Abstract
Phenanthrene derivatives acting as potent PARP1 inhibitors prevented the bi-focal clustering of supernumerary centrosomes in multi-centrosomal human cancer cells in mitosis. The phenanthridine PJ-34 was the most potent molecule. Declustering of extra-centrosomes causes mitotic failure and cell death in multi-centrosomal cells. Most solid human cancers have high occurrence of extra-centrosomes. The activity of PJ-34 was documented in real-time by confocal imaging of live human breast cancer MDA-MB-231 cells transfected with vectors encoding for fluorescent γ-tubulin, which is highly abundant in the centrosomes and for fluorescent histone H2b present in the chromosomes. Aberrant chromosomes arrangements and de-clustered γ-tubulin foci representing declustered centrosomes were detected in the transfected MDA-MB-231 cells after treatment with PJ-34. Un-clustered extra-centrosomes in the two spindle poles preceded their cell death. These results linked for the first time the recently detected exclusive cytotoxic activity of PJ-34 in human cancer cells with extra-centrosomes de-clustering in mitosis, and mitotic failure leading to cell death. According to previous findings observed by confocal imaging of fixed cells, PJ-34 exclusively eradicated cancer cells with multi-centrosomes without impairing normal cells undergoing mitosis with two centrosomes and bi-focal spindles. This cytotoxic activity of PJ-34 was not shared by other potent PARP1 inhibitors, and was observed in PARP1 deficient MEF harboring extracentrosomes, suggesting its independency of PARP1 inhibition. Live confocal imaging offered a useful tool for identifying new molecules eradicating cells during mitosis.
Keywords: Cancer Biology, Issue 78, Medicine, Cellular Biology, Molecular Biology, Biomedical Engineering, Anatomy, Physiology, Genetics, Neoplastic Processes, Pharmacologic Actions, Live confocal imaging, Extra-centrosomes clustering/de-clustering, Mitotic Catastrophe cell death, PJ-34, myocardial infarction, microscopy, imaging
Introduction
Phenanthrene derived PARP1 inhibitors, including PJ-34, were designed to protect quiescent cells from apoptotic cell death induced by the energy consuming PARP1 mediated DNA-repair under stress conditions (stroke or myocardial infarction)1. However, recently we discovered that PJ-34, at twice higher concentration than that inducing PARP1 inhibition, can exclusively cause cell death in human cancer cells2,3. The more rapid the proliferation of the cell was, the more efficient the eradication of the cells was. The cytotoxic activity of PJ-34 was attributed to extra-centrosomes de-clustering in mitosis2. Many human cancer cells harbor multicentrosomes4,5. Incubation of human breast cancer cells MDA-MB-231, which harbor supernumerary centrosomes, with 20 μM PJ-34 efficiently eradicated these cells within 72-96 hr without impairing quiescent cells or some benign proliferating cells harboring two centrosomes in mitosis2,3. Benign cells included human mammary epithelial cells MCF-10, Human endothelial cells (huvec) and primary mesenchymal cells prepared from human thymus. These cells were resistant to the cytotoxic activity of PJ-34. PJ-34 did not interfere with their cell cycle during 96 hr incubation or affect their centrosomes and bi-focal spindle formation2,3.
Bipolar centrosome assembly is crucial for bipolar spindle formation in mitosis4,5. Therefore, cells with more than two centrosomes have developed a scarcely understood molecular mechanism, clustering their extra centrosomes at two poles 4-9. Failure of bipolar assembly of their centrosomes may cause multipolar distorted spindles and aberrant chromosomes segregation that arrests the cell-cycle in G2/M arrest, and leads to cell death attributed to mitotic failure4,5. The molecular mechanisms underlying extra-centrosomes de-clustering are intensively investigated10. Understanding this death mechanism will enable exclusive eradication of cancer cells while sparing healthy tissues 5,10.
Thus, compounds that activate mitotic catastrophe cell death offer a new mode of a selective cancer therapy, which may be efficient in a wide range of human solid cancers.Our results suggest that confocal imaging can be used to identify molecules affecting extra-centrosomes clustering in mitosis2,3, rendering these compounds cancer targeting drug candidates.
We have documented the cytotoxic activity of the phenanthridine PJ-34 by scanning fixed and live human cancer cells (with high occurrence of extra-centrosomes in mitosis) versus normal cells. A step-by step description of the imaging procedures used to identify the cytotoxic activity of PJ-34 in human cancer cells is included below.
Protocol
1. Cell Culture Preparation
MDA-MB-231 cells were purchased from ATCC (American Type Culture Collection) and stored in liquid nitrogen.
Seed 106 MDA-MB-231 cells in 92 mm diameter Petri dish in 10 ml complete medium containing Dulbeco Modified Eagle Medium (DMEM), 10% horse serum, 1% L-glutamine, and 1% Penstrep-Amphotericine B. Allow cells to proliferate to about 80-100% confluence.
Remove culture medium from dish and discard.
Wash the cell layer briefly with 0.25% (w/v) Trypsin-EDTA solution to remove all traces of serum.
Add 2.0 ml of Trypsin-EDTA solution to dish and observe cells by inverted microscope until cell layer is dispersed (usually within 5 to 15 min).
Add 18 ml complete growth medium and gently aspirate the cells by pipette. Transfer to a tube.
Centrifuge the cell suspension at 1,200 rpm.
Re-suspend the cell pellet in 24 ml of culture medium.
Add 2 ml of cell suspension to 35 mm glass bottom dishes (about 25% confluence) and place in incubator (5% CO2 , 37 °C).
- Solutions:
- Complete Medium for cells proliferation: DMEM with 10%FBS, 1% Antibiotics (100 units/ml penicillin G, 100 μg/ml streptomycin, Pen-Strep-Ampho solution) and 2 mM L-glutamine.
- Trypsin-EDTA solution, containing 0.25%Trypsin-EDTA.
Dishes: 92 mm diameter Petri dishes. 35 mm diameter poly-D-lysine coated glass bottom culture dishes.
2. Preparation of Cells for Live Confocal Imaging
Seed 2 x 105 MDA-MB-231 cells in glass bottom culture dishes in 2 ml complete medium as mentioned in section 1. When cell culture reaches confluence of 60-70% (about 3-4 x 105 cells per dish), proceed with transfection.
Transfect the cells with two plasmids encoding the fusion proteins γ-Tubulin-GFP (for fluorescent detection of centrosomes) and Histone-RED (H2b-RED, for fluorescent detection of chromosomes) using the liposomal transfection reagent Jet-PI, following the manufacture protocol. Briefly, mix 2 μg from each plasmid in a tube with 100 μl NaCl (150 mM). Mix the transfection reagent (100 μl) with 100 μl NaCl (150 mM) in a second tube, and incubate 5 min at room temperature (RT). Then combine the two solutions, mix (using mild vortex) and spin-down. Incubate for 30 min at RT.
During the incubation of the transfection mixture, wash the cells once with PBS and replace the cell medium with 2 ml of warm DMEM with no supplements (37 °C).
Gently add the transfection mixture to the cells in DMEM and then return the cells to the incubator (37 °C, 5% CO2) for 8 hr.
After 8 hr of incubation, replace the DMEM with 2 ml complete medium and incubate the cells in the incubator for 24 hr.
24 hr after transfection, replace the medium of the cells with 2 ml complete medium containing 20 μM PJ-34.
Incubate the cells for additional 18 hr (37 °C, 5% CO2).
Subject the cells to live confocal imaging for at least 16 hr in imaging chamber keeping the cells at 5% CO2 and 37 °C.
- In parallel, examine transfection efficacy 36 hr post transfection using fluorescent microscopy as follow:
- Seed 2 x 105 MDA-MB-231 cells in 6-well plate containing 1 coverslip per well in 2 ml complete medium.
- Transfect the cells as mentioned in sections 2.2-2.5.
- 36 hr post transfection fix the transfected cells mounted on a coverslip by incubation in cold methanol:acetone (1:1) solution, 7 min, -20 °C.
- Aspirate the fixation solution and let the coverslip with the mounted cells to dry in a chemical hood.
- Apply ProLong Gold antifade reagent with DAPI and let the coverslip to dry in the dark for 6 hr.
- Examine the slide under fluorescent microscope and calculate the percentage of the transfected cells (red and green signals) from the total population of cells (DNA staining by DAPI). The desired transfection percentage is about 20-40% when 100-200 cells are counted.
3. Technical Parameters of the Live Confocal Imaging Scanner Settings
ScanMode XYZT; Pinhole [airy] 1.00; Zoom 3.5; Resolution 8 bits; Laser DPSS 561 nm; Argon, visible laser 488 nm; Laser He/Ne visible 633 nm; Objective HCX PL APO CS 63X 1.40 OIL UV; Numerical aperture 1.4; Scan speed 700 Hz; Refraction index 1.52.
Image 3-D presentation were prepared by IMARIS imaging software 7.0.
4. Confocal Imaging of Mitosis in Fixed Cells
Seed 2 x 105 MDA-MB-231 breast cancer cells (ATCC), normal mouse embryonic fibroblasts (MEF), or PARP1 deficient MEF (PARP-/-, prepared by Dr Francoise Dantzer) on glass coverslips in 6-well plate in 2 ml complete medium. Coverslips were washed with 96% ethanol, following by wash with sterile DD water, dried for 2 hr, and placed in each well of 6-well dish.
Add PJ-34 (10-30 μM) to the medium and incubate the cells for the required period (usually up to 96 hr).
Wash the coverslips once with PBS (phosphate buffered saline), and fix the cells using incubation in ice-cold methanol:acetone (1:1) solution, 7 min, -20 °C.
Aspirate the fixation solution and let the coverslips to dry in chemical hood (at this stage, the coverslips can be kept in -20 °C for several weeks).
Wash the coverslips once with PBST (PBS supplemented with 0.1% Tween-20) to permeabilize the cell membranes and block the cells with 10% NDS (Normal Donkey Serum) in PBST ('blocking solution') for 1 hr at RT.
Incubate the permeabilized fixed cells with primary antibodies for 2 hr at RT (for spindles and centrosomes staining). The antibodies are diluted in the blocking solution as follow: anti-α tubulin (1:250 dilution) and anti-γ tubulin (1:200 dilution). Primary antibodies are applied as follows: apply 100 μl (in a drop) of a mixture of the antibodies in blocking solution for each coverslip on 6-well plate cover (the cover is upside down). Gently put the coverslip on the antibodies drop, seeded cells facing the drop. Incubate the coverslips facing the antibodies for 2 hr at room temperature.
Place the coverslips back in the wells and wash the cells 3 times with PBST. Then use the same procedure described in 4.6 for labeling cells on the coverslips with the fluorescent secondary antibodies. Incubate the cells on the coverslips with the secondary antibodies for 1 hr, RT, in the dark. The antibodies are diluted in the blocking solution as follow: Alexa Fluor 488 (1:1,000 dilution; green) and Alexa Fluor 568 (1:1,000 dilution; red).
Mount the coverslips using ProLong Gold antifade reagent with DAPI (for chromosomes staining) and incubate overnight at RT in dark to dry.
Examine the cover slips by confocal microscopy.
5. Cell Viability Measured by ATP Production
ATP production is measured by a luminescent ATP detection assay kit.
Seed the cells in 96-well plate, approximately 20,000 cells in 800 μl medium in each well. Three blank wells should be used for determination of the background luminescence of the medium.
Prepare ATP standard dilution series from approximately 10 μM to 100 μM and keep on ice.
Add 50 μl of detergent to each well and shake the plate for 5 min in orbital shaker, 700 rpm.
Reconstitute each vial of the' lyophilized substrate' with 5 ml of 'substrate buffer' in the kit.
Add 50 μl of the reconstituted substrate solution to the wells, and shake the plate for 5 min on orbital shaker, 700 rpm.
Keep the plate in dark for 10 min.
Measure luminiscence of each well by ELISA microplate reader.
Representative Results
PJ-34 is a stable water soluble phenanthridine1 (Figure 1). Our previous results revealed cell death and de-clustered extra-centrosomes in several types of fixed multi-centrosomal cancer cells that were treated with PJ-34. In contrast, normal proliferating cells were not impaired2,3. Centrosomes were identified by double labeling with antibodies directed against centrine1 and γ-tubulin in the fixed extra-centrosomal cells2.
Here, the cytotoxic activity of PJ-34 was documented in these live extra-centrosomal cells at real time by using live confocal microscopy. Live human breast cancer MDA-MB-231 cells, which have a high occurrence (>50%) of extra-centrosomes4,5, were scanned for at least 16 hr by confocal imaging focused on transfected cells with γ-tubulin-GFP (fluorescent labeling of γ-tubulin foci2) and with histone H2b-RED (fluorescent labeling of chromosomes). Six to ten live transfected cells were scanned in parallel in each experiment. Double immunolabeling of the γ-tubulin foci in the transfected cells with centrin1 was technically impossible.
Dispersed γ-tubulin foci and aberrant chromosomes arrangement were rarely detected in randomly selected untreated MDA-MB-231 cells in mitosis. Bifocal clustering of γ-tubulin foci, representing extra-centrosomes bifocal clustering, was documented in the majority of live untreated MDA-MB-231 cells (Figure 2), In contrast, un-clustered centrosomes and aberrant arrangement of chromosomes were detected in live transfected MDA-MB-231 cells incubated with PJ-34 (20 μM), and mitosis in these cells ended by cell death (Figure 3). These cells were incubated with PJ-34 for 18 - 24 hr before scanning and for additional 16 hr during scanning (Figure 3). The real-time documentation of cell death during mitosis strongly supports a previously defined positive correlation between the number of human malignant cells with multi-polar spindles in mitosis and the percentage of cell death in cells incubated with PJ-34 (20 μM)2.
PJ-34 acts as a potent PARP1 inhibitor1. We therefore examined the possibility of PARP1 inhibition causing cell death associated with mitotic failure. Unlike live imaging, imaging of fixed MDA-MB-231 cells enabled examination of a large population of cells in the cell cultures, thereby enabling statistical analysis of the effects of PARP1 inhibitors in a variety of human cancer cell lines. The activity of PJ-34 was compared to the activity of other potent, non-phenanthrene PARP1 inhibitors in normal or PARP1 deficient cells (i.e. normal and PARP1(-/-) mouse embryonic fibroblasts (MEF)) (Figure 4). PARP1 deficient MEF harbor multi-centrosomes in mitosis, but they are not tumor cells11. These cells were prepared by Dr. Francoise Dantzer, Strasbourg, France.
Fixed normal and PARP1(-/-) MEF were immunolabeled for α- and γ-tubulin that labeled their spindles and centrosomes, respectively, as reported before2. Some of the examined cell cultures were treated with PJ-34 or other potent, non-phenanthrene PARP1 inhibitors, including ABT-888 and AG01469, which inhibit the enzymatic activity of PARP1, and BSI-201, a compound that apparently attenuates PARP1 binding to nicked DNA12-14. None of the tested PARP1 inhibitors impaired normal MEF at concentrations inhibiting PARP1 activity (Figure 4). In contrast, PJ-34 dose-dependently caused un-clustering of γ-tubulin foci, distortion of spindles and cell death in PARP1(-/-) MEF (Figures 4A and B). This was not observed in normal MEF treated with PJ-34 (Figure 4B) or in PARP1(-/-) MEF treated with non-phenenthrene PARP1 inhibitors ABT-888 or AG014699 (Figure 4C). It should be noted that PJ-34 at concentrations exceeding 20 μM did impair normal MEF, although normal MEF were more resistant to PJ-34 activity than PARP1(-/-) MEF.
The fact that PJ-34 eradicated PARP1(-/-) MEF despite their PARP1 deficiency, and the correlation between the formation of multi-focal spindles and cell eradication in PARP1(-/-) MEF incubated with PJ-34 at concentrations higher than those required for PARP1 inhibition, were not consistent with a causal linkage between extra-centrosomes de-clustering in PARP1(-/-) MEF and PARP1 inhibition (Figure 4A). The cytotoxic activity of PJ-34 in PARP1(-/-) MEF could be better explained by its activity as an extra-centrosomes de-clustering agent in multi-centrosomal cells2 (Figure 3). Thus, the combination of live confocal imaging and immunocytochemistry methods was useful for identifying cytotoxic mechanisms impairing mitosis.
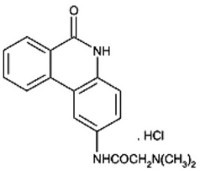 Figure 1. The phenanthridine PJ-34: N-(6-oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethyl-acetamide.
Figure 1. The phenanthridine PJ-34: N-(6-oxo-5,6-dihydro-phenanthridin-2-yl)-N,N-dimethyl-acetamide.
 Figure 2. Bi-focal clustering of extra-centrosomes in a randomly selected live MDA-MB-231 cell in mitosis.A. Upper panel: Labeled centrosomes in a randomly selected live MDA-MB-231 cell transfected with γ-tubulin-GFP. Lower panel: Chromosome re-arrangements during mitosis in a randomly selected MDA-MB-231 cell transfected with histone H2b-RED. B. Bi-focal mitosis with clustered extra-centrosomes identified in a randomly selected cultured MDA-MB-231 cell. Cells were transfected by both γ-tubulin-GFP (labeling γ-tubulin foci; green) and histone H2b-RED (labeling chromosomes; red). 48 hr after transfection, cells were exposed to a live confocal imaging for 16 hr. Six cells were scanned in parallel in each experiment. Four different experiments were performed. See also Supplementary Information. Click here to view larger figure.
Figure 2. Bi-focal clustering of extra-centrosomes in a randomly selected live MDA-MB-231 cell in mitosis.A. Upper panel: Labeled centrosomes in a randomly selected live MDA-MB-231 cell transfected with γ-tubulin-GFP. Lower panel: Chromosome re-arrangements during mitosis in a randomly selected MDA-MB-231 cell transfected with histone H2b-RED. B. Bi-focal mitosis with clustered extra-centrosomes identified in a randomly selected cultured MDA-MB-231 cell. Cells were transfected by both γ-tubulin-GFP (labeling γ-tubulin foci; green) and histone H2b-RED (labeling chromosomes; red). 48 hr after transfection, cells were exposed to a live confocal imaging for 16 hr. Six cells were scanned in parallel in each experiment. Four different experiments were performed. See also Supplementary Information. Click here to view larger figure.
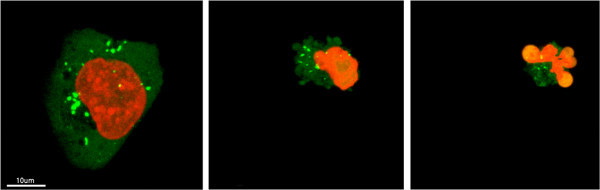 Figure 3. Extra-centrosomes de-clustering preceded cell death in live MDA-MB-231 cells treated with PJ-34. A randomly selected live MDA-MB-231 cell in mitosis with scattered centrosomes (1st frame on left) ended by cell death (2nd and 3rd frames). This cell was randomly selected in a cell culture incubated for 24 hr with PJ-34 (20 μM) applied 24 hr after transfection with vectors expressing γ-tubulin-GFP (labeling γ-tubulin foci including centrosomes; green) and histone H2b-RED (labeling chromosomes; red). The cell was scanned for 16 hr by live confocal imaging. Six cells were scanned in parallel in each experiment. Three different experiments were performed. See also Supplementary Information. Click here to view larger figure.
Figure 3. Extra-centrosomes de-clustering preceded cell death in live MDA-MB-231 cells treated with PJ-34. A randomly selected live MDA-MB-231 cell in mitosis with scattered centrosomes (1st frame on left) ended by cell death (2nd and 3rd frames). This cell was randomly selected in a cell culture incubated for 24 hr with PJ-34 (20 μM) applied 24 hr after transfection with vectors expressing γ-tubulin-GFP (labeling γ-tubulin foci including centrosomes; green) and histone H2b-RED (labeling chromosomes; red). The cell was scanned for 16 hr by live confocal imaging. Six cells were scanned in parallel in each experiment. Three different experiments were performed. See also Supplementary Information. Click here to view larger figure.
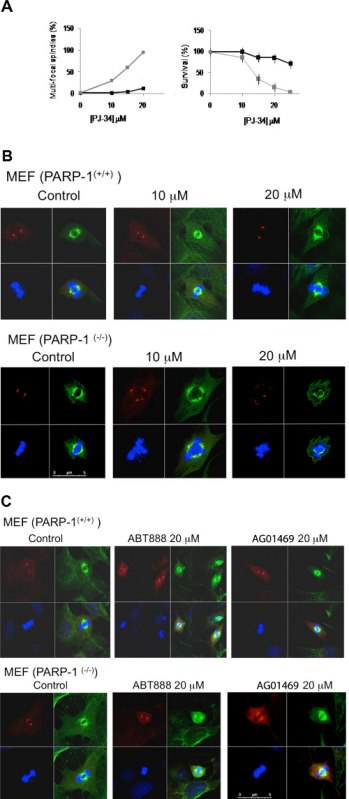 Figure 4. A cytotoxic activity of PJ-34 in PARP1(-/-) mouse embryonic fibroblasts. A. (Left) The percentage of multi-focal spindles calculated in normal (black line) and Parp1-/- (grey line) MEF, incubated for 48 hr with PJ-34 at the indicated concentrations. The percentage of multi-polar spindles was calculated out of 20 total spindles detected in 3 different experiments. (Right) Reduced cell survival detected in cell cultures incubated for 72 hr with PJ-34 (20 μM) relative to the survival of control untreated cells (normal (black line) and Parp1-/- (grey line) MEF). Cell survival was assayed by the cells' ATP production (protocol 5). The mean values of 4 measurements for each cell line in 3 different experiments are presented. B. Spindles in randomly selected fixed normal and Parp1-/- MEF in mitosis, untreated (control) or incubated with PJ-34 for 48 hr at the indicated concentrations. PJ-34 caused multipolar spindles. Cells were fixed, permeabilized and immunolabeled for α- and γ-tubulin (green labeling of spindles and red labeling of centrosomes, respectively). Chromosomes were labeled with DAPI reagent (blue). Representative results of 3 different experiments. C. Potent non-phenanthrene PARP1 inhibitors did not affect centrosomes clustering in PARP1(-/-) MEF. Spindles of randomly selected normal and Parp1-/- MEF are presented; untreated MEF (control) or MEF treated for 48 hr with non-phenanthrene PARP inhibitors, AG01469 (20 μM) or ABT888 (20 μM). Chromosomes are labeled with DAPI reagent (blue). Similar results were obtained in 3 different experiments. Click here to view larger figure.
Figure 4. A cytotoxic activity of PJ-34 in PARP1(-/-) mouse embryonic fibroblasts. A. (Left) The percentage of multi-focal spindles calculated in normal (black line) and Parp1-/- (grey line) MEF, incubated for 48 hr with PJ-34 at the indicated concentrations. The percentage of multi-polar spindles was calculated out of 20 total spindles detected in 3 different experiments. (Right) Reduced cell survival detected in cell cultures incubated for 72 hr with PJ-34 (20 μM) relative to the survival of control untreated cells (normal (black line) and Parp1-/- (grey line) MEF). Cell survival was assayed by the cells' ATP production (protocol 5). The mean values of 4 measurements for each cell line in 3 different experiments are presented. B. Spindles in randomly selected fixed normal and Parp1-/- MEF in mitosis, untreated (control) or incubated with PJ-34 for 48 hr at the indicated concentrations. PJ-34 caused multipolar spindles. Cells were fixed, permeabilized and immunolabeled for α- and γ-tubulin (green labeling of spindles and red labeling of centrosomes, respectively). Chromosomes were labeled with DAPI reagent (blue). Representative results of 3 different experiments. C. Potent non-phenanthrene PARP1 inhibitors did not affect centrosomes clustering in PARP1(-/-) MEF. Spindles of randomly selected normal and Parp1-/- MEF are presented; untreated MEF (control) or MEF treated for 48 hr with non-phenanthrene PARP inhibitors, AG01469 (20 μM) or ABT888 (20 μM). Chromosomes are labeled with DAPI reagent (blue). Similar results were obtained in 3 different experiments. Click here to view larger figure.
Supplementary Information
 Click here to view larger figure.
Click here to view larger figure. Click here to view larger figure. Figure 2B. Supplemental Figure. A randomly selected MDA-MB-231 cell in anaphase with bi-focal clustered γ-tubulin labeled extra-centrosomes was scanned for 16 hr by live confocal imaging, 48 hr after being transfected with vectors expressing γ-tubulin-GFP and H2b-RED (labeling γ-tubulin foci and centrosomes in the fixed cells (green) and H2b histone labeling of the chromosomes (red), respectively).
Click here to view larger figure. Figure 2B. Supplemental Figure. A randomly selected MDA-MB-231 cell in anaphase with bi-focal clustered γ-tubulin labeled extra-centrosomes was scanned for 16 hr by live confocal imaging, 48 hr after being transfected with vectors expressing γ-tubulin-GFP and H2b-RED (labeling γ-tubulin foci and centrosomes in the fixed cells (green) and H2b histone labeling of the chromosomes (red), respectively).
Figure 3. Click here to view movie. Supplemental Figure.
A live confocal imaging documentation of cell death in a randomly selected MDA-MB-231 cell in mitosis with un-clustered γ-tubulin labeled extra-centrosomes. MDA-MB-231 cells were incubated with PJ-34 for 24 hr before scanning and during the 16 hr of live confocal imaging. PJ-34 was applied 24 hr after transfection with vectors expressing γ-tubulin-GFP (green, γ-tubulin foci and centrosomes) and H2b-RED (red, chromosomes).
Discussion
Live confocal imaging provided a real-time documentation of the cytotoxic effect of PJ-34 in live multi-centrosomal cells during mitosis (Figure 3 and Supplementary information). This was the first live documentation attributing the cytotoxicity of PJ-34 in human cancer cells to extra centrosomes de-clustering and cell death, suggesting induction of Mitotic Catastrophe cell death by PJ-345-9. In contrast, bi-focal clustering of super-numerary centrosomes was observed in live untreated MDA-MB-231 cells undergoing normal mitosis with bi-focali clustered extra centrosomes (Figure 2, supplementary information).
According to these results, live confocal imaging of transfected cells could be useful for detecting translocation of proteins implicated in extra-centrosomes clustering in transfected cells during mitosis5,9,15,16. Identification of proteins affected by PJ-34 in multi-centrosomal cells may provide some clues for understanding the death mechanisms activated by extra-centrosomes de-clustering.
The advantage of live confocal imaging in providing real-time information during mitosis in live cells, is also bound to several limitations. The number of cells scanned per experiment is limited. Therefore chances to detect cells in mitosis are low, and several repeating experiments are required for a real-time documentation of mitosis in the scanned cells. In addition, success is highly dependent on high efficiency of transfection of cells with vectors expressing labeled proteins. Thus, despite being reliable, live confocal imaging is time consuming and demands highly experienced hard workers.
In comparison, immunocytochemistry and confocal imaging of fixed cells enable examination of a large number of cells per experiment, which is required for a reliable statistical analysis. We used this method to compare the effects of PJ-34 in mitosis of normal benign cells to its effects in cancer cells undergoing mitosis with supernumerary centrosomes2,3. Similarly, this method was used for comparing the effects of PJ-34 to that of non-phenanthrene PARP1 inhibitors in PARP(-/-) MEF harboring extra-centrosomes (cells with high occurrence of multi-centrosomes11) (Figure 4).
In summary, our results indicate the advantage of combining the most valuable real-time information provided by confocal imaging of live cells in mitosis (Figures 2 and 3) with cytochemistry and confocal analysis of fixed cells. A combination of these methods might be useful for identifying the cytotoxic activity of small molecules that, like PJ-34, target specific mechanisms crucial for cell survival. The unique dependence of many human cancer cells on extra-centrosomes bi-polar clustering for their proliferation and survival renders PJ-34 a possible candidate for cancer therapy.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
Funding sources of this research: a joint fund of Tel Aviv University's technology transfer company, RAMOT and the Sheba-Medical Center (M. C-A. and S.I.), ICRF - Israeli Cancer research foundation (M. C-A.) and Israel science Foundation (S.I.).
References
- Jagtap P, et al. Novel phananthridine inhibitors of poly(adenosine 5'-diphosphate-ribose) synthetase: Potent cytoprotective and antishock agents. Crit. Care Med. 2002;30:1071–1082. doi: 10.1097/00003246-200205000-00019. [DOI] [PubMed] [Google Scholar]
- Castiel A, et al. A small molecule exclusively eradicates human cancer cells: Extra-centrosomes de-clustering agent. BMC Cancer. 2011;11(1):412. doi: 10.1186/1471-2407-11-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbar-Rozensal D, et al. A selective eradication of human nonhereditary breast cancer cells by phenanthridine -derived polyADP-ribose polymerase inhibitors. Breast Cancer Res. 2009;11(6):R78. doi: 10.1186/bcr2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Basto R. Multiple centrosomes: together they stand, divided they fall. Genes Dev. 2008;22:2291–2296. doi: 10.1101/gad.1715208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Canc. Met. Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Doxsey S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2001;2:688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. International Rev. of Cytology. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Ogden A, Rida PCG, Aneja R. Let's huddle to prevent a muddle: centrosome declustering as an attractive anticancer strategy. Cell Death Differ. 2012;19:1255–1267. doi: 10.1038/cdd.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Anderhub S, Maier B. Mechanisms and Consequences of centrosomes clustering in cancer cells. In: Schatten E, editor. The Centrosome: Cell and Molecular mechanisms of functions and disfunctions in disease. Humana Press Springer; 2012. pp. 255–277. [Google Scholar]
- Galimberti F, et al. Anaphase Catastrophe Is a Target for Cancer Therapy. Clin. Cancer Res. 2011;17:1218–1222. doi: 10.1158/1078-0432.CCR-10-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai M, et al. Haploinsufficiency of poly(ADP-ribose) polymerase-1-mediated poly(ADP-ribosyl)ation for centrosome duplication. Biochem. Biophys. Res. Commun. 2007;359:426–430. doi: 10.1016/j.bbrc.2007.05.108. [DOI] [PubMed] [Google Scholar]
- Gartner EM, Burger AM, Lorusso PM. Poly(ADP-ribose) polymerase inhibitors: a novel drug class with a promising future. Cancer J. 2010;16:83–90. doi: 10.1097/PPO.0b013e3181d78223. [DOI] [PubMed] [Google Scholar]
- Wahlberg E, et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nature Biotechnology. 2012;30:283–288. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendze MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nature Rev. Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber B, et al. Proteins Required for Centrosome Clustering in Cancer Cells. Sci. Transl. Med. 2010;2(33):33–38. doi: 10.1126/scitranslmed.3000915. [DOI] [PubMed] [Google Scholar]
- Kwon M, et al. Mechanisms to suppress multipolar division in cancer cells with extra centrosomes. Gene Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]


