Summary
Background
von Willebrand factor (VWF) variant c.2771G>A; p.R924Q has been described as a benign polymorphism or a possible marker for a null allele and been associated with mild bleeding phenotypes. It was identified in several patients in recent type 1 von Willebrand disease (VWD) studies.
Objectives
To determine whether the p.R924Q allele contributes to reduced VWF levels and type 1 VWD.
Methods
1115 healthy controls and 148 index cases from the MCMDM-1VWD study were genotyped for c.2771G>A; VWF and FVIII levels were analysed in ABO blood group stratified individuals and the p.R924Q variant was expressed in 293 EBNA cells.
Results
c.2771G>A was present in six index cases, five of whom had a second VWF variant which likely contributed to phenotype. A common core haplotype identified in families, which included the rare G allele of c.5843-8C>G was present in the majority of 35 c.2771G>A heterozygous controls. c.2771G>A contributed about 10% variance in VWF and FVIII levels in controls and 35% variance when co-inherited with blood group O. Recombinant p.R924Q VWF had no effect on in vitro expression and heterozygous family members had normal VWFFVIII binding and normal clearance of VWF and FVIII.
Conclusions
The allele bearing c.2771A leads to reductions in VWF and FVIII levels particularly in combination with blood group O. Its inheritance alone may be insufficient for VWD diagnosis, but it appears to be associated with further VWF level reduction in individuals with a second VWF mutation and it contributes to population variance in VWF and FVIII levels.
Keywords: ABO blood group, founder effect, factor VIII, R924Q, type 1 von Willebrand disease, von Willebrand factor
Introduction
Three multicentre studies in the European Union (EU), Canada and the United Kingdom (UK) [1-3] have led to increased understanding of the molecular basis of type 1 von Willebrand disease (VWD), a partial quantitative deficiency of plasma von Willebrand factor (VWF). Candidate VWF mutations were identified in approximately 65% of type 1 VWD index cases (IC) and included many different mutations throughout the VWF gene (VWF), mostly missense alterations (80%). A small proportion of sequence variants were seen consistently in all three studies and included c.2771G>A in exon 21, predicted to result in p.Arg924Gln (p.R924Q).
c.2771G>A was first reported in a patient with type 2N VWD, but was thought to be a benign polymorphism as recombinant VWF carrying the p.R924Q change bound normally to FVIII (VWF:FVIIIB) and the patient had a further novel 2N change, p.C1060R which could explain the phenotype [4]. The EU and Canadian studies each reported c.2771G>A as a candidate mutation [1,2], whereas the UK study considered it likely to be a silent variation [3,5]. It results in substitution of a basic by an uncharged amino acid in a highly conserved region of VWF. Analysis of patient mRNA has recently identified a novel truncated transcript resulting from activation of a cryptic splice site in exon 28 in an individual compound heterozygous for the variant (p.[R816W]+[R924Q]) [6].
In this study, the p.R924Q allele appeared to contribute to phenotype in IC with type 1 VWD and to reductions in both VWF and FVIII in heterozygous healthy controls (HC). The majority of heterozygous individuals share a common founder haplotype that likely originated in Eastern Europe. Recurrence of p.R924Q on different haplotypes along with acquisition of further sequence variants on ancestral p.R924Q alleles may contribute to the variable behaviour observed.
Patients and Methods
Study population
Genomic DNA was available from the MCMDM-1VWD study [1] for IC (n=148) historically diagnosed with type 1 VWD, their affected and unaffected family members (AFM; n=278, UFM; n=311) and healthy controls (HC; n=1115). Participants were predominantly Caucasian and were recruited by partners P1-P12, located in nine European countries [1]. FVIII coagulant activity (FVIII:C), VWF antigen (VWF:Ag), ristocetin cofactor activity (VWF:RCo), bleeding score (BS), multimer classification and ABO blood group were determined previously [7-9].
Ability of VWF to bind recombinant FVIII was determined in IC and AFM as described [10]. Results are expressed as the ratio of the slope of test plasma to that of a HC. Normal range (NR; mean ±2SD) was calculated from slope ratios in 63 UFM with no VWF mutation identified.
Genotyping
The c.2771G>A variant (rs33978901) was genotyped using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) using a Sequenom MassARRAY system and iPLEX technology (San Diego, CA, USA). Amplification and primer extension were undertaken according to manufacturer’s instructions and primers listed in supplementary Table 1. Genotype in selected individuals was confirmed by DNA sequencing exon 21 using primers in supplementary Table 1, or by comparison with previous conformation sensitive gel electrophoresis data [1].
Haplotype analysis
Determination of haplotype associated with c.2771G>A was achieved by genotyping eight VWF single nucleotide polymorphisms (SNP) with known population frequencies; c.-2527A>G (promoter, rs7965413), c.1411G>A (exon 12, rs1800377), c.2365A>G (exon 18, rs1063856), c.4141G>A (exon 28, rs216311), c.5843-8C>G (intron 34, rs34444862), c.5844C>T (exon 35, rs216902), c.8116-20A>C (intron 49, rs2270152), c.8155+50C>T (intron 50, rs2270151). Genotyping was performed by allelic discrimination using the 5′ nuclease TaqMan assay (ABI 7900; Applied Biosystems, Warrington, UK) as described [1] or direct DNA sequencing. Primers are listed in supplementary Tables 2 and 3.
Expression of recombinant VWF containing p.R924Q
Site directed mutagenesis of full length VWF cDNA was carried out using the Quick Change kit (Stratagene, Kassel, Germany). Mutagenic primers carrying the required base-exchange at a central position were 40bp long. The expression vector VWF-pcDNA3.1 containing mutant and wild-type (wt) VWF cDNA, was used to transform Top10 supercompetent E coli (Invitrogen, Karlsruhe, Germany). Plasmids were purified by the Endofree Plasmid Maxi Kit (Qiagen, Hilden, Germany). 2×106 293 EBNA cells (Invitrogen, Karlsruhe, Germany) were transiently transfected with 4 μg mutant or wt full length VWF cDNA by liposomal transfer (Lipofectamine 2000, Invitrogen). Cells were grown for 72h (24h in DMEM (Invitrogen) with 10% (v/v) FBS and 48h in serum free IMDM (Sigma) with 2% (v/v) Ultroser G (Invitrogen)). VWF secreted into the medium was concentrated in Centricon tubes to 1/10 of the original volume prior to analysis. To analyze intracellular VWF, transfected cells were lysed by three rounds of freezing (−80°C) and thawing in lysis buffer (0.1 M Tris-HCl pH 8.0; 0.6% (v/v) Triton X-100). Recombinant VWF (rVWF) was analysed as described [11].
Statistical analysis
All internet-based statistical and sequence analysis tools were accessed April 2009. Hardy-Weinberg equilibrium, haplotype prediction and calculation of linkage disequilibrium were assessed using SNPStats (http://bioinfo.iconcologia.net/index.php?module=Snpstats). Allele and genotype frequencies were calculated using SNPStats and confirmed by allele counting. VassarStats (http://faculty.vassar.edu/lowry/VassarStats.html) was used to calculate odds ratios (OR) with 95% confidence intervals (CI). Association between c.2771G>A genotype and phenotype was assessed using Mann-Whitney tests (GraphPad Prism v 5.00, San Diego, CA, USA). p values <0.05 were considered significant in all tests.
In silico analysis
Predicted effect of c.2771G>A and c.5843-8C>G on mRNA splicing was examined using; Splice Site Prediction by Neural Network (http://fruitfly.org:9005/seq_tools/splice.html), WebGene Splice View (http://zeus2.itb.cnr.it/~webgene/wwwspliceview.html), NetGene2 (http://www.cbs.dtu.dk/services/NetGene2/), Human Splicing Finder (http://www.umd.be/SSF/).
Results
c.2771G>A phenotype in MCMDM-1VWD family members
The variant c.2771G>A (p.R924Q) was previously identified in IC from four of 150 MCMDM-1VWD families [1]. Regenotyping all IC for the variant led to its additional identification in IC P8F1II:1 and P9F14II:2. The latter individual was excluded from the previous cohort [1] due to very low plasma VWF level and virtually undetectable multimers. The six IC and their AFM have the variant c.2771G>A in heterozygous and homozygous (P6F11II:4) forms and in compound heterozygous and allelic combinations with further mutations. They originate from five European countries; their mutation and phenotype data are shown in Table 1.
Table 1.
Phenotype in members of six families with c.2771G>A
| Family | Individual | Status | Inheritance | BS | FVIII:C IU/dL |
VWF:RCo IU/dL |
VWF:Ag IU/dL |
FVIII/ VWF:Ag |
VWF:RCo/ VWF:Ag |
Allele 1 | Allele 2 | ABO | VWF:FVIIIB slope |
Multimers† |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P8 F1 | II:1 | IC | Het* | 15 | 61 | 49 | 49 | 1.25 | 1.00 | p.R924Q | - | O/A2 | 0.90 | NM |
| P9 F14 | II:2 | IC | Comp Het | 18 | 7 | 3 | 3 | 2.33 | 1.00 | p.[R924Q+C1927R] | c.1533+1G>T | O/A | 0.85 | Absent |
| P9 F14 | I:2 | AFM | Het | 4 | 63 | 58 | 57 | 1.11 | 1.02 | p.[R924Q+C1927R] | - | A/A | 1.02 | NM‡ |
| P9 F14 | III:1 | AFM | Het | 1 | 25 | 18 | 19 | 1.32 | 0.95 | p.[R924Q+C1927R] | - | O/O | 1.00 | AbM 2M(sm) |
| P9 F14 | I:1 | UFM | Het | 0 | 82 | 79 | 88 | 0.93 | 0.90 | - | c.1533+1G>T | O/B | NT | NM |
| P9 F14 | II:3 | UFM | Het | 5 | 50 | 80 | 57 | 0.88 | 1.40 | p.[R924Q+C1927R] | - | A/B | NT | AbM 2M(sm) |
| P7 F3 | II:1 | IC | Comp Het | 6 | 15 | 38 | 25 | 0.60 | 1.52 | p.R924Q | p.R854Q | O/O | 0.09 | NM |
| P7 F3 | I:2 | AFM | Het* | 8 | 104 | 88 | 81 | 1.28 | 1.09 | p.R924Q | - | O/A | 1.09 | NM |
| P7 F3 | I:1 | UFM | Het | −1 | 77 | 87 | 75 | 1.03 | 1.16 | - | p.R854Q | O/O | 0.65 | NM |
| P6 F5 | II:1 | IC | Comp Het | 18 | 18 | 7 | 11 | 1.64 | 0.64 | p.R924Q | p.R1205H | O/O | 1.15 | NM |
| P6 F5 | III:1 | AFM | Het | 3 | 28 | 17 | 15 | 1.86 | 1.13 | - | p.R1205H | O/O | 1.24 | NM |
| P6 F5 | II:3 | UFM | Het* | -1 | 107 | 95 | 131 | 0.82 | 0.73 | p.R924Q | O/A | NT | NM | |
| P6 F11 | II:1 | IC | Comp Het | 19 | 52 | 34 | 40 | 1.30 | 0.85 | p.R924Q | c.3675-14G>A | O/O | 1.08 | NM |
| P6 F11 | II:4 | AFM | Hom | 3 | 71 | 50 | 49 | 1.45 | 1.02 | p.R924Q | p.R924Q | O/O | NT | NM |
| P6 F11 | III:1 | AFM | Het* | 3 | 68 | 52 | 62 | 1.10 | 0.84 | p.R924Q | - | O/O | 1.22 | NM |
| P6 F11 | II:3 | UFM | Het* | 2 | 77 | 69 | 64 | 1.20 | 1.08 | p.R924Q | - | O/O | NT | NM |
| P6 F11 | III:2 | UFM | Het | 3 | 96 | 82 | 84 | 1.14 | 0.98 | - | c.3675-14G>A | O/A2 | NT | NM |
| P3 F13 | I:1 | IC | Allelic | 10 | 78 | 13 | 42 | 1.86 | 0.31 | p.[R924Q+R1315RL] | - | O/A2 | 0.94 | AbM 2A(IIE) |
| P3 F13 | II:1 | AFM | Allelic | 10 | 35 | 3 | 18 | 1.94 | 0.17 | p.[R924Q+R1315RL] | p.Y1584C | O/O | 0.88 | AbM 2A(IIE) |
|
*Median
(range) of p.R924Q Het with NM |
n=5 | Het * |
3 (-1-15) |
77 (61-107) |
69 (49-95) |
64 (49-131) |
1.2 (0.82-1.28) |
1.0 (0.73-1.09) |
p.R924Q | 40% O/O |
1.07‡ (0.90-1.22) |
NM |
IC, index case; AFM UFM, affected/unaffected family member; HC, healthy control NM AbM, normal/abnormal multimers
Median (range) of 5 indicated heterozygous individuals with NM
Multimer classifications follow Budde et al [7], ‡ minor abnormality, less marked than in relatives sharing allele.
Mean VWF:FVIIIB for 3 indicated (*) heterozygous individuals with NM. Mean VWF:FVIIIB for 63 UFM = 1.06 (Normal range 0.68-1.44)
NT = Not tested
P8F1II:1 was heterozygous for c.2771G>A alone, but had an unusually high bleeding score (BS) of 15 (normal <4) [9] and a further defect in VWF or elsewhere may contribute to her phenotype. Levels of VWF and FVIII:C were lower in compound heterozygous IC P6F5II:1 and P6F11II:1 and BS was considerably higher than in their heterozygous relatives who possessed only the non-c.2771G>A familial mutation. Observations on this limited number of cases suggest that the allele may contribute to reduced VWF and FVIII levels.
Phenotypes in five simple c.2771G>A heterozygotes with normal VWF multimers (1 IC, 2 AFM and 2 UFM) were variable. Median BS was 3, FVIII:C 77 IU/dL, VWF:RCo 69 IU/dL and VWF:Ag 64 IU/dL. Factor VIII binding (VWF:FVIIIB) data was available for three of these c.2771G>A heterozygotes and for 63 UFM with no mutations identified. Mean values for ability of VWF to bind FVIII of 1.07 vs 1.06 was obtained for each group (normal range 0.68-1.44), indicating that a VWF:FVIIIB defect was unlikely in c.2771G>A heterozygotes.
c.2771G>A frequency in healthy controls
The c.2771A allele was present in 35 of 1115 HC. No A allele homozygotes were identified. Frequency of the c.2771A allele did not differ significantly between IC and HC (2.0% vs 1.6%, odds ratio (OR) 1.30 (95% CI 0.54-3.15). It was identified in 1-4 HC from the approximately 100 HC recruited by each EU partner, with the exception of P9 (Czech Republic), where it was present in ten of 98 HC (10.2%). There was no significant difference in frequency between IC and HC with the removal of P9 data. Genotype distributions did not significantly deviate from Hardy-Weinberg equilibrium in IC or HC. However, a comparison of data from Canadian and UK type 1 VWD studies (Table 2) indicated a slight elevation in frequency of c.2771G>A in IC compared with HC when all studies were combined (OR 1.99 (95% CI 1.14-3.45)).
Table 2.
Genotype frequency for c.2771G>A in type 1 VWD studies
The c.2771A allele is associated with a common haplotype
Haplotype analysis using eight SNP was performed in families with c.2771G>A to determine whether a shared common founder was likely. Five haplotypes were identical (Table 3), whilst two differed only in the promoter (P6F11a) or 3′ of exon 35 (P9F14).
Table 3.
VWF haplotype analysis of individuals carrying p.R924Q in type 1 VWD families
| Sequence variant |
Reported allele frequency* |
Family ID | |||||||
|---|---|---|---|---|---|---|---|---|---|
| P3F13 | P6F5 | P6F11a‡ | P6F11b‡ | P7F3 | P9F14 | P8F1 | Common haplotype |
||
|
Promoter c.-2527A>G |
0.68/0.32† | A | A | G | A | A | A | A | A |
|
Exon 12 c.1411G>A p.V411I |
0.45/0.55† | G | G | G | G | G | G | G | G |
|
Exon 18 c.2365A>G p.T789A |
0.66/0.34 | A | A | A | A | NI | A | A | A |
|
Exon 21 c.2771G>A p.R924Q |
0.98/0.02 | A | A | A | A | A | A | A | A |
|
Exon 28 c.4141G>A p.T1381A |
0.62/0.38 | G | G | G | G | G | G | NI | G |
|
Intron 34 c.5843-8C>G |
0.97/0.03† | G | G | G | G | G | G | NI | G |
|
Exon 35 c.5844C>T p.C1948 |
0.63/0.37 | C | C | C | C | C | C | NI | C |
|
Intron 49 c.8116-20A>C |
0.78/0.22 | A | A | A | A | A | C | A | A |
|
Intron 50 c.8155+50C>T |
0.87/0.13 | C | C | C | C | C | T | C | C |
Caucasian frequency, reported on http://www.ncbi.nlm.nih.gov/SNP (Hap-Map European) or
Haplotypes a and b were present in the same family; both parents were c.2771G>A heterozygous, the AFM was homozygous.
NI = non-informative.
Haplotypes were predicted for the c.2771G>A heterozygous HC using SNPStats. The most frequent (22%) predicted haplotype bearing c.2771A was identical to the common haplotype in patients. Visual inspection of genotypes indicated that 30 of 35 were consistent with the common core c.2771A haplotype, whereas in five cases, homozygosity for the alternative allele at ≥1 position made possession of the common haplotype less likely (Supplementary table 4). In two HC, haplotype differed only at promoter variant c.-2527A>G, as seen in P6F11a. c.2771A was in strong linkage disequilibrium (D’=0.997) with the rare G allele of variant c.5843-8C>G located within the exon 35 acceptor splice site. Presence of c.5843-8G, found in only ~3% of the Caucasian population, makes it unlikely that c.2771A and c.5843-8G occur together by chance in the majority of individuals. Absence of c.5843-8G, plus homozygosity for the alternative nucleotide at additional positions made possession of the predicted founder haplotype much less likely in three (9%) remaining HC.
c.2771 genotype association with VWF and FVIII levels
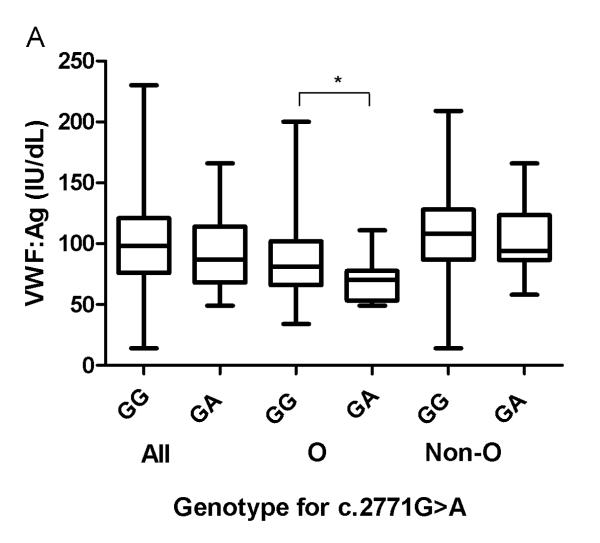
Association of the c.2771A allele with VWF:Ag and VWF:RCo levels was investigated in HC (Fig. 1A; Table 4). VWF:Ag levels only differed significantly in individuals with ABO blood group O, where c.2771G>A heterozygotes had lower median VWF:Ag levels (~14%) than c.2771G homozygotes. c.2771 genotype had no influence on VWF:RCo/VWF:Ag ratios.
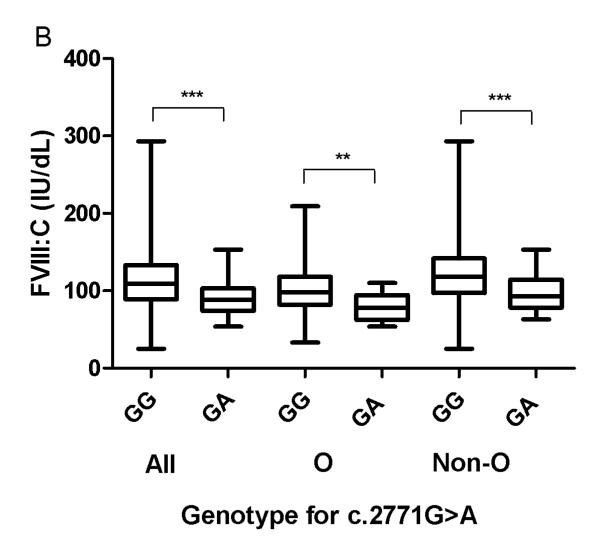
Fig. 1. Association between genotypes at c.2771, VWF:Ag and FVIII:C levels in healthy controls according to ABO blood group.
*, ** and ***indicate significant (p<0.05, p<0.01 and p<0.0001, Mann-Whitney test) differences between median values of (A) VWF:Ag and (B) FVIII:C amongst HC, depending on their c.2771 genotype. “All” refers to the entire HC population, “O” = blood group O and “Non-O” = blood groups other than O.
Table 4.
Association of c.2771 genotype with median levels of VWF and FVIII:C in EU healthy controls
| c.2771G>A genotype & ABO blood group |
FVIII:C | VWF:Ag | VWF:RCo | FVIII:C/VWF:Ag | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | IU/dL | p | n | IU/dL | p | n | IU/dL | p | n | ratio | p | |
| All | ||||||||||||
| GG | 1064 | 109 | ‡ | 1070 | 98 | NS | 1063 | 95 | NS | 1063 | 1.14 | * |
| GA | 35 | 88 | 35 | 87 | 35 | 85 | 35 | 1.09 | ||||
| O | ||||||||||||
| GG | 395 | 98 | † | 399 | 81 | * | 395 | 82 | NS | 395 | 1.18 | NS |
| GA | 13 | 78 | 13 | 70 | 13 | 67 | 13 | 1.20 | ||||
| Non-O | ||||||||||||
| GG | 648 | 118 | ‡ | 650 | 108 | NS | 647 | 103 | NS | 647 | 1.12 | * |
| GA | 21 | 93 | 21 | 94 | 21 | 111 | 21 | 0.96 | ||||
The number of subjects in the “All” group is larger than those included in the other two groups due to inclusion of HC with undetermined blood groups.
indicate degree of significance between GG and GA subjects (p<0.05, p<0.01 and p<0.0001, Mann-Whitney test); NS indicates nonsignificant differences
indicate degree of significance between GG and GA subjects (p<0.05, p<0.01 and p<0.0001, Mann-Whitney test); NS indicates nonsignificant differences
indicate degree of significance between GG and GA subjects (p<0.05, p<0.01 and p<0.0001, Mann-Whitney test); NS indicates nonsignificant differences
Median FVIII:C levels were significantly lower (~20%) in c.2771G>A heterozygotes in comparison with c.2771G homozygotes in both ABO blood group O and non-O HC (Fig. 1B; Table 4). FVIII:C/VWF:Ag ratios were lower in c.2771G>A heterozygotes than in c.2771G homozygotes before ABO blood group stratification and in non-O HC c.2771G>A heterozygotes compared to c.2771G homozygotes (Table 4).
In vitro expression analysis of p.R924Q
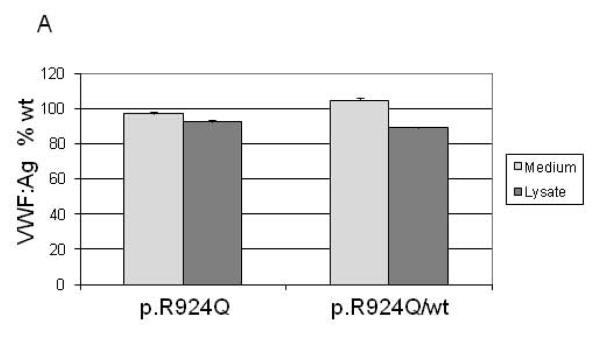
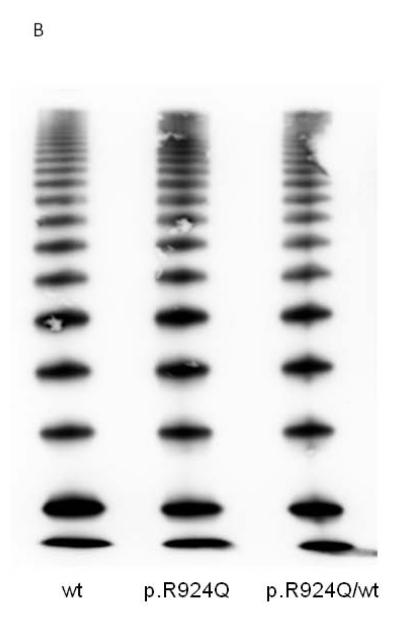
Recombinant VWF (rVWF) comprising wt (c.2771G), mutant (c.2771A) or a 50:50 mix mimicking heterozygosity was transiently transfected into 293 EBNA cells. Secreted and intracellular VWF levels measured three days post-transfection were very similar for all transfections. rVWFR924Q and rVWFwt multimers were identical (Fig. 2). In this model system, p.R924Q as sole genetic variant did not result in altered VWF expression.
Fig. 2. Expression of recombinant VWF in 293 EBNA cells.
(A) rVWF secreted into the medium and retained within the cell relative to rVWFwt alone. (B) Profile of secreted rVWFR924Q multimers electrophoresed on SDS-agarose (1.2%).
In silico assessment of c.2771G>A and c.5843-8C>G variants
None of four in silico splice site analysis tools predicted a significant effect of c.2771G>A on strength of adjacent splice sites. c.5843-8C>G was predicted by each of four splice site prediction tools to lead to a minor reduction in splicing efficiency (~3%), predicting the rare G allele would function less well than the C allele.
The allele bearing p.R924Q appears not to behave as a null allele
Median VWF:Ag levels in 35 HC c.2771G>A heterozygotes and five family members who were heterozygous only for c.2771G>A were 87 IU/dL and 64 IU/dL respectively (Tables 1 and 4). The lowest VWF:Ag level observed among these 41 individuals was 49 IU/dL in 1 HC and 1 IC. Nichols et al [12] compiled data on 190 heterozygous relatives of type 3 VWD patients, the majority of whom would have a null allele [13]. Mean VWF:Ag was 47 IU/dL (range ±2SD 16-140 IU/dL), considerably lower than in p.R924Q heterozygotes. Additionally, the single c.2771A homozygous AFM (P6F11II:4) had VWF and FVIII levels only slightly lower than in heterozygous individuals (VWF:RCo 50 IU/dL; VWF:Ag 49 IU/dL). c.2771G>A behaviour is thus unlike that of a null allele.
Discussion
Previous studies have identified p.R924Q in healthy controls, patients with VWD and their unaffected relatives [2-6,14], but have not reached a conclusion regarding its pathogenic significance. The current study has demonstrated that the p.R924Q allele does have an effect on phenotype in heterozygous HC. The extent of its influence on VWF and FVIII levels is modest and analysis in a larger group of individuals may be required to appreciate the degree of effect on VWF levels. Blood group O c.2771G>A heterozygous HC had VWF:Ag and FVIII:C levels approximately 35% lower than c.2771G homozygotes with non-O blood groups. Approximately 10% variance was attributable to the A allele and the remainder to blood group O. The p.R924Q allele and blood group O displayed an additive effect, similar to that associated with the p.Y1584C variant, where an additive effect (~25%) of the allele with blood group O was observed [15]. Slightly enhanced VWF clearance could contribute to lower levels observed in individuals bearing p.R924Q, as seen for p.Y1584C [15]. Limited data on clearance derived from follow-up of desmopressin administration and ratios of VWF propeptide to VWF:Ag (VWFpp/VWF:Ag) are available. IC P7F3II:1 (p.[R924Q]+[R854Q]) was included in a desmopressin response study [16] and was not amongst cases demonstrating increased clearance (VWF:RCo half-life <4 hr). VWFpp/VWF:Ag ratios have recently been determined. Mean ratio of 1.30 in four simple p.R924Q heterozygous family members was not different from the mean ratio of 1.32 in 376 c.2771G homozygous HC. Therefore, enhanced clearance is unlikely to explain reduced VWF levels.
In EU IC, heterozygous inheritance of p.R924Q alone appeared insufficient for VWD symptoms, although in other type 1 VWD studies, only a small proportion of IC (3 of 10 [5] and 2 of 8 [2]) had further mutations identified. In five of six EU IC heterozygous for p.R924Q, a second VWF sequence variant seems to contribute to phenotype. In P8F1II:1, a further factor may contribute to bleeding. Phenotypic modifiers, for example platelet defects have been shown to influence bleeding propensity in type 1 VWD [17]. Many factors can influence VWF levels including age, stress and hormonal status. The extent of effect on VWF levels attributable to the c.2771G>A allele may be confounded by ABO and other blood groups including Lewis [18] and Secretor [19]. Further loci influencing VWF levels have been sought but await characterisation [20].
In vitro expression of recombinant p.R924Q VWF showed no reduced secretion, increased intracellular retention or alteration in VWF multimers and in silico analysis predicted no effect of c.2771G>A on intron 20 acceptor/intron 21 donor splice sites. A recent study using a different model system also indicated lack of evidence for effect of p.R924Q on VWF expression level [6].
The majority of p.R924Q alleles are likely to be identical by descent. Occurrence of c.5843-8G consistently on the same allele as c.2771A was also reported by Lester et al [5]. Elevated p.R924Q frequency in Czech HC (10.2% vs 2.5% in all other HC) may indicate an Eastern European origin. p.R924Q was absent from 183 unrelated Argentinian blood donors [14], further supporting a European origin. Three (9%) of 35 heterozygous HC were unlikely to share the common haplotype through identity by descent. c.2771G>A occurs at a CpG dinucleotide, a relative mutation hotspot and it is possible that in addition to its occurrence on the founder haplotype, the G>A substitution has arisen independently. Berber et al comment that c.5843-8G was not consistently identified on c.2771A alleles [6], suggesting that some de novo c.2771G>A cases were present among Canadian subjects. Independent occurrence of c.2771G>A, along with additional sequence variants arising on the founder haplotype as seen in both the current and UK studies [3,5], likely helps to explain variable behaviour of p.R924Q alleles. As suggested by Berber et al [6], “not all p.R924Q alleles are created equal.”
Cumming et al [3] analysed platelet VWF mRNA from a patient heterozygous for c.[2771A+5843-8G] and identified a single normal VWF transcript in the region surrounding intron 34, with no evidence of aberrant splicing. As it is improbable given its position relative to exon 34 that c.5843-8C>G results in complete lack of VWF expression, any significant effect on VWF mRNA appears unlikely. Cumming’s result neither supports nor refutes Berber’s identification of abnormal mRNA resulting from exon 28 cryptic splice site activation, as if the p.R924Q allele bears a mutation resulting in a truncated mRNA, aberrant mRNA may not extend as far as exon 34.
In conclusion, the p.R924Q variant marks a VWF allele found in approximately 3% of Caucasians, of which the majority are likely to be identical by descent. The allele is associated with reductions in VWF and FVIII:C levels particularly when co-inherited with blood group O. The missense alteration p.R924Q alone in an in vitro model does not lead to altered protein expression and neither recombinant VWF nor the allele in heterozygous individuals reduces VWF:FVIIIB in a static system nor affect clearance. The effect of the allele may therefore be that of a further, as yet unidentified sequence variant in strong linkage disequilibrium with p.R924Q.
Supplementary Material
Acknowledgements
This work was supported by the European Community Fifth Framework Programme (QLG1-CT-2000-00387) and National Institutes of Health Zimmerman Program for Molecular and Clinical Biology of VWD (HL-081588).
The authors are grateful to Alberto Tosetto, Javier Batlle, Dominique Meyer, Claudine Mazurier, Jorgen Ingerslev, David Habart, Zdena Vorlova, Lars Holmberg, Stefan Lethagen, John Pasi, Frank Hill, Mohammad Hashemi-Soteh, Luciano Baronciani, Christer Hallden, Andrea Guilliatt and Will Lester for contributions to the MCMDM-1VWD study and thank John Anson, Sheffield University for excellent technical support.
Footnotes
Disclosure of Conflict of Interests The authors state that they have no conflict of interest.
Addendum Contribution of Authors
- Study initiation and coordination
- Study design, data collection and laboratory analyses
- Result analysis and interpretation
- Lead authors of initial manuscript
- Draft manuscript revision
- Final manuscript review and approval
Hickson2-6, Hampshire3,5,6, Winship3,5,6, Goudemand2,3,5,6, Schneppenheim2,3,5,6, Budde2,3,6, Castaman1,2,3,6, Rodeghiero1,2,3,6, Federici2,3,6, James3,5,6, Peake1,3,5,6, Eikenboom2,3,5,6, Goodeve1-6.
References
- 1.Goodeve A, Eikenboom J, Castaman G, Rodeghiero F, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Schneppenheim R, Budde U, Ingerslev J, Habart D, Vorlova Z, Holmberg L, Lethagen S, Pasi J, Hill F, Hashemi Soteh M, Baronciani L, et al. Phenotype and genotype of a cohort of families historically diagnosed with type 1 von Willebrand disease in the European study, Molecular and Clinical Markers for the Diagnosis and Management of Type 1 von Willebrand Disease (MCMDM-1VWD) Blood. 2007;109:112–21. doi: 10.1182/blood-2006-05-020784. [DOI] [PubMed] [Google Scholar]
- 2.James PD, Notley C, Hegadorn C, Leggo J, Tuttle A, Tinlin S, Brown C, Andrews C, Labelle A, Chirinian Y, O’Brien L, Othman M, Rivard G, Rapson D, Hough C, Lillicrap D. The mutational spectrum of type 1 von Willebrand disease: Results from a Canadian cohort study. Blood. 2007;109:145–54. doi: 10.1182/blood-2006-05-021105.. [DOI] [PubMed] [Google Scholar]
- 3.Cumming A, Grundy P, Keeney S, Lester W, Enayat S, Guilliatt A, Bowen D, Pasi J, Keeling D, Hill F, Bolton-Maggs PH, Hay C, Collins P. An investigation of the von Willebrand factor genotype in UK patients diagnosed to have type 1 von Willebrand disease. Thromb Haemost. 2006;96:630–41. [PubMed] [Google Scholar]
- 4.Hilbert L, Jorieux S, Proulle V, Favier R, Goudemand J, Parquet A, Meyer D, Fressinaud E, Mazurier C. Two novel mutations, Q1053H and C1060R, located in the D3 domain of von Willebrand factor, are responsible for decreased FVIII-binding capacity. Br J Haematol. 2003;120:627–32. doi: 10.1046/j.1365-2141.2003.04163.x. [DOI] [PubMed] [Google Scholar]
- 5.Lester W, Guilliatt A, Grundy P, Enayat S, Millar C, Hill F, Cumming T, Collins P. Is VWF R924Q a benign polymorphism, a marker of a null allele or a factor VIII-binding defect? The debate continues with results from the UKHCDO VWD study. Thromb Haemost. 2008;100:716–8. [PubMed] [Google Scholar]
- 6.Berber E, James PD, Hough C, Lillicrap D. An assessment of the pathogenic significance of the R924Q von Willebrand factor substitution. J Thromb Haemost. 2009;7:1672–9. doi: 10.1111/j.1538-7836.2009.03551.x. [DOI] [PubMed] [Google Scholar]
- 7.Budde U, Schneppenheim R, Eikenboom J, Goodeve A, Will K, Drewke E, Castaman G, Rodeghiero F, Federici AB, Batlle J, Perez A, Meyer D, Mazurier C, Goudemand J, Ingerslev J, Habart D, Vorlova Z, Holmberg L, Lethagen S, Pasi J, et al. Detailed von Willebrand factor multimer analysis in patients with von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 von Willebrand disease (MCMDM-1VWD) J Thromb Haemost. 2008;6:762–71. doi: 10.1111/j.1538-7836.2008.02945.x. [DOI] [PubMed] [Google Scholar]
- 8.Eikenboom J, Van Marion V, Putter H, Goodeve A, Rodeghiero F, Castaman G, Federici AB, Batlle J, Meyer D, Mazurier C, Goudemand J, Schneppenheim R, Budde U, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Lethagen S, Pasi J, Hill F, et al. Linkage analysis in families diagnosed with type 1 von Willebrand disease in the European study, molecular and clinical markers for the diagnosis and management of type 1 VWD. J Thromb Haemost. 2006;4:774–82. doi: 10.1111/j.1538-7836.2006.01823.x. [DOI] [PubMed] [Google Scholar]
- 9.Tosetto A, Rodeghiero F, Castaman G, Goodeve A, Federici AB, Batlle J, Meyer D, Fressinaud E, Mazurier C, Goudemand J, Eikenboom J, Schneppenheim R, Budde U, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Lethagen S, Pasi J, Hill F, et al. A quantitative analysis of bleeding symptoms in type 1 von Willebrand disease: results from a multicenter European study (MCMDM-1 VWD) J Thromb Haemost. 2006;4:766–73. doi: 10.1111/j.1538-7836.2006.01847.x. [DOI] [PubMed] [Google Scholar]
- 10.Caron C, Mazurier C, Goudemand J. Large experience with a factor VIII binding assay of plasma von Willebrand factor using commercial reagents. Br J Haematol. 2002;117:716–8. doi: 10.1046/j.1365-2141.2002.03488.x. [DOI] [PubMed] [Google Scholar]
- 11.Eikenboom J, Hilbert L, Ribba AS, Hommais A, Habart D, Messenger S, Al-Buhairan A, Guilliatt A, Lester W, Mazurier C, Meyer D, Fressinaud E, Budde U, Will K, Schneppenheim R, Obser T, Marggraf O, Eckert E, Castaman G, Rodeghiero F, et al. Expression of 14 von Willebrand factor mutations identified in patients with type 1 von Willebrand disease from the MCMDM-1VWD study. J Thromb Haemost. 2009;7:1304–12. doi: 10.1111/j.1538-7836.2009.03486.x. [DOI] [PubMed] [Google Scholar]
- 12.Nichols WL, Hultin MB, James AH, Manco-Johnson MJ, Montgomery RR, Ortel TL, Rick ME, Sadler JE, Weinstein M, Yawn BP. von Willebrand disease (VWD): evidence-based diagnosis and management guidelines, the National Heart, Lung, and Blood Institute (NHLBI) Expert Panel report (USA) Haemophilia. 2008;14:171–232. doi: 10.1111/j.1365-2516.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 13.Eikenboom JC. Congenital von Willebrand disease type 3: clinical manifestations, pathophysiology and molecular biology. Best Pract Res Clin Haematol. 2001;14:365–79. doi: 10.1053/beha.2001.0139. [DOI] [PubMed] [Google Scholar]
- 14.Casais P, Carballo GA, Woods AI, Kempfer AC, Farias CE, Grosso SH, Lazzari MA. R924Q substitution encoded within exon 21 of the von Willebrand factor gene related to mild bleeding phenotype. Thromb Haemost. 2006;96:228–30. [PubMed] [Google Scholar]
- 15.Davies JA, Collins PW, Hathaway LS, Bowen DJ. von Willebrand factor: evidence for variable clearance in vivo according to Y/C1584 phenotype and ABO blood group. J Thromb Haemost. 2008;6:97–103. doi: 10.1111/j.1538-7836.2007.02809.x. [DOI] [PubMed] [Google Scholar]
- 16.Castaman G, Lethagen S, Federici AB, Tosetto A, Goodeve A, Budde U, Batlle J, Meyer D, Mazurier C, Fressinaud E, Goudemand J, Eikenboom J, Schneppenheim R, Ingerslev J, Vorlova Z, Habart D, Holmberg L, Pasi J, Hill F, Peake I, et al. Response to desmopressin is influenced by the genotype and phenotype in type 1 von Willebrand disease (VWD): results from the European Study MCMDM-1VWD. Blood. 2008;111:3531–9. doi: 10.1182/blood-2007-08-109231. [DOI] [PubMed] [Google Scholar]
- 17.Daly ME, Dawood BB, Lester WA, Peake IR, Rodeghiero F, Goodeve AC, Makris M, Wilde JT, Mumford AD, Watson SP, Mundell SJ. Identification and characterization of a novel P2Y 12 variant in a patient diagnosed with type 1 von Willebrand disease in the European MCMDM-1VWD study. Blood. 2009;113:4110–3. doi: 10.1182/blood-2008-11-190850. [DOI] [PubMed] [Google Scholar]
- 18.Green D, Jarrett O, Ruth KJ, Folsom AR, Liu K. Relationship among Lewis phenotype, clotting factors, and other cardiovascular risk factors in young adults. J Lab Clin Med. 1995;125:334–9. [PubMed] [Google Scholar]
- 19.Orstavik KH, Kornstad L, Reisner H, Berg K. Possible effect of secretor locus on plasma concentration of factor VIII and von Willebrand factor. Blood. 1989;73:990–3. [PubMed] [Google Scholar]
- 20.Souto JC, Almasy L, Soria JM, Buil A, Stone W, Lathrop M, Blangero J, Fontcuberta J. Genome-wide linkage analysis of von Willebrand factor plasma levels: results from the GAIT project. Thromb Haemost. 2003;89:468–74. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.