Abstract
Here we identify the amino acid transporter AnsP1 as the unique aspartate importer in the human pathogen Mycobacterium tuberculosis. Metabolomic analysis of a mutant inactivated in AnsP1 revealed the transporter is essential for M. tuberculosis to assimilate nitrogen from aspartate. Virulence of the AnsP1 mutant is impaired in vivo, revealing aspartate is a primary nitrogen source required for host colonization by the tuberculosis bacillus.
Mycobacterium tuberculosis, the causative agent of tuberculosis (TB), is still responsible for nearly 1.5 million deaths annually, and new drugs and vaccines are needed to achieve eradication in the decades to come. In this context, a better understanding of M. tuberculosis metabolic requirements during host colonization may help identify targets for novel antimicrobials. For instance, flexible catabolic pathways allowing utilization of host-derived fatty acids, cholesterol and glucose, were reported to provide M. tuberculosis with multiple carbon and energy sources, and to play a key part in mycobacterial physiology and virulence1-5. Inhibitors of such pathways represent today one of the most promising therapeutic approaches for TB in combination with the current set of antibiotics5.
In addition to carbon, nitrogen is an essential component of biomolecules, including amino acids, nucleotides and organic cofactors, and nitrogen sources used by the pathogen during infection have yet to be identified6. Recent studies have substantiated past observations establishing amino acids (asparagine, in particular) as the favorite in vitro nitrogen source for M. tuberculosis in comparison to inorganic nitrogen donors (e.g. ammonia or nitrates)6-9. In the present study, we provide unequivocal evidence that the putative asparagine transporter AnsP1/Rv2127 (ref. 10) is an essential transporter in the assimilation of nitrogen from aspartate, alluding that aspartate-dependent pathways may enhance the fitness of M. tuberculosis during colonization of its host.
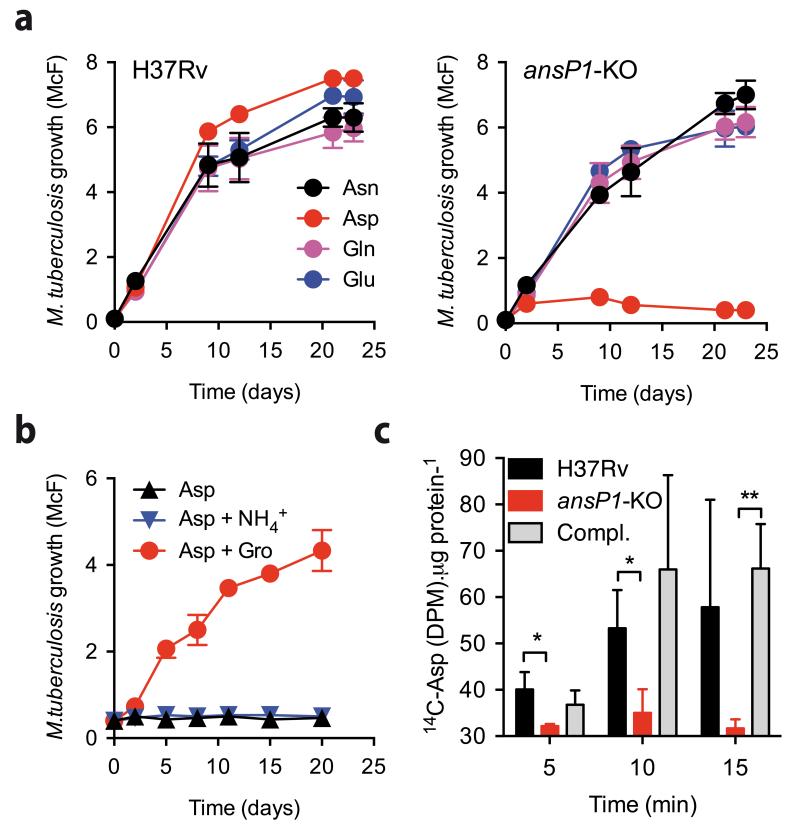
We initially chose to inactivate the AnsP1-encoding gene (Supplementary Results, Supplementary Fig. 1) largely due to the strong potential of AnsP1 to be an asparagine transporter in M. tuberculosis10, as reflected by the 55% identity and 70% similarity to the Salmonella enterica asparagine transporter AnsP11. Yet, when we evaluated the capacity of the ansP1-knock out (KO) mutant to grow in minimal medium containing asparagine or other structurally related amino acids (i.e. glutamine and glutamate) as sole nitrogen sources, there was no significant growth defect as compared to the wild-type strain (Fig. 1a). This result suggested that AnsP1 is either irrelevant for the transport of these amino acids or compensated by unknown transporters with redundant substrate affinities. Notwithstanding, we found that the ansP1-KO strain was strikingly unable to grow with aspartate as sole nitrogen source, a phenotype that was fully reversed upon genetic complementation of the mutant with a cosmid encompassing the ansP1 region of the genome (Supplementary Fig. 2a). This surprising phenotype was observed either when aspartate was given from the start of the culture (Fig. 1a), or when the nitrogen source was switched to aspartate after initial bacterial replication (Supplementary Fig. 2b), indicating AnsP1 is continuously required for growth on aspartate. We excluded a possible toxic effect of aspartate to the ansP1-KO mutant because i/the loss of viability of the mutant over time was not different in the presence of aspartate compared to that in a culture medium void of any nitrogen source (Supplementary Fig. 2c), and ii/the growth of the mutant normally resumed upon addition of a second nitrogen source (i.e. asparagine), to an aspartate-containing culture medium (Supplementary Fig. 2d). Beyond the essential role as a nitrogen source, we tested the capacity of aspartate to support mycobacterial growth when provided as a sole carbon and energy source. In line with a previous report12, we found aspartate failed to foment mycobacterial growth when provided as sole carbon and energy source even in the presence of ammonium as a nitrogen source, unless glycerol was included (as a carbon source) to the medium (Fig. 1b). Nevertheless, our metabolomics analysis of bacteria grown on U-13C-aspartate revealed the presence of 13C in various metabolites, including α-ketoglutarate for instance (Supplementary Fig. 3). In fact, the addition of glycerol enhanced the carbon incorporation from aspartate (Supplementary Fig. 3), indicating a synergistic utilization of carbon sources in M. tuberculosis, as we previously reported2. Finally, as proof-of-principle, we demonstrated AnsP1 is an aspartate importer using 14C-labeled aspartate in a standard amino acid transport assay (Fig. 1c). Collectively, our results show AnsP1 is the unique aspartate importer in M. tuberculosis, and this amino acid transporter is strictly required for growth of the pathogen with aspartate as sole nitrogen source, although aspartate-derived carbon can enter the mycobacterial carbon flux to some extends.
Figure 1. AnsP1 is essential for M. tuberculosis growth on aspartate as sole nitrogen source.
(a,b) Growth of M. tuberculosis H37Rv, the ansP1-KO mutant and the ansP1-KO complemented (Compl.) strains in various conditions. Growth was measured by monitoring turbidity; data represent mean ±s.d. of triplicate samples and are representative of at least three independent experiments. (a) Bacteria were grown in minimal medium containing 5 mM asparagine (Asn), aspartate (Asp), glutamine (Gln) or glutamate (Glu), as sole nitrogen sources. (b) Bacteria were grown in minimal medium containing 50 mM aspartate (Asp) only, 50 mM aspartate and 15 mM ammonium (Asp+NH4+), or 50 mM aspartate and 10 g/L glycerol (Asp+Gro), as nitrogen or carbon sources. (c) 14C-aspartate uptake assay with M. tuberculosis H37Rv, the ansP1-KO mutant and its complemented strains (Compl.). Bacteria previously grown in 7H9 with 5 mM aspartate, were harvested and resuspended in an uptake buffer containing a mix of 14C-radiolabeled and non-labeled aspartate to obtain a final concentration of 20 μM aspartate. Bacteria were incubated at 37°C and samples were removed; bacteria-associated 14C radioactivity was quantified at the indicated time points. Data are expressed as the number of disintegrations per minute (DPM) per total protein concentration (14C-Asp (DPM).μg protein−1), represent mean ±s.d. of triplicate samples, are representative of three independent experiments, and were analyzed using the Student’s t test; *, P<0.05; **, P<0.01.
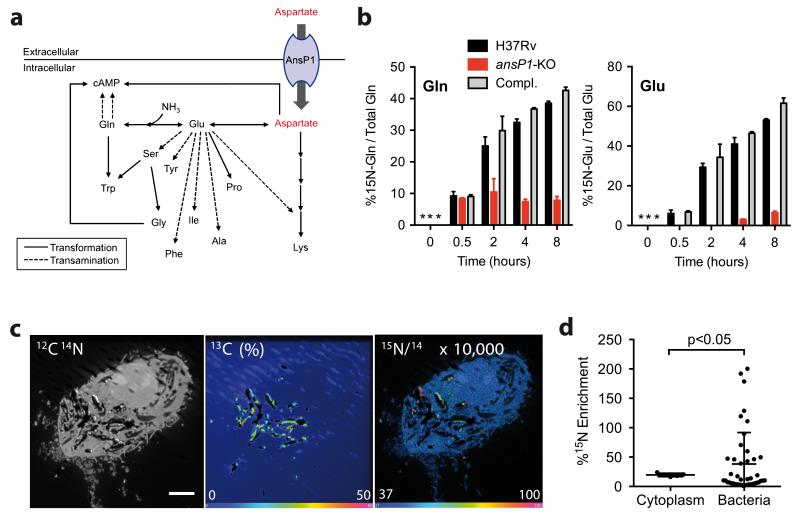
To further explore the contribution of aspartate to nitrogen assimilation in M. tuberculosis, we conducted a series of metabolomic investigations, using stable isotopic labeling. The 15N incorporation from 15N-labeled aspartate into the mycobacterial N-containing metabolome (defined here as all extractable N-containing small molecules present in the bacterium), was followed by liquid chromatography accurate time-of-flight mass spectrometry (TOF-MS; Fig. 2a & Supplementary Fig. 4a). In M. tuberculosis H37Rv, the 15N assimilation was first observed in glutamine and glutamate, two amino acids that play an important role in nitrogen assimilation and storage (Fig. 2b). This result is in agreement with previous studies showing nitrogen incorporation takes place first in these two major nitrogen-acceptor and -donor. In the ansP1-KO mutant, however, the nitrogen transfer from species in bacteria13,14 aspartate into glutamine and glutamate was abolished, a phenotype that was reversed by genetic complementation (Fig. 2b). This deficiency in nitrogen assimilation was specific to aspartate as nitrogen-donor given that the ansP1-KO mutant was able to incorporate it from 15N2-asparagine at a rate similar to those observed for the wild-type and complemented strains (Supplementary Fig. 4b). Similar to glutamine and glutamate, the mutant strain was impaired in nitrogen incorporation into secondary N-containing metabolites resulting either from transamination reactions (Fig. 2a), such as alanine, serine, tyrosine, phenylalanine and isoleucine (Supplementary Fig. 5a), or from other N-incorporation reactions (Fig. 2a), such as proline, glycine, lysine and tryptophan (Supplementary Fig. 5b). As expected, the 15N-labeling of nucleosides, that are biosynthetically more distant from aspartate, such as cyclic adenosine monophosphate (cAMP), could not be detected in the mutant strain (Fig. 2a, Supplementary Fig. 5c). Therefore, our results unequivocally demonstrate that aspartate-derived nitrogen is fully assimilated in M. tuberculosis, and this process is exclusively AnsP1-dependent.
Figure 2. AnsP1 is essential for Mycobacterium tuberculosis nitrogen assimilation from aspartate.
(a) Nitrogen incorporation pathways from aspartate into various N-containing metabolites resulting from transamination (dotted line) and/or transformation (plain lines) reactions. (b) Frequency of 15N-glutamine and 15N-glutamate detected in the presence of 15N-aspartate in either M. tuberculosis wild type (H37Rv), the ansP1-KO mutant, or its complemented strain (Compl.). Data represent mean±s.d. of triplicate samples and are representative of at least two independent experiments. *, no signal detected. Any signal below 5-10% corresponds to background noise. (c) 13C-labeled bacteria were used to infect mouse macrophages. After pulsing the cells with 15N-aspartate, 13C and 15N isotope compositions were analyzed by NanoSIMS. Images display a representative infected cell. Left panel is the as recorded 12C14N− image showing the histological aspect of the cell (scale bar represents 5 μm). The central panel represents the 13C atomic fraction map (in %) of the corresponding area. The right panel shows the 15N/14N ratio image indicating the 15N-aspartate uptake. For enhanced visibility, the ratio was multiplied by 1×104. The 15N/14N ratio at natural 15N abundance appears blue. (d) Quantification of 15N enrichment (compared to the resin) in surface areas chosen in the cell cytoplasm (n=5), and intracellular 13C labeled bacteria (n=43). Data represent mean ±s.d. and were analyzed using the Student’s t test. The 15N-enriched phagosomes, arbitrarily defined as those vacuoles with a 15N enrichment above mean+3s.d. of that observed in the host cell cytoplasm, represent 34.9% of all phagosomes.
In order to assess the physiological relevance of our findings, we next asked whether M. tuberculosis could access aspartate inside its natural cellular niche, namely the macrophage. Indeed, M. tuberculosis is a facultative intracellular pathogen that resists innate antimicrobial mechanisms and thrives inside macrophages and other phagocytes15. To address this question, we used an ionic micro-probe based on dynamic secondary ion mass spectroscopy (NanoSIMS), a powerful technology that allows spatial imaging of elemental and isotopic distribution with high resolution16. Murine macrophages were infected for 20 hrs with 13C-labeled M. tuberculosis H37Rv, and pulsed with 15N-aspartate for 4 hrs before fixation and analysis. The 13C-labeled bacteria were unambiguously localized inside infected cells (Fig. 2c, middle panel). Aspartate-derived 15N signal was found distributed in spots (Fig. 2c, right panel), and a correlation analysis showed every 15N-containing spots co-localized with a spot from a bacterium. Interestingly, a fraction of intracellular bacteria presented a 15N enrichment higher than that observed in the host cell cytoplasm, as compared to the background natural isotopic signal observed in the resin (Fig. 2d, Supplementary Fig. 6). Analysis of multiple cells indicated that at least ~35% of the mycobacterial phagosomes were reproducibly enriched in 15N, as compared to the host cell cytoplasm. These results strongly suggested M. tuberculosis could access aspartate inside host macrophages. The bimodal distribution of 15N-aspartate accumulation within the M. tuberculosis vacuoles (Fig. 2d & Supplementary Fig. 6b) is reminiscent of the distribution observed for various maturation markers to the mycobacterial phagosomes17,18. Such heterogeneity likely reflects variability of the metabolic status of the phagocytosed bacteria. Whether aspartate accumulates differentially to the vacuoles depending on various cellular parameters (e.g. differentiation program, activation status) remains to be elucidated.
To validate the pertinence of our findings during the infection context, we first tested the ability of the ansP1-KO mutant to proliferate inside macrophages. Since aspartate is accessible as a nitrogen source in the mycobacterial phagosome, we were surprised to find no growth defect for the mutant strain in either resting or activated macrophages (Supplementary Fig. 7). To further investigate the role of Ansp1 in survival in vivo, we conducted infection experiments in immune-deficient (SCID) or -competent (C57BL/6) animals. Contrary to our in vitro approach with macrophages, we observed the virulence of the ansP1-KO mutant was severely compromised both in immuno-deficient SCID (Supplementary Fig. 8a) and immuno-competent C57BL/6 (Supplementary Fig. 8b) mice, as compared to the wild-type and complemented strains. Consistent with the reduced bacterial load observed in animals infected with the mutant strain, immuno-histopathology analysis revealed this strain induces smaller granulomas than its wild-type and complemented counterparts (Supplementary Fig. 8c,d) further evidencing that Ansp1 is involved in M. tuberculosis virulence. The apparent discrepancy between our models of infection most likely reflects the complexity of the nutritional environment and immune-mediated pressure encountered by the bacteria in vivo, as compared to the less physiological conditions encountered in the in vitro culture systems.
Collectively, our results indicate AnsP1 is required for M. tuberculosis virulence, most likely because it allows the bacteria to capture aspartate and incorporate nitrogen from host tissues. Importantly, we found that M. tuberculosis can access aspartate inside host cells. In line with our findings, aspartate was found among the most abundant molecules in M. tuberculosis infection sites in a guinea pig model19. While nitrate- and urea-derived ammonium are often considered important potential sources of nitrogen for M. tuberculosis during infection20, amino acids are usually underestimated from this point of view. In this context, our results reveal for the first time that amino acids serve as an important nitrogen source for M. tuberculosis during infection.
A limited number of M. tuberculosis amino acid transporters have been studied so far6. Here we show that AnsP1 is the unique aspartate transporter in this pathogen. This finding is unexpected because AnsP1 belongs to the family of Amino Acid Transporters (AAT, 2.A.3.1 family21) and is not homologous to classical bacterial Dicarboxylate Amino Acid:Cation Symporter (DAACS, 2.A.23 family22) usually involved in aspartate uptake. Of note, we found the AnsP1 homologue AnsP2 (Supplementary Fig. 9a) cannot compensate for AnsP1 deficiency in aspartate transport, since a mutant inactivated in the ansP2 gene is able to grow on aspartate as sole nitrogen source (Supplementary Fig. 9b). The role of AnsP2 and of the other M. tuberculosis putative amino acid transporters in mycobacterial physiology and virulence remains to be elucidated. Furthermore, the AnsP1 transporter is duplicated in the obligate pathogen Mycobacterium leprae, the causative agent of leprosy (Supplementary Fig. 9a), and all the other amino acid transporters found in M. tuberculosis are either deleted or pseudogenized in this species23. This suggests an important role for aspartate utilization in pathogenicity of the leprosy bacillus as well.
In conclusion, we propose aspartate is a primary nitrogen source for M. tuberculosis during infection, and AnsP1 as an essential aspartate transporter that ultimately enhances this pathogen’s fitness within its host. We envision that biosynthesis pathways depending on aspartate as nitrogen provider may constitute a novel “chink in the armor” for TB and possibly leprosy bacilli to be targeted for therapeutic purposes.
Online Methods
Mycobacteria and classic culture conditions
Mycobacteria were grown at 37°C in Middlebrook 7H9 medium (Difco) supplemented with 10% albumin-dextrose-catalase supplement (Difco) and 0.05% Tween-80 (Sigma), or on Middlebrook 7H11 agar medium (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase supplement (OADC, Difco). When required, kanamycine or hygromycine were added to the culture media to a final concentration of 50 μg/ml.
Construction of ansP1-KO and ansP2-KO mutants and complemented strains
ansP1-KO and ansP2-KO mutant strains of Mycobacterium tuberculosis H37Rv containing disrupted ansP1 (Rv2127)::KanR or ansP2 (Rv0346c)::KanR alleles were constructed by allelic exchange using a recombineering method24. H37Rv:pJV53 was grown in 7H9-ADC-Tween80 in the presence of hygromycin until mid-log phase and induced by 0.2% acetamide (Sigma) overnight at 37°C. After induction, electrocompetent bacteria were prepared. Electroporation was performed with linearized digestion fragments of kanamycine resistant cassette-interrupted genes flanking with homologous regions (400-700 pb length). After 72 h incubation at 37 °C, bacteria were plated onto 7H11-OADC agar medium in the presence of kanamycin. For complemented strain construction, the pYUB412-derived integrative cosmid I257 encompassing the region 2,377.9-2,415.5 kb in the genome of M. tuberculosis H37Rv, and containing a hygromycin resistance cassette25 was used. Mutation and complementation events were verified by PCR and sequencing using the following primers: P1, 5′-TTCGTCGGCGAACCCACCTTTCCCG-3′, and P2, 5′-TCATCAAGTCACCGGCACCGTGGAA-3′, for the ansP1-KO strain; P3, 5′-CGGGTACTTGGGATGGGCCG TCACA-3′, and P4, 5′-GTGACCAGCACTATGGGCAGCAGA-3′, for the ansP2-KO strain.
Macrophages & infection procedure
Bone marrow cells were flushed from the femurs and tibias of 6-8 weeks old female C57BL/6 mice, and cultured in Petri dishes (2.106 cells/dish) in RPMI 1640 GlutaMax (GIBCO) supplemented with 10% fetal calf serum (FCS, Pan-Biotech) and 20 ng/ml macrophage colony-stimulating factor (M-CSF, Peprotech) at 37°C in the presence of 5% CO2. At day 6, cells were transferred to 24-well plastic plates (2.105 cells/well). For macrophage activation, cells were incubated with 10 ng/ml interferon-gamma (IFNγ, Peprotech) over night prior infection. Infection was performed in triplicate at a multiplicity of infection of 0.1 bacterium per cell for 4 h at 37°C. Cells were then washed 2 times with DPBS before addition of fresh medium. At day 0, 2, 5 and 7, cells were lysed in 0.01% Triton X-100 (Sigma), and serial dilutions of the lysates were plated onto 7H11-OADC agar medium for CFU scoring.
Mouse infection
All animal experiments were performed in animal facilities that meet all legal requirements in France and by qualified personnel in such a way to minimize discomfort for the animals. All procedures including animal studies were conducted in strict accordance with French laws and regulations in compliance with the European community council directive 68/609/EEC guidelines and its implementation in France. All protocols were reviewed and approved by the Comité d’Ethique Midi-Pyrénées (reference MP/04/26/07/03). Six- to eight-week-old female C57BL/6 or SCID mice were anesthetized with a cocktail of ketamine (60 mg/kg; Merial) and xylasine (10 mg/kg; Bayer) and infected intranasally with 500 CFUs of the various mycobacterial strains in 25 μl of PBS-0.01% Tween 80. After infection, SCID mice were sacrificed when first signs of hurt were observed. For C57BL/6 mice, at time 21 and 42 days post-infection, mice were sacrificed and lung and spleen homogenates were plated onto 7H11 agar plates for CFU scoring. In parallel, lungs of infected mice were filled with 2% low melting agarose and fixed for 24 hours in a Periodate-Lysine-Paraformaldehyde (PLP) fixative. After fixation, lungs were incubated in serial incubations in sucrose solutions (10, 20 and 30%). Finally, lungs were sectioned (30 μm) and stained for confocal microscopy with F4/80 Pacific Blue, CD4 Alexa 488 and Gr1 Alexa 647 (Biolegend). Z stacks were acquired in an Olympus FV1000 confocal microscope and analyzed in Imaris software (Bitplane).
M. tuberculosis growth in minimal media
Bacteria were grown in Sauton’s modified medium containing 0.05% Tween-80, 0.5 g/L KH2PO4, 0.5 g/L MgSO4, 2 g/L citric acid, 10 g/L glycerol and 5 mM aspartate, asparagine, glutamate, glutamine or ammonium sulfate (pH 6.5-7.0). In media prepared for carbon assimilation, aspartate and ammonium sulfate were added at a final concentration of 50 mM and 15 mM, respectively, Tween-80 was replaced by 0.05 % tyloxapol (Sigma) and citrate was omitted. Cultures were performed in triplicate in glass tubes and bacterial growth was monitored by plating onto 7H11-OADC agar medium and CFU scoring, or by measuring turbidity (in McFarland units) over time using a Densimat apparatus (BioMerieux).
14C-Aspartate uptake experiment
Aspartate uptake experiments were carried out as described before for asparagine with minor modification8. Bacteria were grown in Middlebrook 7H9 containing 0.05% Tween 80 and aspartate (5mM) at 37°C. Bacteria were harvested by centrifugation when an OD600 of 0.5 was reached. Bacterial pellets were washed twice in uptake buffer [50 mM Tris-HCl pH 6.9, 15 mM KCl, 10 mM (NH4)2SO4, 0.05% Tween 80] and resuspended in the same buffer. Radiolabeled 14C-aspartate (PerkinElmer, Purity: 97 atom % 14C) and non-labeled aspartate (Sigma) were mixed and added to cell suspensions to obtain a final concentration of 20 μM aspartate. The mixtures were incubated at 37°C and 250 μl of samples were removed at the indicated time points. Bacteria were collected on a 0.45 μm Spin-X centrifuge tube filter (Costar) by mixing with an equal volume of 10% paraformaldehyde (PFA, Polyscience, Inc) containing 0.1 M LiCl (Sigma). Filters radioactivity was determined in a liquid scintillation counter (Packard). The uptake rate was expressed in desintegration per minute (DPM) per total protein concentration (14C-Asp (DPM).μg protein−1).
Metabolite extraction experiments
Bacteria were cultivated to an OD600 of 1 in 7H9-0.05% Tween-80. Bacteria were centrifuged and resuspended in DPBS (3-fold concentration). 1 mL was transferred to a filter (Fisher) mounted on a filtration device (Fisher) and connected to a trap and vacuum line. Filters were transferred to solid media containing 0.5 g/L KH2PO4, 0.5 g/L MgSO4, 2 g/L citric acid, 10 g/L glycerol, aspartate (2mM) and 1.5% agar (Invitrogen) prepared in tap water and neutralized to pH 6.5-7.0 with NaOH before autoclaving. Plates were incubated for 5 days at 37°C. Three filters were used per strain and time point. For labeling experiments, filters were transferred on equivalent plates where aspartate was replaced by 15N-aspartate (2 mM, Sigma, Purity 98 atom % 15N) and incubated for 0.5, 2, 4 or 8 hours at 37°C. For U-13C incorporation experiments, bacteria were cultivated in the same conditions without citric acid and with or without glycerol (10 g/L); next, aspartate was replaced by 13C aspartate (2 mM, Sigma, Purity 98 atom % 13C). 15N incorporation from asparagine was evaluated following the same procedure, except that aspartate was replaced by asparagine and that 7H10 was used instead of solid minimal medium; in this case, 15N2-asparagine (Sigma, Purity 98 atom % 15N) was used. At each time point, filters were plunged into 1 mL acetonitrile/methanol/water (2:2:1, v/v/v) mixture at −40°C. Bacteria were then broken by glass beads using a bead-beater (5 min at 30 m/s). After centrifugation, supernatants were collected and filtered through a Spin-X column 0.2 μm at 14,000 rpm for 15 minutes. Extracts were stored at −80°C before analysis.
Liquid chromatography-mass spectrometry
Aqueous normal phase liquid chromatography was performed using an Agilent 1200 LC system equipped with a solvent degasser, binary pump, temperature-controlled auto-sampler (set at 4°C) and temperature-controlled column compartment (set at 20°C), containing a Cogent Diamond Hydride Type C silica column (150 mm × 2.1 mm; dead volume 315 μl), from Microsolv Technology Corporation. Flow-rate of 0.4 ml/min was used. Elution of polar metabolites was carried out using gradient 3 (ref. 26). Briefly, solvent A consists in deionized water (Resistivity ~ 18 MΩ cm), 0.2% acetic acid and solvent B consists in acetonitrile and 0.2% acetic acid, and the gradient as follows: 0 min 85% B; 0-2 min 85% B; 2-3 min to 80% B; 3-5 min 80% B; 5-6 min to 75% B; 6-7 min 75% B; 7-8 min to 70% B; 8-9 min 70% B; 9-10 min to 50% B; 10-11 min 50% B; 11-11.1 min to 20% B; 11.1-14 min hold 20% B. Accurate mass spectrometry was carried out using an Agilent Accurate Mass 6230 TOF apparatus. Dynamic mass axis calibration was achieved by continuous infusion, post-chromatography, of a reference mass solution using an isocratic pump connected to a Multimode ionization source, operated in the positive-ion mode. ESI capillary and fragmentor voltages were set at 3,500 V and 100 V, respectively. The nebulizer pressure was set at 40 psi and the nitrogen drying gas flow rate was set at 10 L/min. The drying gas temperature was maintained at 250°C. The MS acquisition rate was 1.5 spectra/sec and m/z data ranging from 80-1,200 were stored. This instrument routinely enabled accurate mass spectral measurements with an error of less than 5 parts-per-million (ppm), mass resolution ranging from 10,000-25,000 over the m/z range of 121-955 atomic mass units, and a 100,000-fold dynamic range with picomolar sensitivity. Data were collected in the centroid mode in the 4 GHz (extended dynamic range) mode. Detected m/z were deemed to be identified metabolites on the basis of unique accurate mass-retention time identifiers for masses exhibiting the expected distribution of accompanying isotopomers2. Typical variation in abundance for most of the metabolites stayed between 5 and 10% under these experimental conditions.
Stable isotope labeling analysis
Under the experimental conditions described above, M+1 arising from 15N incorporation can be readily distinguished from M+1 arising from natural abundance 13C or from 13C labeling, therefore allowing direct monitoring of 15N labeling and 13C labeling. The extent of 15N or 13C labeling for each metabolite was determined by dividing the summed peak height ion intensities of all 15N or 13C labeled species by the ion intensity of both labeled and unlabeled species, expressed in percent.
Secondary ion mass spectrometry
For 13C labeling, bacteria were grown in minimal medium containing 0.5 g/L KH2PO4, 0.5 g/L MgSO4, 15 mM NH4SO4, 10 g/L 13C3-glycerol (Sigma, Purity: 99 atom % 13C) supplemented with 0.05% tyloxapol (Sigma) and neutralized to pH 6.5-7.0 with NaOH before filtration. IFNγ-activated macrophages, plated on cover glasses, were infected at a multiplicity of infection of 10 bacteria per cell with 13C labeled-bacteria for 4 hours. At 20h post-infection, the culture medium was replaced by fresh RPMI containing 10% FCS and 2 mM 15N-aspartate. After 4h at 37°C, cells were fixed with 4% PFA, 2.5% Glutaraldehyde in a 0.1 M cacodylate buffer (pH 7.4) and dehydrated through successive 30%, 50%, 70%, 90% and 100% ethanol baths. Cells were then directly included in Epoxy resin prior to NanoSIMS analysis. Epoxy resin was prepared by mixing 4.63 mL of EMBed-812 (Sigma) and 3.70 mL of Epoxy embedding medium-hardener DDSA (Sigma) at room temperature under agitation until homogeneity was freached. Then, 1.39 mL of Epoxy embedding medium-hardener MNA (Sigma) was added to the mix prior the addition of 280 μL N,N-Dimethylbenzylamine (Sigma). Once dehydrated, cover glasses containing cells were incubated with Ethanol:Epoxy resin (2:1) for one hour, Ethanol:Epoxy resin (1:2) for one hour, before final incubation in pure Epoxy resin for one hour. Epoxy resin was then replaced by fresh Epoxy resin before incubation of the samples overnight at room temperature. Polymerization of Epoxy resin was performed at 65°C during 12 hours. 150-nm thin section were deposited on clean Silicon chips and were inserted into a NanoSIMS-50 Ion microprobe (CAMECA, Gennevilliers, France) operating in scanning mode16. For the present study, by using a Cs+ primary ion beam tightly focused to a typical probe size of ≈85 nm (distance between 16%-84% of peak intensity from a line scan), five secondary ion species (12C−, 13C−, 12C14N−, 12C15N−, 31P−) were monitored simultaneously. The primary beam steps over the surface of the sample to create images of the selected ion species. The identification of bacteria location was highlighted by high 13C content while the aspartate uptake was revealed by 15N enrichment. After careful Cs+ ion implantation to get steady state ion emission, the acquisition was carried out using multiframe mode. The primary beam intensity was 1 pA and the raster size was from 20 to 35 μm in order to image a whole cell with an image definition of 512 × 512 pixels. With a Dwell time of 2 ms per pixel, up to 15 frames were acquired and the total analysis time was 2 hours. Image treatment was performed using ImageJ software27. First, multiframe images were properly aligned using CN− images as reference before a summed image was obtained for each ion species. A map of 13C atomic fraction was deduced from 12C− and 13C− images. In parallel, ROI’s (region of interest) were manually defined based on the 13C− map so as to outline individual bacterium for data extraction. For 15N/14N ratio quantification, a sample containing no labelled cells was used as working reference for adjusting the detectors. Further, a check was performed on the resin area surrounding the observed cells. The final 13C map, as well as the one for 15N/14N ratio, are displayed in Hue-Saturation-Intensity (HSI) mode. These HSI color images were generated using OpenMIMS, an ImageJ plugin developped by Claude Lechene’s Laboratory28.
Supplementary Material
Acknowledgments
We thank Claude Gutierrez and Denis Hudrisier for carefully reading the manuscript and helpful suggestions. We thank Laurence Lepourry and Flavie Moreau for technical assistance with mouse infections. We thank Alan Benard for providing mouse cells, and Roland Brosch for providing the I257 cosmid. We thank Yannick Verkindère (YV Photographiste, Toulouse, France) for help with graphical work. We thank the PICT-IBiSA imaging facility in the Institut Curie. This work was supported by Agence Nationale de la Recherche (ANR, Contracts SLC-TB & TB-HITS), MRC (MC_UP_A253_1111), and the EU FP7 programme NEWTBVAC (Contract n° 241745). This work also benefited from the TRI RIO Optical Imaging Platform at IPBS (Genotoul, Toulouse, France) supported by grants from the Région Midi-Pyrénées (CPER), the Grand Toulouse community, the ARC (ARC Equipement N°8505), the CNRS and the EU through the FEDER program. AG and GLV hold a fellowship from the Fondation pour la Recherche Médicale (FRM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
References
- 1.McKinney JD, et al. Nature. 2000;406:735–8. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 2.de Carvalho LP, et al. Chem Biol. 2010;17:1122–31. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Marrero J, Trujillo C, Rhee KY, Ehrt S. PLoS Pathog. 2013;9:e1003116. doi: 10.1371/journal.ppat.1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pandey AK, Sassetti CM. Proc Natl Acad Sci U S A. 2008;105:4376–80. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhee KY, et al. Trends Microbiol. 2011;19:307–14. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook GM, et al. Adv Microb Physiol. 2009;55:81–182. 318–9. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyon RH, Hall WH, Costas-Martinez C. J Bacteriol. 1974;117:151–6. doi: 10.1128/jb.117.1.151-156.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song H, et al. Mol Microbiol. 2011;80:900–18. doi: 10.1111/j.1365-2958.2011.07619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song H, Niederweis M. J Bacteriol. 2012;194:956–64. doi: 10.1128/JB.06132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole ST, et al. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 11.Jennings MP, Anderson JK, Beacham IR. Microbiology. 1995;141(Pt 1):141–6. doi: 10.1099/00221287-141-1-141. [DOI] [PubMed] [Google Scholar]
- 12.Youmans AS, Youmans GP. J Bacteriol. 1953;65:96–9. doi: 10.1128/jb.65.1.96-99.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reitzer L. Annu Rev Microbiol. 2003;57:155–76. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 14.Yuan J, Fowler WU, Kimball E, Lu W, Rabinowitz JD. Nat Chem Biol. 2006;2:529–30. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]
- 15.Russell DG. Immunol Rev. 2011;240:252–68. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerquin-Kern JL, Wu TD, Quintana C, Croisy A. Biochim Biophys Acta. 2005;1724:228–38. doi: 10.1016/j.bbagen.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Clemens DL, Horwitz MA. J Exp Med. 1995;181:257–70. doi: 10.1084/jem.181.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S, et al. J Immunol. 1994;153:2568–78. [PubMed] [Google Scholar]
- 19.Somashekar BS, et al. J Proteome Res. 2011;10:4186–95. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 20.Amon J, Titgemeyer F, Burkovski A. J Mol Microbiol Biotechnol. 2009;17:20–9. doi: 10.1159/000159195. [DOI] [PubMed] [Google Scholar]
- 21.Jack DL, Paulsen IT, Saier MH. Microbiology. 2000;146(Pt 8):1797–814. doi: 10.1099/00221287-146-8-1797. [DOI] [PubMed] [Google Scholar]
- 22.Saier MH., Jr. Microbiology. 2000;146(Pt 8):1775–95. doi: 10.1099/00221287-146-8-1775. [DOI] [PubMed] [Google Scholar]
- 23.Cole ST, et al. Nature. 2001;409:1007–11. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 24.van Kessel JC, Hatfull GF. Nat Methods. 2007;4:147–52. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 25.Bange FC, Collins FM, Jacobs WR., Jr. Tuber Lung Dis. 1999;79:171–80. doi: 10.1054/tuld.1998.0201. [DOI] [PubMed] [Google Scholar]
- 26.Pesek JJ, Matyska MT, Fischer SM, Sana TR. J Chromatogr A. 2008;1204:48–55. doi: 10.1016/j.chroma.2008.07.077. [DOI] [PubMed] [Google Scholar]
- 27.Schneider CA, Rasband WS, Eliceiri KW. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lechene C, et al. J Biol. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.