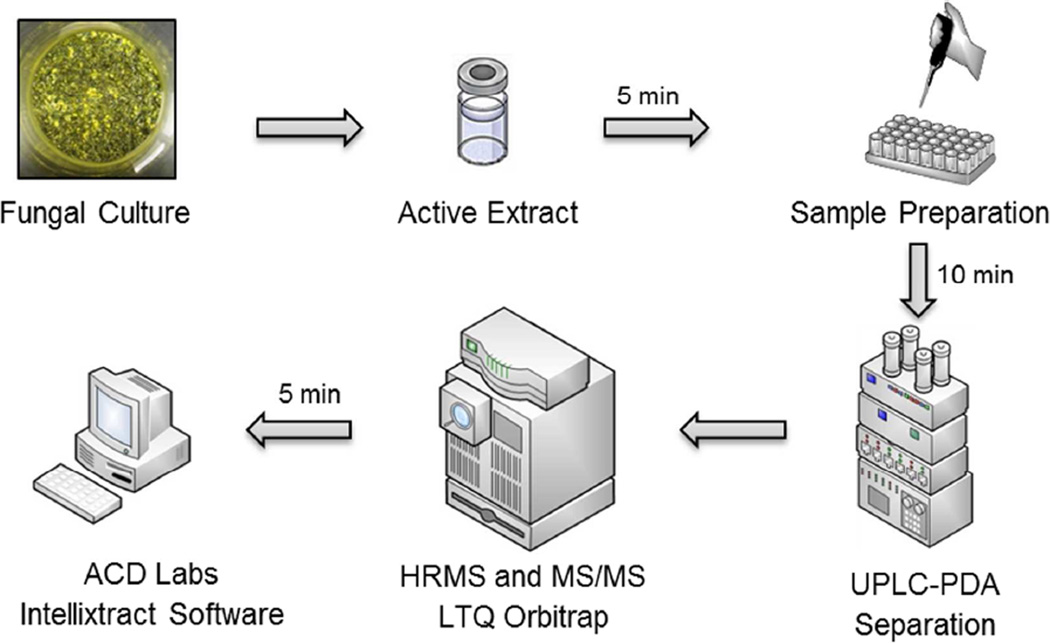
To initiate the dereplication process, small-scale cultures of fungi were grown on a solid medium and extracted using previously described protocols;30,32–34 the extracts were then evaluated for cytotoxicity against a panel of human cancer cell lines in culture, including MCF-7, H-460, and SF-268 cells.35,36 Samples deemed active were prepared for analysis by dissolving a sub-milligram aliquot of the bioactive crude extracts in equal volumes of MeOH and dioxane to obtain a final concentration of 2 mg/mL. The subsequent dereplication strategy utilized a three-step approach (Figure 1). First, UPLC-HRMS was used to acquire the TIC (total ion chromatogram) of the extract and hence measure the accurate masses of all ions detectable. This TIC was then uploaded into ACD MS Manager with add-in software Intellixtract, which crossreferenced the molecular ion peaks with the database that was constructed as a list of compound names, corresponding molecular weights, and retention times (Table S1, Supporting Information). These results were then confirmed manually using HRMS data, and the MS/MS and UV spectra were compared with the database to verify the identity of any hits. The use of the MS/MS data, retention times, and UV spectra confirmed the identity of the compounds in the extract; the only exception would be for the possible, albeit rare, case of isomers. Moreover, the ACD/IntelliXtract software, which has the capability to extract all chromatographic components in the LC/MS datasets, expedited the identification process, due to its capability of resolving overlapping and co-eluting components. Finally, using IntelliXtract, it was possible to reconstruct pure component chromatograms for each chromatographic component and annotate spectral peaks with the mass of the protonated or deprotonated molecule.37
Figure 1.
Schematic of the workflow for the proposed dereplication protocol. Aliquots of active fungal extracts were evaluated using an UPLC-PDA-HRESIMS-MS/MS method, the data were analyzed using ACD Labs Intellixtract software and compared with an in-house database, and extracts producing known compounds were excluded.
Using these procedures, 106 small-scale culture extracts were dereplicated. All of these displayed sufficiently potent cytotoxicity against a small cancer cell-line panel to warrant further investigation. However, based on the dereplication results, 55 samples were ruled out as containing known compounds. By doing so, resources were prioritized on samples most likely to yield new compounds. The three examples shown below demonstrate the work flow of the dereplication methodology, illustrating its utility for a broad range of structural classes of bioactive natural products.
Dereplication of Verticillins (Epipolythiodioxopiperazine Alkaloids)
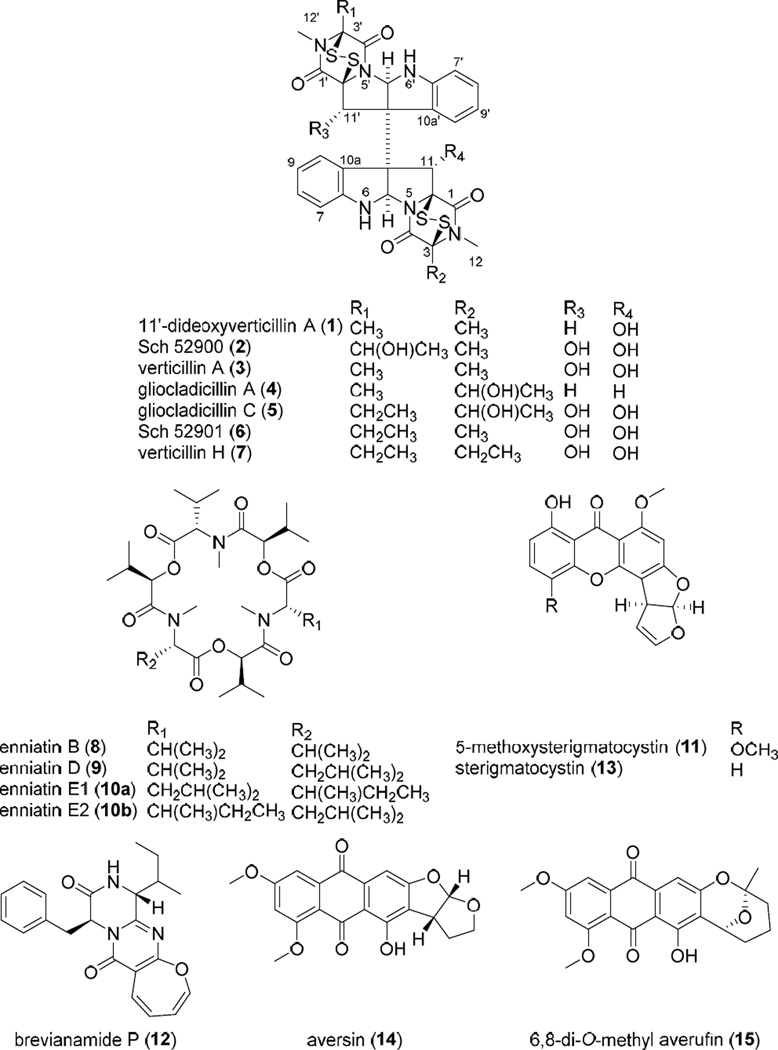
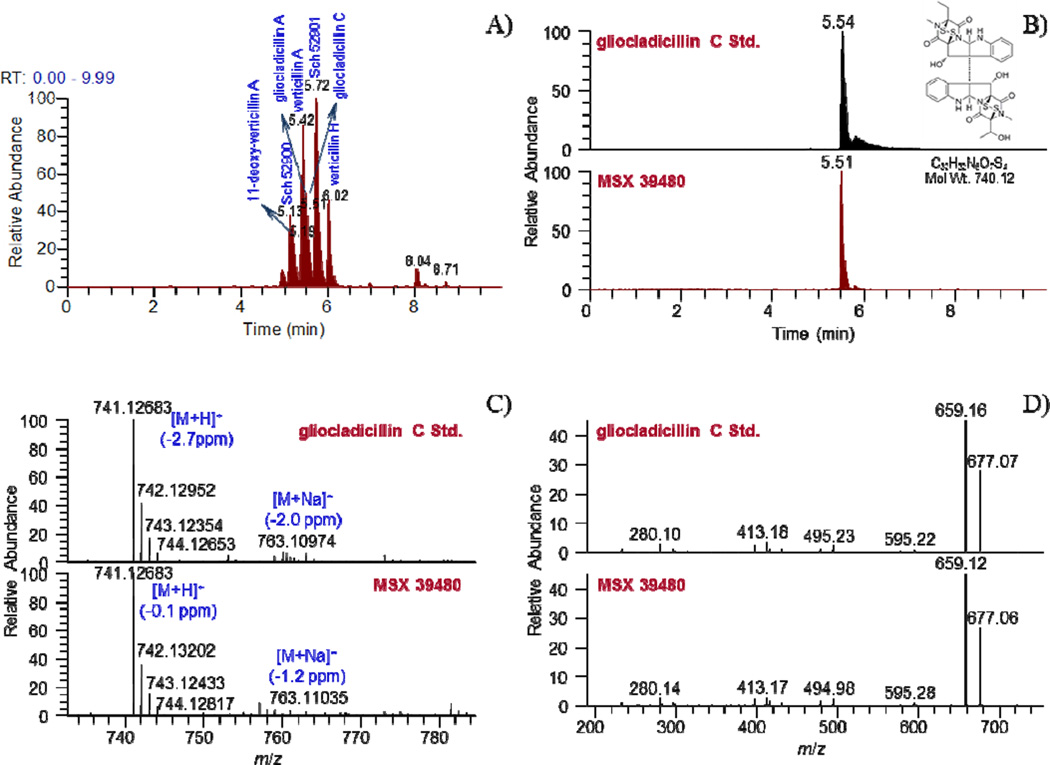
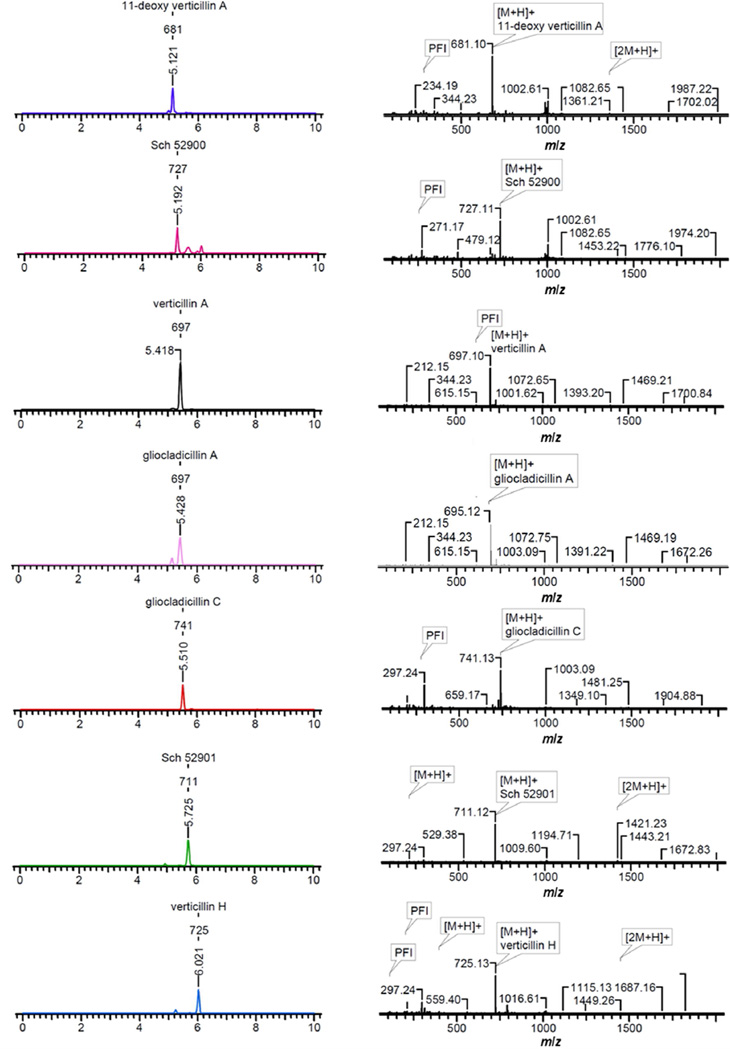
An extract of the filamentous fungus MSX 39480 displayed potent cytotoxic activity for the H460 cell line (94 and 88% inhibition of cell growth when tested at 20 and 2 µg/mL, respectively) and thus was subjected to the dereplication protocol. Seven epipolythiodioxopiperazine (ETP) alkaloids were dereplicated by matching retention times, HRMS, and MS/MS data, namely, 11'-deoxyverticillin (1), Sch 52900 (2), verticillin A (3), gliocladicillin A (4), gliocladicillin C (5), Sch 52901 (6), and verticillin H (7). To illustrate the value of the protocol, a compound eluting at 5.51 min with a m/z of 741.1285 was detected in the extract. The isotopic pattern of the molecular ion peak, in conjunction with intensity-ratio calculations, suggested the presence of four sulfur atoms. ACD/Intellixtract analysis of the UPLC-HRMS data identified this as compound 5, a dimeric ETP alkaloid, and the accurate mass and retention time data matched that of a standard of 5 (Figures 2 and 3). As a confirmation, excellent agreement between the MS/MS spectra of the standard and unknown was achieved (Figure 2). The same methodology was used to verify the identity of compounds 1–4, 6, and 7 (Figure 3). Dimeric ETP alkaloids are bioactive secondary metabolites reported to have potent cytotoxic38–47 and antibacterial activities,48–53 along with antiparasitic,54 nematicidal,55 antiviral,56 and immunosuppressive properties.57 Hence, this class of compounds has been well-studied, including previously in our laboratory,33 and, thus, their rapid dereplication allowed us to focus on other leads with a greater potential to yield new substances.
Figure 2.
A. (+)-ESI SIC of crude MSX 39480 extract (m/z: 681, 695, 697, 711, 725, 727, 741), B. Overlay chromatographic peaks of gliocladicillin C, C. (+)-HRESIMS of gliocladicillin C, and D. MS/MS CID fragmentation spectra of gliocladicillin C.
Figure 3.
ACD/Intellixtract analysis of the TIC of an extract of MSX 39480.
Dereplication of Enniatins
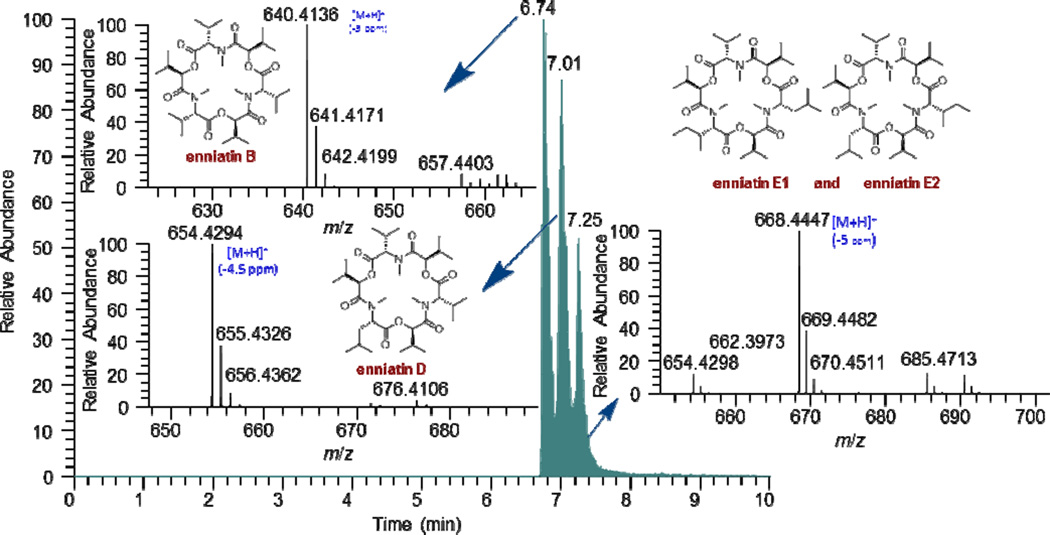
On the small scale, an extract of the fungus MSX 44407 showed potent cytotoxicity (96 and 71% inhibition when tested against H460 cells at 20 and 2 µg/mL, respectively). The TIC of the crude extract revealed the presence of three major compounds eluting at 6.74, 7.01, and 7.25 min (Figure 4). ACD/Intellixtract analysis of the UPLC-HRMS data matched the second and third compounds to the cyclodepsipepetides, enniatins D (9) and the unresolved isomers, enniatins E1 (10a) and E2 (10b), respectively; HRMS and MS/MS data confirmed these assignments (Figure 4). While the mass spectrometric data of the first eluting compound did not match any substance in the database, the HRMS data yielded a molecular formula of C33H57N3O9 (m/z 640.4138 [M+H]+, calcd for 640.4168). Searching the Dictionary of Natural Products6 using the molecular formula resulted in one lead, enniatin B (8). Although 8 was not in the database, given its structurally similarity to compounds 9, 10a, and 10b, the extract was assigned as not of further interest. Twenty-nine enniatins, which are cyclohexadepsipeptides, have been characterized in the literature, either as a single compounds or mixtures of unresolved isomers, and a recent review discusses their range of biological activities.58
Figure 4.
(+)-ESI SIC of the crude extract of MSX 44407 (m/z: 668, 654, 640).
Dereplication of Aflatoxins
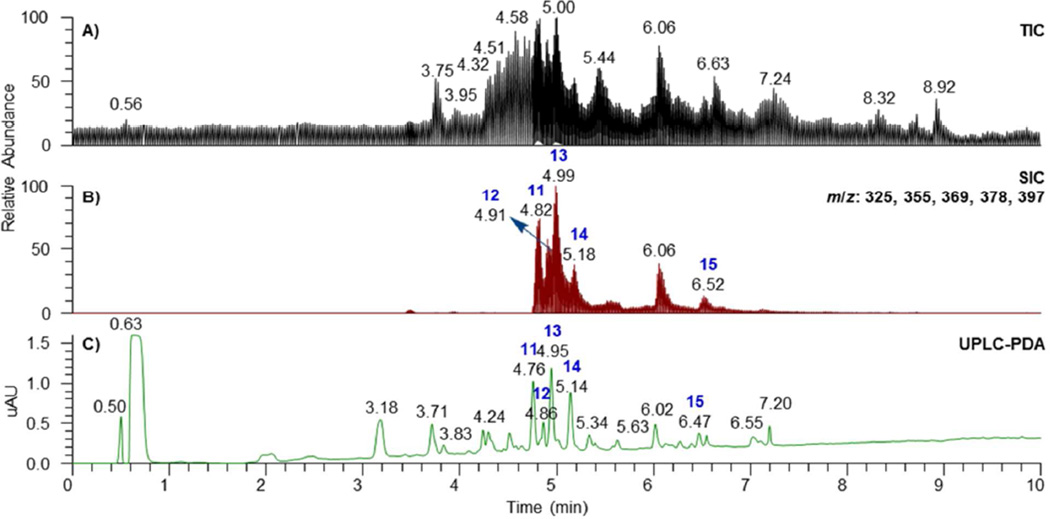
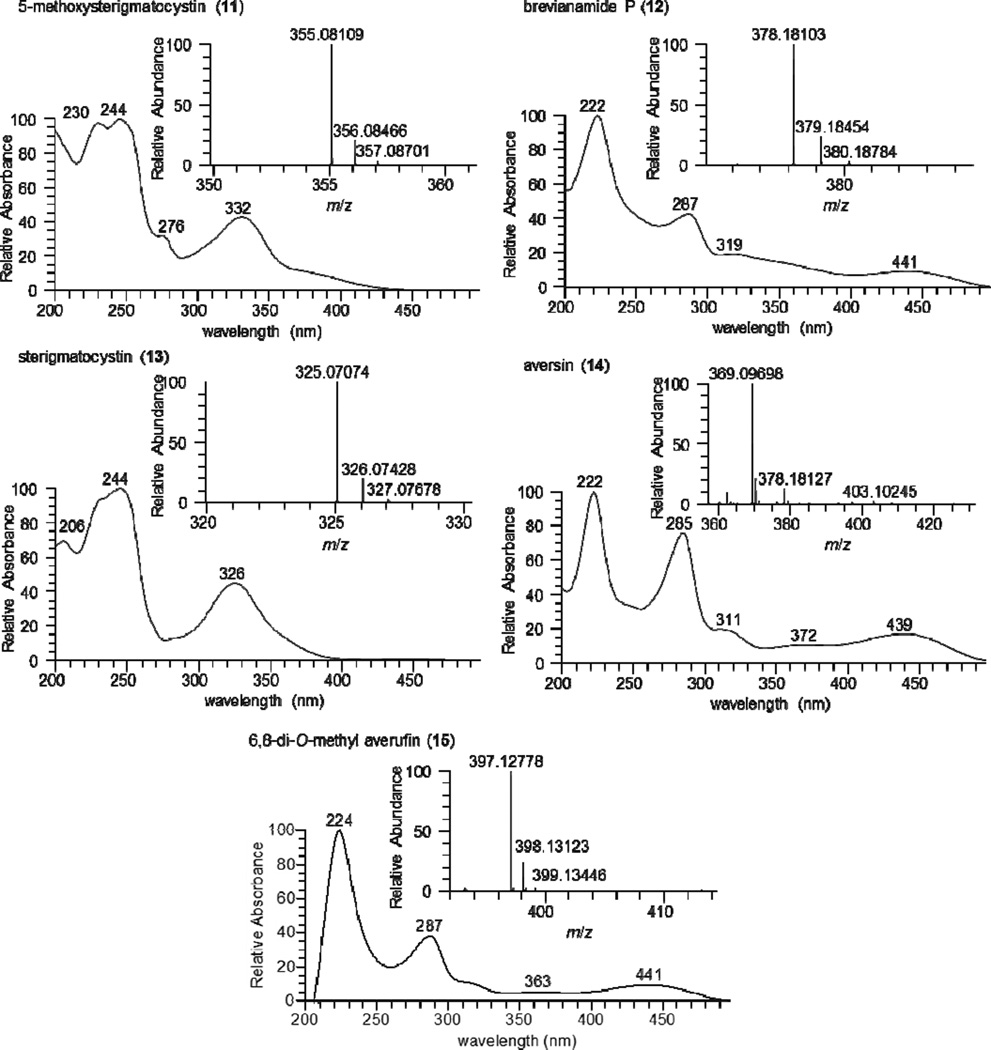
Another culture, MSX 40080, showed potent cytotoxic activity when tested against H460 cells (95 and 76% growth inhibition when evaluated at 20 and 2 µg/mL, respectively). The TIC and PDA data of the crude extract suggested a series of structurally related compounds with distinctive UV absorption patterns. ACD/Intellixtract analysis of the UPLC-HRMS data lead to the identification of five compounds, 5- methoxysterigmatocystin (11), brevianamide P (12), sterigmatocystin (13), aversin (14), and 6,8-di-O-methyl averufin (15). These assignments were confirmed by checking the HRMS and MS/MS spectra and UV absorption maxima with those in the database (Figures 5 and 6). Aflatoxins are regarded as nuisance mycotoxins with potent biological activity; hence their facile and rapid dereplication is important. These toxins have been investigated extensively due to their carcinogenic effects,59 particularly in contaminated grains.60
Figure 5.
A. (+)-ESI TIC, B. SIC (m/z: 325, 355, 369, 378, and 397 for peaks 13, 11, 14, 12, and 15, respectively), and C. UPLC-PDA of the crude extract of MSX 40080.
Figure 6.
(+)-HRESIMS and PDA spectra of compounds 11–15.
In conclusion, UPLC-PDA-HRMS-MS/MS methodology has been developed and implemented for the dereplication of cytotoxic secondary metabolites in crude extracts of fungal cultures. This methodology has several attributes that distinguish it from previous systems. First, the database, which includes full-scan high-resolution mass spectra and MS/MS spectra from both the positive- and negative-ionization modes coupled with UV-absorption maxima and retention times, is the first to be constructed based on cytotoxic fungal secondary metabolites. Furthermore, the use of UPLC enables a rapid (10 min) chromatographic method, the fastest utilized in a comprehensive natural product dereplication strategy. This in turn facilitates the rapid nature of the protocol, such that less than 25 min, including the time for sample preparation, is needed to acquire and interpret the data. Moreover, the use of HRMS and MS/MS data imparts a high degree of confidence in the structure of the dereplicated leads. Coupling these data with the use of the ACD/IntelliXtract, which extracts all chromatographic components in the LC-MS datasets and automatically assigns each as [M+H]+ or [M-H]−, expedites the identification process. In particular, the capability of the ACD/IntelliXtract to deconvolute overlapping and co-eluting components makes it possible to dereplicate trace compounds and enables the application of a short (10-min) chromatographic run time. Finally, the method was designed as a qualitative tool, and as such, the sensitivity for individual compounds was not measured a priori, especially since sensitivity is compound specific. However, the coupling of the resolving power of the UPLC with the enhanced sensitivity of mass spectrometry enabled the detection of trace amounts of compounds in crude extracts. In using this protocol for over two years on hundreds of samples, compounds present in as low as 0.22 mg/g of extract were detected readily.
A promising side application of the dereplication methodology developed could be the detection and identification of mycotoxins in food commodities and agricultural products. Mycotoxins have attracted world-wide attention due to their profound negative economic consequences. It has been estimated that one quarter of the world’s crops are contaminated with mycotoxins. Losses in the USA and Canada due to mycotoxins on the feed and livestock industries are of the order of $5 billion annually.61,62 The protocol described could be utilized in any laboratory having a mass spectrometer coupled with a UPLC, using the same analytical column, thereby enabling the identification of a suite of mycotoxins, even without reference standards.
EXPERIMENTAL SECTION
General Experimental Procedures
HRESIMS was performed on a Thermo LTQ Orbitrap XL mass spectrometer (ThermoFisher, San Jose, CA) equipped with an electrospray ionization source. Source conditions in the positive-ionization mode were set at 275 °C for the capillary temperature, 4.5 kV for the source voltage, 20 V for capillary voltage, and 95 V for the tube lens. For the negative-ionization mode, the source conditions for temperature were 3.5 kV for the source voltage, 42 V for capillary voltage, and 110 V for the tube lens. Nitrogen was utilized for the sheath gas and set to 25 and 20 arb for the positive and negative modes, respectively. For the negative-ionization mode, nitrogen was also used as an auxiliary gas and set at 10 arb. In both modes, two scan events were carried out, full-scan (100–2000) and ion-trap MS/MS of the most intense ion from the parent mass list utilizing CID with a normalized collision energy of 30. External instrument calibration was performed using an LTQ ESI positive-ion calibration solution consisting of caffeine (20 µg/mL), MRFA (1 µg/mL) and Ultramark 1621 (0.001%) in an aqueous solution of CH3CN (50%), MeOH (25%) and acetic acid (1%). For the ESI negativeion calibration, sodium dodecyl sulfate (2.9 µg/mL) and sodium taurocholate (5.4 µg/mL) were added to the LTQ ESI calibration solution instead of caffeine and MRFA. Thermo Scientific Xcalibur 2.1 software was used for instrument control and data analysis. UPLC was carried out on a Waters Acquity system [using a BEH C18 (2.1 × 50 mm, 1.7 µm) column (Waters Corp., Milford, MA, USA) equilibrated at 40 °C]. A mobile phase consisting of CH3CN-H2O (acidified with 0.1% formic acid) was used, starting with 15:85 then increasing linearly to 100% CH3CN within 8 min, holding for 1.5 min and then returning to the starting conditions within 0.5 min. An Acquity UPLC photodiode array detector was used to acquire PDA spectra, which were collected from 200–500 nm with 4 nm resolution.
Reference Standards
Secondary metabolites were isolated and characterized from the Mycosynthetix Inc. library of filamentous fungi, as described previously.19,30,32–34,58,63–66
Extraction Procedure
Small-scale cultures of fungi were grown on a solid, grain-based medium in 250 mL Erlermeyer flasks, as described in detail previously.30,33,34 To each flask were added 60 mL of 1:1 MeOH-CHCl3. The culture was chopped with a spatula and shaken overnight (~16 h; at ~100 rpm) at rt. Each sample was filtered using vacuum, and the remaining residues were washed with small volumes of 1:1 MeOH-CHCl3. To the filtrate, 90 mL CHCl3 and 150 mL H2O were added; the mixture was stirred for 2 h, and then transferred into a separatory funnel. The bottom layer was drawn off into a round-bottomed flask and evaporated to dryness. The dried organic extract was re-constituted in 100 mL of 1:1 MeOH-CH3CN and 100 mL of hexanes. The biphasic solution was stirred for 1 h and then transferred to a separatory funnel. The MeOH-CH3CN layer was drawn off and evaporated to dryness under vacuum. This defatted material was then submitted for cytotoxic activity evaluation using a set of three cancer cell lines, as described previously.35,36 Extracts displaying potent cytotoxic activities (i.e., less than 20% survival when tested at 20 µg/mL) were dereplicated. A sub-milligram aliquot of the bioactive extracts was used for dereplication analysis by dissolving the extract in equal volumes of MeOH and dioxane to obtain a final concentration of 2 mg/mL in a total volume of 150 µL.
Data Analysis
Data from 172 fungal secondary metabolites are in the Supporting Information (Table S1), which lists the metabolites, molecular formula, retention time, UV-absorption maxima, high-resolution full-scan mass spectra, and MS-MS data (top ten most intense peaks), utilizing both the positive- and negative-ionization modes. ACD MS Manager with add-in software IntelliXtract, Advanced Chemistry Development, Inc. (Toronto, Canada), was used for the primary analysis of the LC-MS data.
Cytotoxicity Assay
The cytotoxicity measurements against the MCF-767 human breast carcinoma (Barbara A. Karmanos Cancer Center), NCI-H46068 human large cell lung carcinoma (HTB-177, American Type Culture Collection (ATCC), and SF-26869 human astrocytoma (NCI Developmental Therapeutics Program) cell lines were performed as described previously.35,36
Supplementary Material
ACKNOWLEDGMENT
This research was supported by program project grant P01 CA125066 from the National Cancer Institute, National Institutes of Health, Bethesda, MD. Partial support was also provided by aCollaborative Funding Grant (2011-CFG-8008) from the North Carolina Biotechnology Center and the Kenan Institute for Engineering, Technology & Science. The authors thank Drs. Arlene Sy-Cordero and Sloan Ayers for isolating secondary metabolites from filamentous fungi. The high-resolution mass spectrometric data were acquired in the Triad Mass Spectrometry Laboratory at the University of North Carolina at Greensboro.
Footnotes
ASSOCIATED CONTENT
Supporting Information
(+)-ESI TIC and SIC of a mixture of ten standard compounds and their structures. A database of 172 fungal secondary metabolites including molecular formula, retention time, UV-absorption maxima, HRMS, MS/MS (top ten intense peaks), utilizing both positive and negative ionization modes. This information is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest
REFERENCES
- 1.Ghisalberti EL. In: Detection, Isolation and Structural Determination. Colegate SM, Molyneux RJ, editors. Boca Raton, FL: CRC Press Inc; 1993. pp. 9–57. [Google Scholar]
- 2.Kingston DG. In: The Practice of Medicinal Chemistry. Meyer P, editor. London: 1996. pp. 101–116. [Google Scholar]
- 3.Wani MC, Taylor HL, Wall ME, Coggon P, McPhail AT. J. Am. Chem. Soc. 1971;93:2325–2327. doi: 10.1021/ja00738a045. [DOI] [PubMed] [Google Scholar]
- 4.Wall ME, Wani MC, Cook CE, Palmer KH, McPhail AT, Sim GA. J. Am. Chem. Soc. 1966;88:3888–3890. [Google Scholar]
- 5.Oberlies NH, Kroll DJ. J. Nat. Prod. 2004;67:129–135. doi: 10.1021/np030498t. [DOI] [PubMed] [Google Scholar]
- 6.Dictionary of Natural Products Online 21.2. London: Taylor & Francis Group; 2013. [Google Scholar]
- 7.Blunt J, Munro M, Upjohn M. In: Fattorusso E, Gerwick WH, Taglialatela-Scafati O, editors. Dordrecht, The Netherlands: Springer; 2012. pp. 389–421. [Google Scholar]
- 8.Hostettmann K, Wolfender JL. Terreaux, C. Pharm. Biol. 2001;39(Suppl. 1):18–32. doi: 10.1076/phbi.39.s1.18.0008. [DOI] [PubMed] [Google Scholar]
- 9.Wolfender JL, Ndjoko K, Hostettmann K. J. Chromatogr. A. 2003;1000:437–455. doi: 10.1016/s0021-9673(03)00303-0. [DOI] [PubMed] [Google Scholar]
- 10.Wolfender J-L, Queiroz EF, Hostettmann K. Exp. Opin. Drug Discov. 2006;1:237–260. doi: 10.1517/17460441.1.3.237. [DOI] [PubMed] [Google Scholar]
- 11.Alali FQ, Gharaibeh A, Ghawanmeh A, Tawaha K, Oberlies NH. Phytochem. Anal. 2008;19:385–394. doi: 10.1002/pca.1060. [DOI] [PubMed] [Google Scholar]
- 12.LeVen J, Schmitz-Afonso I, Lewin G, Laprévote O, Brunelle A, Touboul D, Champy P. J. Mass Spectrom. 2012;47:1500–1509. doi: 10.1002/jms.3092. [DOI] [PubMed] [Google Scholar]
- 13.Funari CS, Eugster PJ, Martel S, Carrupt P-A, Wolfender J-L, Silva DHS. J. Chromatogr. A. 2012;1259:167–178. doi: 10.1016/j.chroma.2012.03.069. [DOI] [PubMed] [Google Scholar]
- 14.Musharraf SG, Goher M, Shahnaz S, Choudhary MI, Atta-ur-Rahman Rapid Commun. Mass Spectrom. 2013;27:169–178. doi: 10.1002/rcm.6441. [DOI] [PubMed] [Google Scholar]
- 15.Qiu F, Imai A, McAlpine JB, Lankin DC, Burton I, Karakach T, Farnsworth NR, Chen SN, Pauli GF. J. Nat. Prod. 2012;75:432–443. doi: 10.1021/np200878s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lang G, Mayhudin NA, Mitova MI, Sun L, vanderSar S, Blunt JW, Cole ALJ, Ellis G, Laatsch H, Munro MHG. J. Nat. Prod. 2008;71:1595–1599. doi: 10.1021/np8002222. [DOI] [PubMed] [Google Scholar]
- 17.Mitova MI, Murphy AC, Lang G, Blunt JW, Cole ALJ, Ellis G, Munro MHG. J. Nat. Prod. 2008;71:1600–1603. doi: 10.1021/np800221b. [DOI] [PubMed] [Google Scholar]
- 18.Orjala J, Oberlies NH, Pearce CJ, Swanson SM, Kinghorn AD. In: Bioactive Compounds from Natural Sources. Natural Products as Lead Compounds in Drug Discovery. 2nd. Tringali C, editor. London, UK: Taylor & Francis; 2012. pp. 37–63. [Google Scholar]
- 19.El-Elimat T, Zhang X, Jarjoura D, Moy FJ, Orjala J, Kinghorn AD, Pearce CJ, Oberlies NH. ACS Med. Chem. Lett. 2012;3:645–649. doi: 10.1021/ml300105s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackwell M. Am. J. Bot. 2011;98:426–438. doi: 10.3732/ajb.1000298. [DOI] [PubMed] [Google Scholar]
- 21.Hawksworth DL. Mycol. Res. 1991;95:641–655. [Google Scholar]
- 22.Gloer JB. In: Environmental and Microbial Relationships. Kubicek CP, Druzhinina IS, editors. Vol. 4. Berlin-Heidelberg: Springer; 2007. pp. 257–283. [Google Scholar]
- 23.Murphy PA, Hendrich S, Landgren C, Bryant CM. J. Food Sci. 2006;71:R51–R65. [Google Scholar]
- 24.Kabak B, Dobson AD, Var I. Crit. Rev. Food Sci. Nutr. 2006;46:593–619. doi: 10.1080/10408390500436185. [DOI] [PubMed] [Google Scholar]
- 25.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. Fungal Barcoding Consortium. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen KF, Smedsgaard J. J. Chromatogr. A. 2003;1002:111–136. doi: 10.1016/s0021-9673(03)00490-4. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen KF, Månsson M, Rank C, Frisvad JC, Larsen TO. J. Nat. Prod. 2011;74:2338–2348. doi: 10.1021/np200254t. [DOI] [PubMed] [Google Scholar]
- 28.Fredenhagen A, Derrien C, Gassmann E. J. Nat. Prod. 2005;68:385–391. doi: 10.1021/np049657e. [DOI] [PubMed] [Google Scholar]
- 29.Larsen TO, Smedsgaard J, Nielsen KF, Hansen ME, Frisvad JC. Nat. Prod. Rep. 2005;22:672–695. doi: 10.1039/b404943h. [DOI] [PubMed] [Google Scholar]
- 30.Ayers S, Graf TN, Adcock AF, Kroll DJ, Matthew S, Carcache de Blanco EJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:1126–1131. doi: 10.1021/np200062x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sy-Cordero AA, Graf TN, Wani MC, Kroll DJ, Pearce CJ, Oberlies NH. J. Antibiot. 2010;63:539–544. doi: 10.1038/ja.2010.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sy-Cordero AA, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2011;74:2137–2142. doi: 10.1021/np2004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Figueroa M, Graf TN, Ayers S, Adcock AF, Kroll DJ, Yang J, Swanson SM, Munoz-Acuna U, Carcache de Blanco EJ, Agrawal R, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. J. Antibiot. 2012;65:559–564. doi: 10.1038/ja.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Elimat T, Figueroa M, Raja HA, Graf TN, Adcock AF, Kroll DJ, Day CS, Wani MC, Pearce CJ, Oberlies NH. J. Nat. Prod. 2013;76:382–387. doi: 10.1021/np300749w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alali FQ, El-Elimat T, Li C, Qandil A, Alkofahi A, Tawaha K, Burgess JP, Nakanishi Y, Kroll DJ, Navarro HA, Falkinham JO, 3rd, Wani MC, Oberlies NH. J. Nat. Prod. 2005;68:173–178. doi: 10.1021/np0496587. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Lee D, Graf TN, Phifer SS, Nakanishi Y, Riswan S, Setyowati FM, Saribi AM, Soejarto DD, Farnsworth NR, Falkinham JO, 3rd, Kroll DJ, Kinghorn AD, Wani MC, Oberlies NH. J. Nat. Prod. 2009;72:1949–1953. doi: 10.1021/np900572g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balogh MP. LCGC North America. 2006;24:762–769. [Google Scholar]
- 38.Chen Y, Guo H, Du Z, Liu XZ, Che Y, Ye X. Cell Proliferation. 2009;42:838–847. doi: 10.1111/j.1365-2184.2009.00636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Blood. 2007;109:2579–2588. doi: 10.1182/blood-2006-07-027326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang YX, Chen Y, Guo XN, Zhang XW, Zhao WM, Zhong L, Zhou J, Xi Y, Lin LP, Ding J. Anti-Cancer Drugs. 2005;16:515–524. doi: 10.1097/00001813-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi C, Numata A, Matsumura E, Minoura K, Eto H, Shingu T, Ito T, Hasegawa T. J. Antibiot. 1994;47:1242–1249. doi: 10.7164/antibiotics.47.1242. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi C, Numata A, Ito Y, Matsumura E, Araki H, Iwaki H, Kushida K. J. Chem. Soc., Perkin Trans. 1. 1994:1859–1864. [Google Scholar]
- 43.Takahashi C, Minoura K, Yamada T, Numata A, Kushida K, Shingu T, Hagishita S, Nakai H, Sato T, Harada H. Tetrahedron. 1995;51:3483–3498. [Google Scholar]
- 44.Chen Y, Zhang Y-X, Li M-H, Zhao W-M, Shi Y-H, Miao Z-H, Zhang X-W, Lin L-P, Ding J. Biochem. Biophys. Res. Commun. 2005;329:1334–1342. doi: 10.1016/j.bbrc.2005.02.115. [DOI] [PubMed] [Google Scholar]
- 45.Saito T, Suzuki Y, Koyama S, Natori S, Iitaka Y, Kinosita T. Chem. Pharm. Bull. 1998;36:1942–1956. [Google Scholar]
- 46.Son BW, Jensen PR, Kauffman CA, Fenical W. Nat. Prod. Lett. 1999;13:213–222. [Google Scholar]
- 47.Feng Y, Blunt JW, Cole AL, Munro MHG. J. Nat. Prod. 2004;67:2090–2092. doi: 10.1021/np030326w. [DOI] [PubMed] [Google Scholar]
- 48.Zheng CJ, Park SH, Koshino H, Kim YH, Kim WG. J. Antibiot. 2007;60:61–64. doi: 10.1038/ja.2007.8. [DOI] [PubMed] [Google Scholar]
- 49.Zheng CJ, Kim CJ, Bae KS, Kim YH, Kim WG. J. Nat. Prod. 2006;69:1816–1819. doi: 10.1021/np060348t. [DOI] [PubMed] [Google Scholar]
- 50.Bertinetti BV, Rodriguez MA, Godeas AM, Cabrera GM. J. Antibiot. 2010;63:681–683. doi: 10.1038/ja.2010.103. [DOI] [PubMed] [Google Scholar]
- 51.Argoudelis AD. J. Antibiot. 1972;25:171–178. doi: 10.7164/antibiotics.25.171. [DOI] [PubMed] [Google Scholar]
- 52.Joshi BK, Gloer JB, Wicklow DT. J. Nat. Prod. 1999;62:730–733. doi: 10.1021/np980530x. [DOI] [PubMed] [Google Scholar]
- 53.Minato H, Matsumoto M, Katayama T. J. Chem. Soc. D. 1971:44–45. [Google Scholar]
- 54.Watts KR, Ratnam J, Ang KH, Tenney K, Compton JE, McKerrow J, Crews P. Bioorg. Med. Chem. 2010;18:2566–2574. doi: 10.1016/j.bmc.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong JY, He HP, Shen YM, Zhang KQ. J. Nat. Prod. 2005;68:1510–1513. doi: 10.1021/np0502241. [DOI] [PubMed] [Google Scholar]
- 56.Ovenden SP, Sberna G, Tait RM, Wildman HG, Patel R, Li B, Steffy K, Nguyen N, Meurer-Grimes BM. J. Nat. Prod. 2004;67:2093–2095. doi: 10.1021/np0497494. [DOI] [PubMed] [Google Scholar]
- 57.Erkel G, Gehrt A, Anke T, Sterner O. Z. Naturforsch., C. 2002;57:759–767. doi: 10.1515/znc-2002-7-834. [DOI] [PubMed] [Google Scholar]
- 58.Sy-Cordero AA, Pearce CJ, Oberlies NH. J. Antibiot. 2012;65:541–549. doi: 10.1038/ja.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heathcote JG, Hibbert JR. Aflatoxins: Chemical and Biological Aspects. Amsterdam: Elsevier Scientific Publishing. Co; 1978. [Google Scholar]
- 60.Rodrigues I, Naehrer K. Toxins (Basel) 2012;4:663–675. doi: 10.3390/toxins4090663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett JW, Klich M. Clin. Microbiol. Rev. 2003;16:497–516. doi: 10.1128/CMR.16.3.497-516.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anonymous. Manual on the Application of the HACCP System in Mycotoxin Prevention and Control. Rome: Food and Agriculture Organization of the United Nations; 2001. [Google Scholar]
- 63.Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Wani MC, Pearce CJ, Oberlies NH. Tetrahedron Lett. 2011;52:5733–5735. doi: 10.1016/j.tetlet.2011.08.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayers S, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Matthew S, Carcache de Blanco EJ, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. J. Antibiot. 2012;65:3–8. doi: 10.1038/ja.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ayers S, Graf TN, Adcock AF, Kroll DJ, Shen Q, Swanson SM, Wani MC, Darveaux BA, Pearce CJ, Oberlies NH. Tetrahedron Lett. 2011;52:5128–5230. doi: 10.1016/j.tetlet.2011.07.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ayers S, Ehrmann BM, Adcock AF, Kroll DJ, Carcache de Blanco EJ, Shen Q, Swanson SM, Falkinham JO, Wani MC, Mitchell SM, Pearce CJ, Oberlies NH. J. Pept. Sci. 2012;18:500–510. doi: 10.1002/psc.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soule HD, Vazguez J, Long A, Albert S, Brennan M. J. Natl. Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 68.Carney DN, Gazdar AF, Bunn PA, Jr, Guccion JG. Stem Cells. 1982;1:149–164. [PubMed] [Google Scholar]
- 69.Rosenblum ML, Gerosa MA, Wilson CB, Barger GR, Pertuiset BF, de Tribolet N, Dougherty DV. J. Neurosurg. 1983;58:170–176. doi: 10.3171/jns.1983.58.2.0170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.