Abstract
Elevated blood pressure (BP) and chronic kidney disease (CKD) are complex traits representing major global health problems1,2. Multiple genome-wide association studies (GWAS) identified common variants giving independent susceptibility for CKD and hypertension in the promoter of the UMOD gene3-9, encoding uromodulin, the major protein secreted in the normal urine. Despite compelling genetic evidence, the underlying biological mechanism is not understood. Here, we demonstrate that UMOD risk variants directly increase UMOD expression in vitro and in vivo. We modeled this effect in transgenic mice and showed that uromodulin overexpression leads to salt-sensitive hypertension and to age-dependent renal lesions that are similarly observed in elderly subjects homozygous for UMOD risk variants. We demonstrate that the link between uromodulin and hypertension is caused by activation of the renal sodium co-transporter NKCC2. This very mechanism is relevant in humans, as pharmacological inhibition of NKCC2 is more effective in lowering BP in hypertensive patients homozygous for UMOD risk variants. Our findings establish a link between the genetic susceptibility to hypertension and CKD, the control of uromodulin expression and its role in a salt-reabsorbing tubular segment of the kidney. These data point to uromodulin as a novel therapeutic target to lower BP and preserve renal function.
Our current understanding of the complex genetic architecture of hypertension and CKD stems on the identification of mutations causing rare inherited disorders10,11 and of several susceptibility loci through population-based association studies12-14. However, understanding the biological mechanisms underlying these genetic associations has proven to be a major challenge.
Recent GWAS in more than 200,000 individuals of European ancestry have identified susceptibility variants for renal function, CKD and hypertension in the UMOD gene encoding uromodulin3-9. Uromodulin (or Tamm-Horsfall protein) is the most abundant urinary protein, specifically produced and secreted by the epithelial cells lining the thick ascending limb (TAL) of Henle’s loop in the kidney15. Studies in Umod knock-out mice revealed that uromodulin may protect against urinary tract infection16 and kidney stones17, and modulate electrolyte tubular transport18. Recent evidence suggested that uromodulin regulates the activity of NKCC2 and ROMK, the two main ion transporters involved in NaCl reabsorption by the TAL segment19,20. Mutations in UMOD have been associated with rare, dominantly inherited disorders causing kidney damage and CKD21. The fact that susceptibility variants in the UMOD gene have high frequency (about 0.8) in the general population and confer about 20% increased risk for CKD and 15% for hypertension emphasizes the pressing need to understand the nature of their associated risk and how they affect the function of uromodulin22.
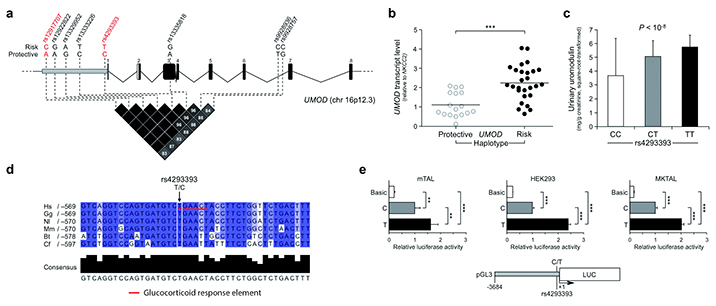
Given the localization of top SNPs identified by GWAS in a linkage disequilibrium block that includes the UMOD gene promoter (Fig. 1a), we hypothesized that these variants could be associated with an effect on gene expression. We tested our hypothesis in vivo, by measuring UMOD transcript levels in nephrectomy samples from individuals homozygous for either the risk or protective alleles at top variants rs12917707 and rs4293393. Carriers of the UMOD risk haplotype showed a two-fold increase in UMOD transcript in kidney samples (Fig. 1b). We confirmed the remarkable association of UMOD risk variants with higher uromodulin expression by showing a similar, dose-dependent increase in urinary uromodulin levels in a large, population-based cohort (Fig. 1c). In silico analysis revealed that among the top SNPs from GWAS, the rs4293393 SNP maps to the best-conserved region of the UMOD promoter (Supplementary Fig. 1) and is predicted to be part of a glucocorticoid response element that is disrupted by the protective allele (Fig. 1d). We hence tested whether risk or protective alleles at this SNP had an effect on the transcriptional activity of the human UMOD gene promoter. We performed a standard in vitro luciferase reporter assay employing a 3.7 kb UMOD promoter fragment in three different renal cell lines, including highly differentiated mouse TAL primary cells that retain high Umod expression. Notably, the presence of the risk allele increased the expression of the luciferase gene, relative to the protective allele, by about two-fold in all cell lines, confirming the in vivo findings (Fig. 1e). The predicted effect of the rs4293393 risk allele on the promoter response to glucocorticoids was also confirmed (Supplementary Fig. 2). Of note, glucocorticoids induced transcription from both constructs, consistent with the presence of additional predicted glucocorticoid receptor elements in UMOD promoter (data not shown) and suggesting a complex hormonal regulation of uromodulin expression. Overall, these data demonstrate that the effect of top rs4293393 UMOD risk variant together with likely others within the same linkage disequilibrium block, is associated with higher uromodulin expression.
Figure 1. Biological effect of UMOD SNP variants.
(a) Schematic representation of human UMOD gene structure. The position of top SNPs in the UMOD promoter associated with hypertension and CKD is shown. All variants are within the same linkage disequilibrium block, as shown by the LD plot (r2 values, data from HapMap CEU, release #28). Genotyped SNPs are shown in red. (b) UMOD expression levels (Real-Time qRT-PCR) on human nephrectomy samples homozygous for either the protective (n = 17) or risk (n = 27) UMOD variants. We normalized UMOD expression to NKCC2 to account for TAL content in the different renal samples. Distribution and mean values of expression levels are shown. ***P < 0.001 (Mann-Whitney test). (c) Urinary uromodulin concentrations in individuals of the SKIPOGH cohort by genotype at rs4293393 (n = 18 CC, 214 CT, 532 TT). Bars indicate average ± s.e.m. The P value reflects significant association of rs4293393 genotype with square-root-transformed daytime urinary uromodulin-to-creatinine ratio in a mixed linear model. (d) Partial alignment of human (Hs), gorilla (Gg), gibbon (Nl), macaque (Mm), cow (Bt) and dog (Cf) UMOD promoter sequences. The intensity of the blue color shading corresponds to nucleotide conservation. The rs4293393 SNP is predicted to be part of a glucocorticoid response element that is lost in the presence of the protective (C) allele. (e) Quantitative analysis of the relative effect of protective (C) and risk (T) alleles of SNP rs4293393 on the transcriptional activity of UMOD promoter in mTAL, highly differentiated mouse primary TAL cells retaining uromodulin expression, immortalized MKTAL (mouse kidney TAL) and HEK293 (human embryonic kidney) cells. A UMOD promoter fragment (3.7 kb) was cloned upstream of LUC (firefly luciferase) gene. We assessed the effect of variants at SNP rs4293393 on the risk haplotype by measuring luciferase activity. Graphs represent 4 independent experiments. Bars indicate average ± s.e.m. **P < 0.01; ***P < 0.001 (ANOVA followed by Bonferroni’s test).
In order to model this effect in vivo, we took advantage of a mouse transgenic line expressing HA-tagged wild type uromodulin (TgUmodwt)23. We also generated a line homozygous for the transgene (TgUmodwt/wt). Transgenic animals from both lines were viable, apparently healthy and undistinguishable from control non-transgenic mice. The two transgenic lines showed increasing levels of uromodulin expression and secretion, comparable to the expression increase in subjects homozygous for UMOD risk variants (about +80% in TgUmodwt/wt, relative to control non-transgenic mice) (Supplementary Fig. 3a-c). As for the endogenous uromodulin, transgenic protein is exclusively expressed in TAL segments of the nephron (Supplementary Fig. 3d-f; see also23).
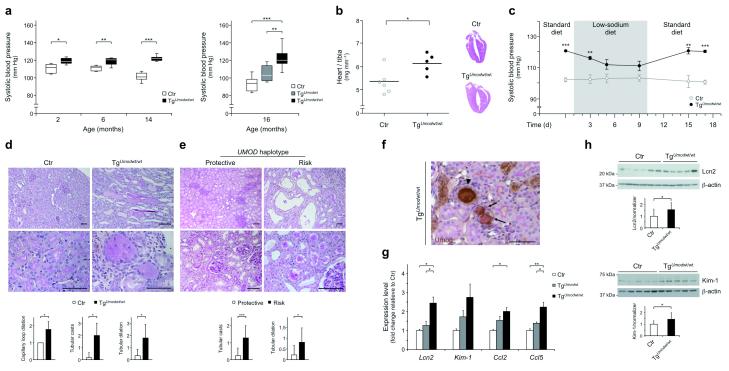
As uromodulin is expressed in the TAL, a tubular segment that has been involved in rare inherited disorders characterized by defective NaCl reabsorption and decreased BP10, we first tested whether its overexpression could affect blood pressure by similar mechanisms. Blood pressure was indeed remarkably increased in transgenic mice in an Umod dosage-dependent fashion (P < 0.0001, ANOVA post test for linear trend) (Fig. 2a). Although significantly higher as soon as 2 months of age, the different BP in TgUmodwt/wt mice relative to controls increased with age. This was partly due to decreased BP in aging control mice, as already described24,25 (Fig. 2a). Increased BP in TgUmodwt/wt mice was associated with a significant increase of the total heart mass and left ventricular hypertrophy, consistent with a chronic hypertensive state (Fig. 2b). Hypertension in transgenic mice was salt-sensitive, as BP could be normalized to control levels by low-NaCl diet, while returning to elevated levels when the mice were fed again with standard NaCl diet (Fig. 2c). The changes in dietary salt exposure were reflected by expected modifications in aldosterone levels and NaCl handling that were similar in control and transgenic animals (Supplementary Fig. 4).
Figure 2. Uromodulin overexpression leads to hypertension and renal damage.
(a) Box and whiskers plot showing the systolic BP at baseline in control and Umod transgenic mice at different age (n = 6–10 per group). Bars represent min and max values. (b) Average value and distribution of heart weight in 16 month-old control and TgUmodwt/wt mice (left panel) and heart histology showing left ventricular hypertrophy in TgUmodwt/wt mice (right panel) (Hematoxylin-Eosin; 1×). (c) Average systolic BP in 14 month-old control and TgUmodwt/wt mice on standard (1% NaCl) or low-sodium (0.01% NaCl) diet (n = 7 per group). Error bars indicate ± s.e.m. (d) Representative renal histological images of 16 month-old control and TgUmodwt/wt mice (n = 5). Control tissue displays normal features. Kidneys from TgUmodwt/wt mice show numerous dilated tubules (upper panel), mostly filled by casts (upper and lower panel). (e) Representative renal tissue from elderly subjects homozygous for the protective variants of rs4293393 and rs12917707 (left panels) showing normal interstitial compartment (upper panel) and mild focal tubular damage with increased thickness of the tubular basement membrane (lower panel) (n = 9). Renal tissue from subjects homozygous for the risk variants (right panels) displays dilated tubules, with detachment of the tubular epithelium (upper panel) and presence of tubular casts (lower panel) (n = 15) (PAS, bar = 100 μm). The graphs below d and e panels show average quantification of the histological analysis (only classes reaching statistical significance are shown). Error bars indicate ± s.d. (f) Tubular casts in renal tissue from a TgUmodwt/wt mouse are present in TALs (uromodulin-positive) or more distal tubules (uromodulin-negative) and are formed mostly by uromodulin (arrowhead) or by PAS-positive uromodulin-negative material (black arrows). Bar = 100 μm. (g) Transcript level (Real-Time qRT-PCR) of renal damage markers and chemokines in kidneys from 16 month-old mice: Lcn2 (Lipocalin 2); Kim-1 (Kidney injury molecule-1); Ccl2 (C-C motif chemokine 2); Ccl5 (C-C motif chemokine 5) (n = 5). Bars indicate average ± s.e.m. (h) Representative immunoblot showing Lcn2 and Kim-1 protein levels in kidney lysates of 16 month-old control and TgUmodwt/wt mice (n = 13 per group). Beta actin is shown as a loading control. Bars indicate average ± s.d. *P < 0.05; **P < 0.01; ***P < 0.001 determined by unpaired t test (a (left), b, c, h), ANOVA followed by Bonferroni’s test (a (right), g) or Mann-Whitney test (d, e).
Transgenic mice had similar body weight, as well as baseline urine and plasma parameters relative to controls up to 16 months of age (Supplementary Table 1, and data not shown). Renal function, measured by FITC-sinistrin clearance26, was also normal (Supplementary Table 1). However, histological analysis of kidneys from aging TgUmodwt/wt mice revealed signs of renal damage, mainly localized and affecting distal segments, with segmental dilation and increased tubular casts (Fig. 2d). We could detect similar focal lesions in human nephrectomy samples from individuals above 65 years of age and they were increased in homozygous carriers of UMOD risk variants relative to homozygous carriers of the protective ones (Fig. 2e). Further investigation in the mouse model revealed that casts were often but not always positive for uromodulin and localized to TALs and to more distal segments (Fig. 2f). Renal damage in transgenic mice was also supported by increased renal expression of established markers of tubule damage (Lcn2 and Kim-1) and chemokines (Ccl2 and Ccl5) (Fig. 2g,h), and substantial microalbuminuria (data not shown). Of note, we observed significant dilations of glomerular capillary loops (Fig. 2d and Supplementary Fig. 5a). There were no signs of interstitial vascular remodeling in the kidney that could be ascribed to chronic hypertension (Supplementary Fig. 5b), in line with the fairly rapid BP response induced by changes of dietary salt (see above).
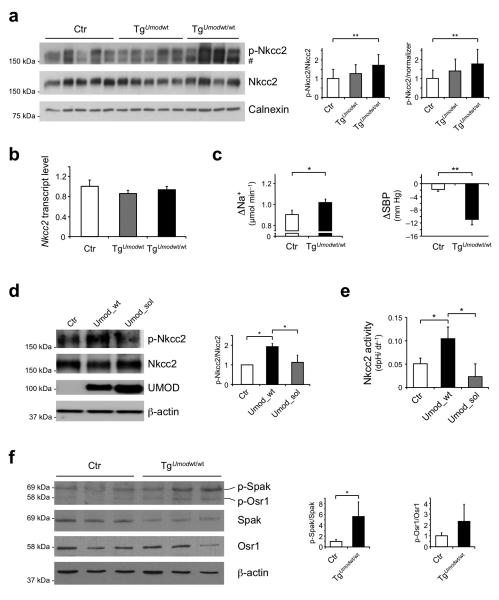
In the context of normal renal function and a trend for decreased renin expression (Supplementary Fig. 6), we hypothesized that salt-sensitive hypertension in transgenic mice could be caused by abnormal activation of Nkcc2, the main sodium transporter in the TAL. Consistent with our hypothesis, we identified a significant increase of Nkcc2 phosphorylation at activating sites (Thr96 and Thr101)27,28 in TgUmodwt/wt mice relative to control animals (Fig. 3a). The specificity of this effect on Nkcc2 rather than on Nkcc1, that can also be recognized by the available antibodies, is supported by the almost exclusive localization of the phosphorylated protein signal on the apical membrane of TAL cells, where Nkcc1 is not expressed, and by the 40-fold higher global expression of Nkcc2 in the kidney (Supplementary Fig. 7a,b). In line with a post-translational regulation of Nkcc2, the level of Nkcc2 transcript was not different (Fig. 3b). Romk expression and membrane localization were similar in transgenic and control mice (Supplementary Fig. 8a-c). We hence assessed the response to furosemide, a loop diuretic that specifically targets NKCC2, as an index of the co-transporter activity. Treatment with a single dose of furosemide induced both a significantly enhanced natriuretic effect and a significant reduction of blood pressure in TgUmodwt/wt mice (Fig. 3c). This greater response is due to increased Nkcc2 activity and increased sodium reabsorption in the TAL rather than to adaptive down-regulation of NaCl reabsorption in more distal nephron segments, as demonstrated by the comparable levels of Ncc (distal convoluted tubules) or ENaC (collecting ducts) (Supplementary Fig. 8d,e) and the similar levels of aldosterone (Supplementary Fig. 4b) in control and transgenic mice.
Figure 3. Increased activation of Nkcc2 co-transporter and Spak kinase in TgUmodwt/wt mice.
(a) Representative immunoblot analysis showing levels of phosphorylated (Thr96 and Thr101) and total Nkcc2 in kidney lysates from 16 month-old mice. Densitometric analysis (average ± s.d. of 4 independent experiments) shows increased p-Nkcc2 in TgUmodwt/wt mice (n = 10–13 per group). # unspecific signal. The increase of p-Nkcc2 is linearly correlated with Umod gene dosage (P < 0.01, ANOVA post test for linear trend). Calnexin was used as a loading control. (b) Transcript level of Nkcc2 (Real-Time qRT-PCR) on total kidney extracts from 16 month-old mice (n = 5). Bars indicate average ± s.e.m. (c) Change in sodium excretion (left panel) and systolic BP (right panel) in 16 month-old TgUmodwt/wt and control mice, 2 hours after treatment with furosemide (n = 8 Ctr, 6 TgUmodwt/wt (sodium excretion), 5 per group (systolic BP)). Bars indicate average ± s.e.m. (d) Representative immunoblot analysis showing levels of phosphorylated and total Nkcc2 and uromodulin in HEK293 cells stably expressing Nkcc2 and transfected with either wild type (Umod_wt) or soluble (Umod_sol) uromodulin. Beta actin was used as a loading control. Densitometric analysis (average ± s.d. of 3 independent experiments) is shown. (e) Quantification of Nkcc2 activity as assessed by dpHi dt−1. Bars indicate average ± s.e.m. (f) Representative immunoblot analysis showing levels of phosphorylated and total Spak and Osr1 in kidney lysates from 16 month-old control and TgUmodwt/wt mice (n = 4 per group). Bars indicate average ± s.d. *P < 0.05; **P < 0.01 determined by ANOVA followed by Bonferroni’s test (a, b, d, e) or unpaired t test (c, f).
We investigated the direct role of uromodulin on NKCC2 activation in renal cells stably expressing Nkcc2 and transiently transfected with either wild type uromodulin (Umod_wt) or with a soluble uromodulin isoform (Umod_sol), truncated at the GPI-anchoring site (S614X)29. Co-expression of wild-type uromodulin in this cellular system led to a significant increase of Nkcc2 phosphorylation that correlated with an increase of its activity (Fig. 3d,e). This effect was completely lost in cells transfected with soluble uromodulin. These findings expand previous knowledge19, confirming the role of uromodulin on NKCC2 phosphorylation and activity in a mammalian system and strongly suggesting that this role is exerted by the cellular membrane-anchored protein.
NKCC2 phosphorylation is, at least in part, mediated by STE20/SPS1-related proline/alanine-rich kinase (SPAK) and by oxidative stress response 1 kinase (OSR1)30,31. The relative levels of Spak phosphorylated at Thr243, i.e. active, were significantly increased in TgUmodwt/wt mice, and there was a trend for increased relative levels of phosphorylated (Thr185) Osr1, demonstrating upregulation of this regulatory network by uromodulin overexpression (Fig. 3f and Supplementary Fig. 7c). Consistent with this effect, the transcript level of kidney specific (KS)-Spak, an isoform mainly expressed in the TAL and acting as a negative regulator of NKCC2 phosphorylation32, but not the one of full-length Spak, was reduced in transgenic mice (Supplementary Fig. 7d,e).
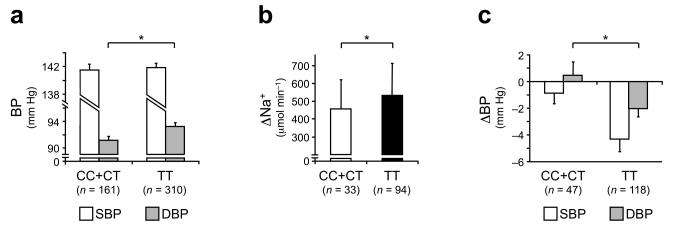
We finally investigated whether the involved UMOD variants could play a role in modulating BP in humans, by taking advantage of a well-characterized cohort of naïve (never treated) hypertensive subjects stratified a posteriori for the UMOD SNP rs4293393 (Supplementary Table 2). The baseline mean diastolic blood pressure was significantly higher in hypertensive individuals homozygous for the risk allele relative to carriers of the protective one (Fig. 4a). For a subset of these subjects, data from furosemide test were also available. Patients homozygous for the risk allele showed an increased response to the diuretic, with significantly higher increase of natriuresis over baseline values (Fig. 4b) and more marked response of BP (Fig. 4c and Supplementary Table 2). Despite the limitations of a retrospective and relatively small sized cohort, these results suggest that the mechanism causing hypertension in transgenic mice (i.e. increased NKCC2 activity linked to overexpression of uromodulin) may play a role in human hypertension and could represent a new therapeutic target.
Figure 4. Increased blood pressure and response to furosemide in hypertensive patients homozygous for UMOD risk variants.
(a) Twenty-four hour ambulatory BP monitoring, average values for systolic (SBP) and diastolic (DBP) blood pressure are shown. DBP is significantly higher in hypertensive subjects homozygous for the UMOD risk genotype. (b, c) Response to furosemide treatment in hypertensive patients with different rs4293393 genotype. Changes in sodium excretion (b), and systolic (SBP) and diastolic (DBP) blood pressure (c) 4 hours after treatment are shown. The loop diuretic is more effective in lowering BP in hypertensive patients homozygous for rs4293393 risk variant, with a significant difference in the decrease of DBP and a similar trend for SBP (P = 0.06). This effect is associated with higher natriuretic effect. Bars indicate average ± s.e.m. (a, c) or ± s.d. (b) *P < 0.05 (ANOVA test).
This study identifies the causal effect underlying a major risk locus for CKD and hypertension evidenced by multiple GWAS. Current knowledge of the functional relationship between NaCl handling in the kidney and BP regulation has been significantly contributed by studies on rare monogenic disorders33. Loss of function mutations impairing sodium reabsorption have been associated with salt wasting and reduced BP in monogenic diseases affecting the TAL (Bartter syndrome) or more distal segments (Gitelman syndrome and pseudohypoaldosteronism type 1). Thus far, increased sodium reabsorption leading to hypertension was exclusively linked to mutations affecting these more distal segments (Liddle syndrome and pseudohypoaldosteronism type 2)10,14. The new gain-of-function mechanism here described completes the paradigm by linking renal transport of NaCl in the TAL with blood pressure regulation. This mechanism is of potential wide relevance, being contributed by variants present in the homozygous state in about 60% of Caucasians (Supplementary Table 3). Further prospective studies will be necessary to confirm our findings in hypertensive patients and fully explore the contribution of the newly identified mechanism to hypertension.
Through evidence obtained in mouse and cellular models, this study establishes the importance of uromodulin in modulating salt handling in the TAL. It is of note that both the expression and urinary excretion of uromodulin are increased by high sodium intake in rats34 and humans35. Here we demonstrate that UMOD promoter activity is also modulated by glucocorticoids, known to play a central role in ion homeostasis and BP regulation and already shown to act on TAL cells36.
Both in aging TgUmodwt/wt mice and in elderly individuals homozygous for UMOD risk variants, the upregulation of uromodulin associates with focal renal lesions, as tubular dilations and presence of casts, which are detected in the context of normal kidney function. The focal kidney damage in TgUmodwt/wt mice seems unlikely to be secondary to chronic hypertension, since no such lesions were detected in other rodent models of hypertension37. Rather, these lesions and the upregulation of lipocalin 2 and Kim-1 are reminiscent of changes observed in aging kidneys38,39. These results suggest that uromodulin overexpression is unlikely to lead to renal failure per se. Instead, it could predispose to CKD when additional conditions harm the kidney, a risk that increases with age. This hypothesis is supported by recent evidence that the association of UMOD promoter risk variants with CKD is stronger in the older age groups and with additional comorbid conditions7,9.
Our study provides a rare example of the demonstration of the causal role of high-effect common variants of genes causing monogenic disorders, which are linked to complex traits in the general population. The UMOD risk variants show high frequency in all ethnic groups (Supplementary Table 3), suggesting the action of selective pressure. This is atypical but not unprecedented, an established example being APOL1 variants that were strongly associated with kidney disease in African Americans and shown to be protective factors improving resistance to Trypanosoma brucei rhodesiense40. What could have been the selective advantage of UMOD risk variants? We speculate that the protective role of uromodulin against urinary tract infections as well as its function in salt reabsorption in the kidney could have favored variants associated with increased expression and urinary levels. With increased life expectancy, better hygienic conditions and higher salt intake, the same variants could also express their deleterious effect. Accordingly, therapies targeting uromodulin expression or function may be relevant for controlling blood pressure and preserving renal function.
Online Methods
Ethical considerations
The MI_HPT study was approved by the ethical committee of the San Raffaele Hospital. The SKIPOGH study was approved by the ethical committees of Lausanne University Hospital, Geneva University Hospitals and University Hospital of Bern (Inselspital). All participants provided informed written consent.
Hypertensive patient cohort (MI_HPT)
In this study we enrolled a cohort of 471 (391 males and 80 females) never treated hypertensive patients, afferent to Outpatient Clinical at San Raffaele Hospital41. Patients were between 20 and 65 years old, they had a body mass index minor than 30 kg m−2 and systolic and diastolic blood pressure (SBP/DBP) >140/90 mm Hg and <160/110 mm Hg. We excluded from the study patients with chronic or acute pathologies, clinical history for ischemic cardiomyopathy, cardiac decompensation, cerebral vasculopathy, creatinine clearance <80 ml min−1, liver failure, diabetes, severe essential hypertension or secondary arterial hypertension. We also excluded subjects under anti-hypertensive or estrogen or progestin therapies, or with addiction and alcohol abuse history. We gave patients dietary indications and verified compliancy by assessing 24-h urinary sodium excretion.
Due to the low frequency of the rs4293393 protective allele, we grouped patients carrying one (CT) or two (CC) copies of the protective variant and compared them with homozygous for the risk one (TT). The two groups did not significantly differ for age, sex, BMI and renal function (see Supplementary Table 2).
SKIPOGH cohort
The Swiss Kidney Project on Genes in Hypertension (SKIPOGH) is a family-based cross-sectional study exploring the role of genes in blood pressure and kidney function regulation. We recruited participants from December 2009 until April 2013 in three centers (Bern, Geneva and Lausanne, Switzerland), as previously described42. Participants collected urine during daytime, which had a median duration of 16 h (interquartile range, 2) hours. We included all available samples in the uromodulin analysis.
Human kidney samples
The study on nephrectomy samples was approved by the ethical committee of the San Raffaele Hospital. All participants provided informed written consent. Human renal samples were from individuals who underwent nephrectomy at the San Raffaele Hospital because of renal carcinoma. We dissected samples from the healthy part of the explanted kidney and properly stored for histological and molecular analysis. We considered eligible for the analysis only kidneys from patients with normal renal function (i.e. eGFR > 90 ml min−1, calculated by using the 4-variable Modification of Diet in Renal Disease formula43) and under no anti-tumor treatment at the time of explant. We matched individuals homozygous for rs12917707 and rs4293393 protective alleles (n = 17; 41.2% females; age at the time of nephrectomy = 67.7 ± 12.7 years) for age and sex with at least one individual homozygous for the risk alleles (n = 27; 33.3% females; age at the time of nephrectomy = 67.9 ± 9.6 years).
Genotyping
We carried out genotyping in the MI_HPT cohort and in individuals who underwent nephrectomy on genomic DNA extracted from blood samples or renal tissue with QIAamp DNA Mini Kit (Qiagen) by using the 5′ nuclease allelic discrimination assays with allele-specific MGB probes (TaqMan, Applied Biosystems) for marker SNPs rs4293393 (Assay ID C__27865986_10) and rs12917707 (Assay ID C__31122302_10) according to manufacturer’s instructions (call rate 97.2%).
Genotyping of SNP rs4293393 in SKIPOGH individuals was done on genomic DNA extracted from white blood cells at LGC Genomics (formerly KBioscience) using Competitive Allele Specific PCR technique (KASPar v4.0) (call rate 96.4%). Additional quality control criteria included inter- and intra-plate duplicate testing and clear separation of signal clusters.
Reproducibility test on randomly selected duplicates (10% in the MI_HPT and 4.5% in the SKIPOGH cohorts) showed 100% concordance. The frequency of the rs4293393 minor allele C was 0.180 in the MI_HPT cohort and 0.163 in the SKIPOGH cohort. The P values for Hardy-Weinberg equilibrium were 0.048 (MI_HPT) and 0.34 (SKIPOGH). The deviation in the MI_HPT is driven by underrepresentation of individuals homozygous for the protective allele C (observed 9 vs. expected 15.3). A result that is in line with GWAS studies showing association of the risk allele T with hypertension3.
Patient Ambulatory Blood Pressure Monitoring and furosemide test
Patients (n = 471) underwent 24-h ambulatory blood pressure monitoring (Spacelab 90207; Spacelab Medical Inc.) on a day chosen for typical weekly activity. Recordings were performed every 10 min during awake hours (day-time) and every 30 min during nighttime.
A subgroup of 165 patients underwent furosemide test, as follows. After 2 hours equilibration, we orally administered 25 mg of furosemide. We collected urines immediately before and 4 hours after furosemide administration. We measured BP every 60 min during equilibration period and every 30 min after furosemide treatment. Reported values are the average of 3 measurements done every minute at each time. Due to the low frequency of the rs4293393 protective allele, we grouped patients carrying one (CT, n = 44) or two (CC, n = 3) copies of the protective variant and compared them with homozygous for the risk one (TT, n = 118). The two groups did not significantly differ for age, sex, BMI and renal function.
We measured urinary Na+ by flame photometry, creatinine by picric acid test and plasma renin activity by commercial radioimmunoassay (Sorin Laboratories).
Human urinary uromodulin measurement
We measured urinary uromodulin concentration by ELISA based on a sheep anti-human uromodulin antibody (Meridian Life Science, K90071C) as the capture antibody; a mouse monoclonal anti-human THP antibody (Cedarlane, CL1032A) as primary antibody; and a goat anti-mouse IgG (H+L) horseradish peroxidase conjugated (BioRad, 1721011) as secondary antibody44. We used human uromodulin (Millipore, stock solution: 100 μg ml−1) to establish the standard curve45. The uromodulin ELISA assay has a sensitivity of 2.8 ng ml−1, a linearity of 1.0, an inter-assay variability of 3.28% and an intra-assay variability of 5.5%. We measured urinary creatinine levels using Beckman Coulter Synchron® System Creatinin Assay (Unicell DxC Synchron® clinical System) 2446, following the manufacturer instructions. We indexed uromodulin to creatinine (uromodulin-to-creatinine ratio) to compensate variations in urine concentrations.
Transgenic mice
We generated TgUmodwt mice as described previously23. Briefly, we injected in FVB mouse strain the transgenic uromodulin construct, composed of a 2.9 Kb fragment of the Umod gene promoter, the first non-coding exon, the first intron, the coding sequence from exon 2 to 11 and the entire 3′ UTR. An HA-tag was inserted at the uromodulin N-terminus. We generated TgUmodwt/wt mice by breeding TgUmodwt mice and verified transgene homozygosity by Southern blot. We carried all animal procedures on female mice at San Raffaele Scientific Institute, Milan, Italy, and at Université Catholique de Louvain, Brussels, Belgium, according to protocols approved by the San Raffaele Institutional Animal Care and Use Committee and by the Belgian National Research Council Guide for the Care and Use of Laboratory Animals/Animal Ethics Committee, respectively.
Metabolic collections, plasma and urine analyses
We obtained urine and plasma on age- and gender-matched transgenic and control mice. They were housed in light- and temperature-controlled room with ad libitum access to tap water and standard chow (Diet AO3, SAFE; 25/18 GR Mucedola Srl) or low-sodium chow (E15430-24, SSNIFF®). We obtained urine collections using individual metabolic cages (14 h overnight for baseline measurement and 2 h after furosemide administration, 10 mg Kg−1), after appropriate training. We collected blood by venous puncture or at time of sacrifice by decapitation. The sampling procedures were identical in all groups. We measured urinary electrolytes, creatinine and albumin, plasma electrolytes, creatinine (enzymatic determination), urea and uric acid on a Synchron CX5 analyzer (Beckman Coulter), and aldosterone by using a validated Aldosterone EIA kit (Cayman Chemical), according to the manufacturer’s instructions and normalizing to urinary creatinine (Beckman Coulter Synchron® System Creatinine Assay). We measured osmolality on a Fiske osmometer (Advanced Instruments).
Mouse blood pressure measurement
We measured systolic BP by the tail-cuff method (Physiograph Narco or BP-2000, Visitech Systems) on two different days in conscious animals, after appropriate training. We averaged four to six successive measurements.
Tissue collection and preparation
To collect organs (kidney, heart, spleen, brain, adrenal glands, aorta and testis), we sacrificed mice by decapitation or after anesthesia with Sevoflurane (Abbott). We immediately homogenized organs for protein or RNA extraction. We fixed renal and cardiac tissue in 4% paraformaldehyde and paraffin-embedded for histological, immunohistochemical and immunofluorescence (IF) analysis.
Cell cultures
We maintained human embryonic kidney (HEK) 293 cells in DMEM media supplemented with 10% fetal bovine serum (Invitrogen) and 1% penicillin/streptomycin. MKTAL cells are an immortalized TAL line that was isolated and characterized by Bourgeois et al47. These cells retain the main properties of TAL cells. We cultured MKTAL cells in DMEM:F12 (1:1) with the SingleQuots Kit (Lonza) containing hydrocortisone, hEGF, fetal bovine serum, epinephrine, insulin, triiodothyronine, transferrin, gentamicin/amphotericin. We obtained TAL primary cultures (mTAL) from microdissected tubules of the outer medulla of 5 week-old collagenase-treated mouse kidneys, according to the method described by Terryn et al48. We cultured the selected tubules, specifically expressing uromodulin and Nkcc2, on permeable filter supports for 14 days, allowing the formation of well-polarized confluent monolayers. These monolayers are characterized by morphological, functional and structural properties similar to those of the TAL segment in vivo, including a high level of endogenous uromodulin expression49.
We kept cells at 37 °C in a humidified atmosphere containing 5% CO2.
Constructs
We cloned the 3.7 kb fragment of the human UMOD gene promoter containing the risk allele at the top SNP rs4293393 (nucleotide −551 T) into the pGL3-basic Luciferase Reporter vector (Promega) in two steps. We obtained the promoter region from nucleotide +1 to −1092 by PCR by using primers with MluI (forward, 5′ TTACGCGTCACGTTGTGCACTTGTACCC 3′) and XhoI (reverse, 5′ TCTCGAGTGGTCATGATGTGCCTCATAC 3′) tails, and the promoter region from nucleotide −1093 to −3684 by PCR by using primers with SacI (forward, 5′ TTGAGCTCATTCAGGACACGGTGTAAGG 3′) and MluI (reverse, 5′ TTACGCGTCAGGTTTGTTACATATGTATAC 3′) tails. We generated the promoter construct containing the rs4293393 protective allele (nucleotide: −551 C) by site-directed mutagenesis using QuickChange Lightning Site-Directed Mutagenesis Kit (Stratagene, Agilent) following the manufacturer’s protocol. We confirmed all plasmids by sequencing.
We cloned HA-tagged human wild-type uromodulin (Umod_wt) and truncated uromodulin soluble isoform (Umod_sol), lacking the GPI-anchoring site (S614X), in expression vector pcDNA3.1 (+) (Invitrogen) as previously described29.
Measurement of UMOD promoter activity in vitro
We tested the effect of rs4293393 protective and risk alleles on the transcriptional activity of UMOD promoter in mTAL, MKTAL and HEK293 cells. While mTAL cells retain high uromodulin expression consistent with their fully differentiated state, weak or no uromodulin expression was detected in MKTAL and HEK293 cells respectively (Real-time qRT-PCR, data not shown).
We transiently transfected mTAL, MKTAL, and HEK293 cells with 2 μg of firefly luciferase reporter plasmid and 10 ng of Renilla luciferase vector using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions.
We evaluated luciferase activity 48 hours after transfection with Dual-Luciferase Reporter Assay System (Promega), using a GloMax 96 luminometer (Promega) with 10 s of integration time for each luciferase reaction. Firefly luciferase activity was corrected for transfection efficiency by using Renilla luciferase measurements. We used the promoter-less pGL3-Basic vector as a negative control. We assessed the response of UMOD promoter to glucocorticoids 24 h after transfection. We assayed extracts from each transfection in duplicate for at least 4 independent transfection experiments.
Measurement of Nkcc2 co-transporter activity in vitro
We generated HEK293 cells stably expressing Nkcc2 by transfecting a construct containing the cDNA for mouse Nkcc2 N-terminally fused to Myc tag and cloned in pTarget expression vector (Promega). We selected transfected cells for resistance to geneticin (G418, 500 μg ml−1) and isolated single clones. We performed all experiments on the same clone. We carried out transient transfection of plasmids expressing uromodulin or empty vector by using Lipofectamine and Plus Reagent (Invitrogen) according to the manufacturer’s instructions. Equal expression of the two transfected uromodulin isoforms was verified by qRT-PCR.
We measured Nkcc2 cotransport activity as bumetanide sensitive NH4 influx, as previously described50. Briefly, we measured cytoplasmic pH (pHi) in cells grown to confluence on coverslips using the intracellularly trapped pH sensitive dye BCECF. We measured baseline pHi in cells bathed at 37 °C in a CO2 free Hepes/Tris buffered medium. We then added NH4Cl (20 mM) to the medium to induce cellular alkalinization and measured the initial rate of intracellular pH recovery (dpH i dt−1) over the first 20 s of records. We used the dpHi caused by NH4Cl addition to calculate the cell buffer capacity. The Nkcc2 co-transporter activity is therefore expressed as dpH i dt−1.
Retrotranscription and Real-time qRT-PCR
We used about 50-200 mg of human renal tissue for total RNA extraction using mirVana miRNA Isolation kit (Ambion, Life technologies), following the manufacturer’s protocol.
We extracted total RNA from mouse whole kidney by homogenization in TRIzol reagent (Invitrogen) and reverse-transcribed extracted RNA using iScript kit (BioRad) according to the manufacturer’s instructions. We analyzed expression of target genes by qRT-PCR on LightCycler 480 (Roche) using qPCR Core kit for SYBR Assay (Eurogentec SA). We designed specific primers by using Primer 351 (primer sequences and PCR conditions available upon request). We determined amplification efficiency by dilution curves. We normalized UMOD expression in human samples to NKCC2 (SLC12A1), to account for TAL segments content in each renal tissue sample, and mouse genes to Hprt1. We calculated the relative mRNA expression of genes of interest following the ΔΔCT method52.
Histology
We performed routine staining (PAS, Trichrome and Hematoxylin-Eosin) according to standard techniques.
We stained heart sections with Hematoxylin-Eosin for evaluation by light microscopy, capturing digital images with a Mirax Midi digital camera (Zeiss).
We used PAS-stained renal sections (three sections per mouse or patient) for the quantification of histological features that was performed by two independent observers unaware of the mouse or patient genotype. Sections were viewed under a Zeiss Axioscope 40FL microscope (Carl Zeiss), equipped with AxioCam MRc5 digital video camera. Images were recorded using AxioVision software 4.3 (Carl Zeiss). In mouse renal sections, we scored vascular damage as present or absent and quantitatively determined tunica media width in at least 6 vessels (average value of 5–6 measurements per vessel) per organ (n = 5 per group). We assessed mesangial expansion and capillary loop dilation semiquantitatively (0 = absent; 1 = 1–50%; 2 = 51–100%), as for interstitial inflammation and fibrosis, presence of tubular casts and tubular dilation (0 = absent; 1 = 1–30%; 2 = 31–60%; 3 = 61–100%). We quantified presence of segmental and global glomerulosclerosis as % of the total number of glomeruli.
Immunofluorescence and immunohistochemistry
We carried out immunofluorescence using standard protocol on 4–5 μm-thick deparaffinized kidney sections after antigenic retrieval with citrate buffer. After blocking, we incubated slides for 1 h with primary antibody followed by appropriate AlexaFluor-labeled secondary antibody (Life Technologies). All slides were viewed under a DM 5000B fluorescence upright microscope (Leica DFC480 camera, Leica DFC Twain Software, 40×/0.75 lens; Leica Microsystems) or under an LSM510Meta Confocal microscope (Zeiss), equipped with a 40×/1.4 Plan-Apochromat oil-immersion objective (Zeiss). We used identical acquisition parameters for the same antibody in different kidney sections.
We performed immunohistochemistry on 5 μm-thick kidney sections following standard protocol and analyzed sections under a Zeiss Axioscope 40FL microscope, equipped with AxioCam MRc5 digital video camera (Carl Zeiss).
Immunoblot
We homogenized mouse tissues from 6–12 weeks old animals at 4 °C in lysis buffer (NaCl 150 mM, n-octylglucoside 60 mM, NaF 10 mM, Na3VO4 1 mM, glycerophosphate 1 mM, Protease-Inhibitor Cocktail 1:1,000 (Sigma-Aldrich) and Tris-HCl 20mM, pH 7.4). We obtained protein lysates from 16 month-old mice by solubilization in lysis buffer, followed by sonication and centrifugation at 16,000 g for 1 min at 4 °C, or from the organic phase of TRIzol (Invitrogen) homogenates that was dialyzed against 0.1% SDS, precipitated with acetone and resuspended in PBS containing SDS 1%, NaF 10 mM, Na3VO4 1 mM, glycerophosphate 1 mM and Protease-Inhibitor Cocktail 1:1,000 (Sigma-Aldrich) (adapted from53). We determined Ncc and ENaC from kidney membrane preparations of 16 month-old mice as previously described54. Briefly, we homogenized kidneys in ice-cold lysis buffer (200 mM mannitol, 80 mM HEPES, 41 mM KOH, pH 7.5) with protease and phosphatase inhibitor cocktails (Complete ULTRA and PhosSTOP, Roche), obtained membrane preparations by ultracentrifugation (100,000 × g, 1 h, 4 °C) and solubilized in Laemmli buffer.
We obtained protein lysates from HEK cells 48 h post-transfection through solubilization in lysis buffer (NaCl 0.4 M; EGTA 0.5 mM; MgCl2 1.5 mM, Hepes 10 mM, pH 7.9; glycerol 5% (v/v); Nonidet P-40 0.5% (v/v) and protease inhibitors (Complete, Roche Diagnostics)). We assessed total and phosphorylated Nkcc2 levels following immunoprecipitation with anti-Myc antibody followed by affinity purification using protein G-agarose beads (Dynal). The purified immunocomplex was washed three times in PBS (Invitrogen).
Urinary proteins were precipitated with acetone, resuspended in PBS and loaded according to creatinine levels.
Protein lysates, immunoprecipitated proteins or urinary proteins were separated on 8–12% SDS-PAGE gel in non-reducing (total uromodulin) or in reducing (all other experiments) condition and transferred onto nitrocellulose membrane (GE Healthcare). We performed Western blot (WB) following standard protocols. Quantification was performed using the gel analysis option of ImageJ software.
Antibodies
We used the following antibodies: sheep polyclonal (Abcam, ab9029-1, 1:200 for IF) and goat polyclonal (MP Biomedicals, 55140, 1:1,000 for WB) antibodies against uromodulin, rat monoclonal antibody against HA (Roche, 11 867 423 001, 1:500 for IF and 1:1,000 for WB), rabbit polyclonal antibody against NKCC2 (Merck Millipore, AB3562P, 1:250 for IF and 1:1,000 for WB), rabbit polyclonal antibody R5 (ref.55) (kind gift of B. Forbush>) against phospho-NKCC2 (phosphorylated Thr96 and Thr101) (1:100 for IF and 1:750 for WB), sheep antibody against SPAK (1:1,000 for WB) and sheep polyclonal antibody against phospho-SPAK (Thr243) / phospho-OSR1 (Thr185) (ref.56) (1:200 for IF and 1:2,000 for WB) (provided by Prof. Dario Alessi), mouse monoclonal against OSR1 (OXSR1) (Abnova, H00009943-M09, 1:500 for WB), rabbit polyclonal against total NCC54 (1:5,000 for WB), rabbit polyclonal against pT53NCC54 (1:5,000 for WB), rabbit polyclonal against pT58NCC54 (1:5,000 for WB), rabbit polyclonal against α-ENaC54 (1:2,000 for WB), or rabbit polyclonal against γ-ENaC57 (1:20,000 for WB), goat polyclonal antibody against renin (Santa Cruz Biotechnology, sc-27318, 1:500 for WB), rabbit polyclonal against ROMK (Chemicon, AB5196, 1:50 for IF), goat polyclonal against ROMK (Santa Cruz Biotechnology, sc-10692, 1:500 for WB), rabbit polyclonal antibody against aquaporin 2 (Sigma, A7310, 1:200 for IF), goat polyclonal against Lipocalin 2/NGAL (R & D Systems, AF1857, 1:500 for WB), rabbit polyclonal against KIM-1 (TIM-1) (Novus Biologicals, NBP1-76701, 1:500 for WB), rabbit polyclonal antibody against calnexin (Sigma, C4731, 1:20,000 for WB), mouse monoclonal antibody against beta actin (Sigma, A2228, 1:20,000 for WB).
In silico analysis
We analyzed UMOD promoter sequence by using Transcription Element Search Software (TESS) (http://www.cbil.upenn.edu/cgi-bin/tess/tess) or PATCH 1.0 (http://www.gene-regulation.com/pub/programs.html) and the associated Transfac database (Computational Biology and Informatics Laboratory, University of Pennsylvania). We aligned promoter sequences with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and displayed with Jalview (http://www.jalview.org/)58.
Statistical analysis
We performed comparisons between groups by using two-tailed unpaired Student’s t test, two-tailed non-parametric Mann-Whitney test or one-way ANOVA test followed by Bonferroni’s post hoc test (when three groups were compared). Only statistically significant pairwise comparisons are indicated in the figures. We used a mixed linear model to explore the association of UMOD rs4293393 with square-root-transformed daytime urinary uromodulin-to-creatinine ratio, in order to better approximate a normal distribution of the residuals and to take familial correlations into account. We expressed continuous measures as the mean ± the standard deviation; averages of measurements as the mean ± the standard error of the mean. We set significance level to P < 0.05.
Supplementary Material
Acknowledgements
We thank M. Azizi, A. Blanchard, G. Capasso, Y. Cnops, A. Creatore, S. Delli Carpini, P. Houillier, X. Jeunemaitre, N. Morel, S. Terryn, R. Latini, M. Carrel for help, technical assistance and fruitful discussions. We are grateful to S. Bourgeois (University of Zurich) for providing MKTAL cells, to D. Alessi (University of Dundee) and B. Forbush (Yale University) for antibodies, to D. Schock-Kusch and N. Gretz (University of Heidelberg) for FITC-sinistrin clearance reagents and technical assistance.
This work was supported by Telethon-Italy (TCR08006), the Italian Ministry of Health (grant RF-2010-2319394), ABN (Associazione per il Bambino Nefropatico), the Belgian agencies FNRS and FRSM, a Concerted Research Action (10/15-029), an Inter-university Attraction Pole (IUAP) program initiated by the Belgian Science Policy Office, the Gebert Rüf Stiftung (Project GRS-038/12), the National Centre of Competence in Research (NCCR) Kidney.CH (Swiss National Science Foundation), the Swiss National Science Foundation project grant 310030_146490 and the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 246539 (Marie Curie) and grant 305608 (EURenOmics). The SKIPOGH project is funded by the Swiss National Science Foundation (33CM30-124087/1; 33CM30_140331). L. R. is an Associate Telethon Scientist.
The SKIPOGH team
Murielle Bochud12, Michel Burnier13, Olivier Devuyst2, Pierre-Yves Martin14, Markus Mohaupt15, Fred Paccaud12, Antoinette Pechére-Bertschi16, Bruno Vogt15, Daniel Ackermann15, Georg Ehret17, Idris Guessous12,18, Belen Ponte14, Menno Pruijm13
12Institute of Social and Preventive Medicine (IUMSP), Lausanne University Hospital (CHUV), Lausanne, Switzerland
13Department of Nephrology, Lausanne University Hospital (CHUV), Lausanne, Switzerland
14Department of Nephrology, Geneva University Hospitals, Geneva, Switzerland
15Department of Nephrology, Hypertension and Clinical Pharmacology, Inselspital, University Hospital and University of Bern, Berne, Switzerland
16Hypertension Unit, Geneva University Hospitals, Geneva, Switzerland
17Cardiology, Department of Specialties of Internal Medicine, Geneva University Hospitals, Geneva, Switzerland
18Unit of Population Epidemiology, Division of Primary Care Medicine, Department of Community Medicine and Primary Care and Emergency Medicine, Geneva University Hospitals, Geneva, Switzerland
Footnotes
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Kearney PM, et al. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Levey AS, et al. Chronic kidney disease as a global public health problem: approaches and initiatives - a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007;72:247–259. doi: 10.1038/sj.ki.5002343. [DOI] [PubMed] [Google Scholar]
- 3.Padmanabhan S, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1001177. doi:10.1371/journal.pgen.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kottgen A, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat. Genet. 2009;41:712–717. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattaro C, et al. A meta-analysis of genome-wide data from five European isolates reveals an association of COL22A1, SYT1, and GABRR2 with serum creatinine level. BMC Med. Genet. 2010 doi: 10.1186/1471-2350-11-41. doi:10.1186/1471-2350-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, et al. Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet. 2010 doi: 10.1371/journal.pgen.1001039. doi:10.1371/journal.pgen.1001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boger CA, et al. Association of eGFR-Related Loci Identified by GWAS with Incident CKD and ESRD. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002292. doi:10.1371/journal.pgen.1002292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pattaro C, et al. Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet. 2012 doi: 10.1371/journal.pgen.1002584. doi:10.1371/journal.pgen.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 11.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482:98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckardt KU, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet. 2013 doi: 10.1016/S0140-6736(13)60439-0. doi:10.1016/S0140-6736(13)60439-0. [DOI] [PubMed] [Google Scholar]
- 13.O’Seaghdha CM, Fox CS. Genome-wide association studies of chronic kidney disease: what have we learned? Nat. Rev. Nephrol. 2012;8:89–99. doi: 10.1038/nrneph.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padmanabhan S, Newton-Cheh C, Dominiczak AF. Genetic basis of blood pressure and hypertension. Trends Genet. 2012;28:397–408. doi: 10.1016/j.tig.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Rampoldi L, Scolari F, Amoroso A, Ghiggeri G, Devuyst O. The rediscovery of uromodulin (Tamm-Horsfall protein): from tubulointerstitial nephropathy to chronic kidney disease. Kidney Int. 2011;80:338–347. doi: 10.1038/ki.2011.134. [DOI] [PubMed] [Google Scholar]
- 16.Bates JM, et al. Tamm-Horsfall protein knockout mice are more prone to urinary tract infection: rapid communication. Kidney Int. 2004;65:791–797. doi: 10.1111/j.1523-1755.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, et al. Progressive renal papillary calcification and ureteral stone formation in mice deficient for Tamm-Horsfall protein. Am. J. Physiol. Renal Physiol. 2010;299:F469–478. doi: 10.1152/ajprenal.00243.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bachmann S, et al. Renal effects of Tamm-Horsfall protein (uromodulin) deficiency in mice. Am. J. Physiol. Renal Physiol. 2005;288:F559–567. doi: 10.1152/ajprenal.00143.2004. [DOI] [PubMed] [Google Scholar]
- 19.Mutig K, et al. Activation of the bumetanide-sensitive Na+,K+,2Cl− cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J. Biol. Chem. 2011;286:30200–30210. doi: 10.1074/jbc.M111.222968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Renigunta A, et al. Tamm-Horsfall glycoprotein interacts with renal outer medullary potassium channel ROMK2 and regulates its function. J. Biol. Chem. 2011;286:2224–2235. doi: 10.1074/jbc.M110.149880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart TC, et al. Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J. Med. Genet. 2002;39:882–892. doi: 10.1136/jmg.39.12.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy AA. Scraping fibrosis: UMODulating renal fibrosis. Nat. Med. 2011;17:553–555. doi: 10.1038/nm0511-553. [DOI] [PubMed] [Google Scholar]
- 23.Bernascone I, et al. A transgenic mouse model for uromodulin-associated kidney diseases shows specific tubulo-interstitial damage, urinary concentrating defect and renal failure. Hum. Mol. Genet. 2010;19:2998–3010. doi: 10.1093/hmg/ddq205. [DOI] [PubMed] [Google Scholar]
- 24.Simpson RU, Hershey SH, Nibbelink KA. Characterization of heart size and blood pressure in the vitamin D receptor knockout mouse. J. Steroid Biochem. Mol. Bio. 2007;103:521–524. doi: 10.1016/j.jsbmb.2006.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han J, et al. Age-related changes in blood pressure in the senescence-accelerated mouse (SAM): aged SAMP1 mice manifest hypertensive vascular disease. Lab. Anim. Sci. 1998;48:256–263. [PubMed] [Google Scholar]
- 26.Schreiber A, et al. Transcutaneous measurement of renal function in conscious mice. Am. J. Physiol. Renal Physiol. 2012;303:F783–F788. doi: 10.1152/ajprenal.00279.2012. [DOI] [PubMed] [Google Scholar]
- 27.Giménez I, Forbush B. Regulatory phosphorylation sites in the NH2 terminus of the renal Na-K-Cl cotransporter (NKCC2) Am. J. Physiol. Renal Physiol. 2005;289:F1341–F1345. doi: 10.1152/ajprenal.00214.2005. [DOI] [PubMed] [Google Scholar]
- 28.Ponce-Coria J, et al. Regulation of NKCC2 by a chloride-sensing mechanism involving the WNK3 and SPAK kinases. Proc. Natl. Acad. Sci. U.S.A. 2008;105:8458–8463. doi: 10.1073/pnas.0802966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaeffer C, Santambrogio S, Perucca S, Casari G, Rampoldi L. Analysis of uromodulin polymerization provides new insights into the mechanisms regulating ZP domain-mediated protein assembly. Mol. Biol. Cell. 2009;20:589–599. doi: 10.1091/mbc.E08-08-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piechotta K, Lu J, Delpire E. Cation chloride cotransporters interact with the stress-related kinases Ste20-related proline-alanine-rich kinase (SPAK) and oxidative stress response 1 (OSR1) J. Biol. Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- 31.Richardson C, et al. Regulation of the NKCC2 ion cotransporter by SPAK-OSR1-dependent and -independent pathways. J. Cell. Sci. 2011;124:789–800. doi: 10.1242/jcs.077230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormick JA, et al. A SPAK isoform switch modulates renal salt transport and blood pressure. Cell Metab. 2011;14:352–364. doi: 10.1016/j.cmet.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Devuyst O. Salt wasting and blood pressure. Nat. Genet. 2008;40:495–496. doi: 10.1038/ng0508-495. [DOI] [PubMed] [Google Scholar]
- 34.Ying WZ, Sanders PW. Dietary salt regulates expression of Tamm-Horsfall glycoprotein in rats. Kidney Int. 1998;54:1150–1156. doi: 10.1046/j.1523-1755.1998.00117.x. [DOI] [PubMed] [Google Scholar]
- 35.Torffvit O, Melander O, Hulten UL. Urinary excretion rate of Tamm-Horsfall protein is related to salt intake in humans. Nephron Physiol. 2004;97:31–36. doi: 10.1159/000077600. [DOI] [PubMed] [Google Scholar]
- 36.Stubbe J, Madsen K, Nielsen FT, Skott O, Jensen BL. Glucocorticoid impairs growth of kidney outer medulla and accelerates loop of Henle differentiation and urinary concentrating capacity in rat kidney development. Am. J. Physiol. Renal Physiol. 2006;291:F812–822. doi: 10.1152/ajprenal.00477.2005. [DOI] [PubMed] [Google Scholar]
- 37.Ferrandi M, et al. alpha- and beta-Adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J. Mol. Med. 2010;88:203–217. doi: 10.1007/s00109-009-0549-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulder WJ, Hillen HF. Renal function and renal disease in the elderly: Part I. Eur. J. Intern. Med. 2001;12:86–97. doi: 10.1016/s0953-6205(01)00121-2. [DOI] [PubMed] [Google Scholar]
- 39.Chen G, et al. Increased susceptibility of aging kidney to ischemic injury: identification of candidate genes changed during aging, but corrected by caloric restriction. Am. J. Physiol. Renal Physiol. 2007;293:F1272–1281. doi: 10.1152/ajprenal.00138.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manunta P, et al. Physiological interaction between alpha-adducin and WNK1-NEDD4L pathways on sodium-related blood pressure regulation. Hypertension. 2008;52:366–372. doi: 10.1161/HYPERTENSIONAHA.108.113977. [DOI] [PubMed] [Google Scholar]
- 42.Pruijm M, et al. Heritability, determinants and reference values of renal length: a family-based population study. Eur. Radiol. doi: 10.1007/s00330-013-2900-4. doi:10.1007/s00330-013-2900-4. [DOI] [PubMed] [Google Scholar]
- 43.K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am. J. Kidney Dis. 2002;39:S1–266. [PubMed] [Google Scholar]
- 44.Youhanna S, et al. Determination of uromodulin in human urine: influence of storage and processing. Nephrol. Dial. Transplant. 2013 doi: 10.1093/ndt/gft345. Accepted for publication in. [DOI] [PubMed] [Google Scholar]
- 45.Dahan K, et al. A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J. Am. Soc. Nephrol. 2003;14:2883–2893. doi: 10.1097/01.asn.0000092147.83480.b5. [DOI] [PubMed] [Google Scholar]
- 46.Barr DB, et al. Urinary creatinine concentrations in the U.S. population: implications for urinary biologic monitoring measurements. Environ. Health Perspect. 2005;113:192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bourgeois S, et al. Differentiated thick ascending limb (TAL) cultured cells derived from SV40 transgenic mice express functional apical NHE2 isoform: effect of nitric oxide. Pflugers Arch. 2003;446:672–683. doi: 10.1007/s00424-003-1108-x. [DOI] [PubMed] [Google Scholar]
- 48.Terryn S. A primary culture of mouse proximal tubular cells, established on collagen-coated membranes. Am. J. Physiol. Renal. Physiol. 2007;293:F476–F485. doi: 10.1152/ajprenal.00363.2006. [DOI] [PubMed] [Google Scholar]
- 49.Glaudemans B, et al. A primary culture system of mouse thick ascending limb cells with preserved function and uromodulin processing. Pflugers Arch. 2013 doi: 10.1007/s00424-013-1321-1. doi: 10.1007/s00424-013-1321-1. [DOI] [PubMed] [Google Scholar]
- 50.Zaarour N, Demaretz S, Defontaine N, Mordasini D, Laghmani K. A highly conserved motif at the COOH terminus dictates endoplasmic reticulum exit and cell surface expression of NKCC2. J. Biol. Chem. 2009;284:21752–21764. doi: 10.1074/jbc.M109.000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 52.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hummon AB, Lim SR, Difilippantonio MJ, Ried T. Isolation and solubilization of proteins after TRIzol extraction of RNA and DNA from patient material following prolonged storage. Biotechniques. 2007;42:467–470. 472. doi: 10.2144/000112401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sorensen MV, et al. Rapid dephosphorylation of the renal sodium chloride cotransporter in response to oral potassium intake in mice. Kidney Int. 2013;83:811–824. doi: 10.1038/ki.2013.14. [DOI] [PubMed] [Google Scholar]
- 55.Flemmer AW, Gimenez I, Dowd BFX, Darman RB, Forbush B. Activation of the Na-K-Cl cotransporter NKCC1 detected with a phospho-specific antibody. J. Biol. Chem. 2002;277:37551–37558. doi: 10.1074/jbc.M206294200. [DOI] [PubMed] [Google Scholar]
- 56.Rafiqi FH, et al. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol. Med. 2010;2:63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner CA, et al. Mouse model of type II Bartter’s syndrome. II. Altered expression of renal sodium- and water-transporting proteins. Am. J. Physiol. Renal Physiol. 2008;294:F1373–F1380. doi: 10.1152/ajprenal.00613.2007. [DOI] [PubMed] [Google Scholar]
- 58.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaplan MR, Plotkin MD, Brown D, Hebert SC, Delpire E. Expression of the mouse Na-K-2Cl cotransporter, mBSC2, in the terminal inner medullary collecting duct, the glomerular and extraglomerular mesangium, and the glomerular afferent arteriole. J. Clin. Invest. 1996;98:723–730. doi: 10.1172/JCI118844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.