Abstract
Blood-group antigens, such as those containing fucose and bearing the ABO(H)- and Lewis-type determinants expressed on the carbohydrate chains of glycoproteins and glycolipids, and also on unconjugated free oligosaccharides in human milk and other secretions, are associated with various biological functions. We have previously shown the utility of negative-ion electrospay ionization tandem mass spectrometry with collision-induced dissociation (ESI-CID-MS/MS) for typing of Lewis (Le) determinants, e.g. Lea, Lex, Leb, and Ley on neutral and sialylated oligosaccharide chains. In the present report we extended the strategy to characterization of blood-group A-, B- and H-determinants on type 1 and type 2, and also on type 4 globoside chains to provide a high sensitivity method for typing of all the major blood-group antigens, including the A, B, H, Lea, Lex, Leb, and Ley determinants, present in oligosaccharides. Using the principles established we identified two minor unknown oligosaccharide components present in the products of enzymatic synthesis by bacterial fermentation. We also demonstrated that the unique fragmentations derived from the D- and 0,2A-type cleavages observed in ESI-CID-MS/MS, which are important for assigning blood-group and chain types, only occur under the negative-ion conditions for reducing sugars but not for reduced alditols or under positive-ion conditions.
INTRODUCTION
There has been a considerable resurgence of interest in the biological roles of the diverse oligosaccharide sequences that ‘decorate’ glycoproteins, proteoglycans, glycolipids and polysaccharides. Carbohydrate sequences as a vast source of information can be used as a ‘glyco-code’ waiting to be deciphered in various contexts of biomedical importance, e.g. through carbohydrate-protein interactions that involve specific oligosaccharide chains.
Blood-group antigens, such as those containing fucose and bearing the ABO(H)- and Lewis-type determinants, expressed on the carbohydrate chains of glycoproteins (N-glycans1 and O-glycans2) and glycolipids3, and also on milk oligosaccharides4,5, are associated with various biological functions. The ABH antigens have been identified as ligands for bovine norovirus6, rabbit hemorrhagic disease virus7, and most strains of Helicobacter pylori8. The Lewisx (Lex) antigen appears at specific stages of embryonic development, cellular differentiation and in oncogenesis9, and an H antigen on a globo chain (Globo-H) is highly expressed on the surface of various cancer cells as glycolipids or glycoproteins10. The important roles of Lex/Lea and related sialylated and sulfated oligosaccharide sequences as ligands for the selectins have been well recognized11-13.
Unconjugated free oligosaccharides are the major components of mammalian milk4,5 although they are also present in other human secretions14. Knowledge of the human milk glycome is important to the understanding of its impact on the infant gastrointestinal microbiota4. Milk oligosaccharides with specific sequences are known to be receptors for pathogens. The presence of such sequences in milk implies a defensive strategy against viral, bacterial and parasite infections using the oligosaccharides to prevent binding of pathogens to epithelial cells, thereby protecting infants from disease. It has been shown that human milk oligosaccharides can reduce HIV-1-gp120 binding to DC-SIGN15, block Pseudomonas aeruginosa lectin16, inhibit cholera toxin17 and rotavirus replication, and prevent experimental gastroenteritis18.
To detect and to assign the specificities of the carbohydrate-protein interactions involved in these immunological and pathogenic processes are challenging topics in modern cell biology, and rapid and sensitive methods are highly desirable to determine oligosaccharide sequences in the biological context in order to derive structure/function relationships. Conventionally, blood-group types of oligosaccharides are determined by glycosidase treatment19, inhibition of hemagglutination20, and antibody binding assays21. This information can also be obtained from detailed structural analysis involving mass spectrometry (MS), GC-MS methylation analysis and NMR spectroscopy22. With small amounts of material (e.g. low picomole), no single analytical technique is capable of the complete characterization of an oligosaccharide structure. As a consequence, structure elucidation is usually performed by using several different techniques, of which MS and NMR are two of the most powerful.
Recently, MS has become a primary technique in carbohydrate structural analysis23 for both profiling of glycan mixtures24 and detailed sequencing of individual oligosaccharide chains25. Negative-ion electrospray ionization tandem MS with collision-induced dissociation (ESI-CID-MS/MS) can be carried out directly for sialylated26 or sulfated27 oligosaccharides. We have developed a strategy using ESI-CID-MS/MS for sequence determination of various types of carbohydrate molecules, including linear and branched neutral25,28-31, sialylated32, sulfated33,34, and hexuronated oligosaccharides35-37. We have demonstrated that high sensitivity detection and sequence information can be obtained without the need of derivatization even for neutral oligosaccharides, and blood-group Lea, Lex, Leb and Ley can be readily identified28,29.
We have identified several characteristic fragmentations derived from different types of cleavages at certain monosaccharide residues with specific linkages under negative-ion ESI-CID-MS/MS conditions and have found that these can provide important information on sequence, branching pattern and in certain cases on linkages28,29. We have shown an important double glycosidic cleavage which is unique to 3-linked GlcNAc and Glc residues, and we referred it as a D-type fragmentation28,29. We also observed 0,2A-type cleavage (nomenclature used to define the cleavage based on that introduced previously38) occurring with 4-linked GlcNAc28, Glc29 and unsubstituted glucosamine35. These fragmentations, particularly the D-type fragmentation, were used for identification of important structural features on underivatized neutral and sialylated oligosaccharides, including differentiation of Lea [Galβ1-3(Fucα1-4)GlcNAc] and Lex [Galβ1-4(Fucα1-3)GlcNAc] determinants, occurring naturally on the carbohydrate chains of glycoproteins and glycolipids and comprising recognition motifs for cell-cell and cell-matrix interactions25,28,30-32.
Method for characterization of specific recognition motifs on oligosaccharide chains is important. Lebrilla and colleagues have reported a MALDI tandem MS-based ‘catalog-library’ approach for identification of some specific structural motifs on mucin-type O-glycan chains39. However, there has been no MS-based method aimed specifically for blood-group and chain typing. .
In the present study we extend the strategy to characterization of blood-group A-, B- and H-containing oligosaccharides on either type 1 or type 2 chains, or on globoside type 4 chains, to provide a high sensitivity method for typing of all the major blood-group determinants including the A, B, H, Lea, Lex, Leb, and Ley. We also demonstrate that the unique fragmentations observed in ESI-CID-MS/MS only occur under the negative-ion conditions for reducing sugars.
EXPERIMENTAL SECTION
Materials
Neutral oligosaccharides (see Table 1) lacto-N-fucopentaose I (LNFP-I), II (LNFP-II) and III (LNFP-III) were from Dextra (Reading, UK), and lacto-N-neofucopentaose I (LNnFP-I), blood-group A antigen hexaose type 1 (A-Hexa-T1) and type 2 (A-Hexa-T2), blood-group B antigen hexaose type 1 (B-Hexa-T1) and type 2 (B-Hexa-T2), the globo-H hexasaccharide (Globo-H-Hexa), globo-A and globo-B heptasaccharides (Globo-A-Hepta and Globo-B-Hepta, respectively) were produced by enzymatic synthesis using bacterial fermentation40 and purchased from Elicityl (Crolles, France).
Table 1.
Oligosaccharides used for negative-ion ESI-CID-MS/MS.
| Oligosaccharides | Sequences | Chain types | Blood groups |
|---|---|---|---|
| LNFP-II | Galβ1-3GlcNAcβ1-3Galβ1-4Glc ∣1,4 Fucα |
1 | Lea |
| LNFP-III | Galβ1-4GlcNAcβ1-3Galβ1-4Glc ∣1,3 Fucα |
1 | Lex |
| LNFP-I | Fucα ∣1,2 Galβ1-3GlcNAcβ1-3Galβ1-4Glc |
1 | H |
| LnNFP-I | Fucα ∣1,2 Galβ1-4GlcNAcβ1-3Galβ1-4Glc |
2 | H |
| A-Hexa-T1 | Fucα ∣1,2 GalNAcα1-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc |
1 | A |
| A-Hexa-T2 | Fucα ∣1,2 GalNAcα1-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc |
2 | A |
| B-Hexa-T1 | Fucα ∣1,2 Galα1-3Galβ1-3GlcNAcβ1-3Galβ1-4Glc |
1 | B |
| B-Hexa-T2 | Fucα ∣1,2 Galα1-3Galβ1-4GlcNAcβ1-3Galβ1-4Glc |
2 | B |
| Globo-A-Hepta | Fucα ∣1,2 GalNAcα1-3Galβ1-3GalNAcβ1-3Galα1-4Galβ1-4Glc |
4 (globo) | A |
| Globo-B-Hepta | Fucα ∣1,2 Galα1-3Galβ1-3GalNAcβ1-3Galα1-4Galβ1-4Glc |
4 (globo) | B |
| Globo-H-Hexa | Fucα ∣1,2 Galβ1-3GalNAcβ1-3Galα1-4Galβ1-4Glc |
4 (globo) | H |
HPLC
HPLC was carried out for purity analysis of the fermentation products and for isolation of the minor components using a normal-phase amide column (X-Bridge Amide, 3.5μm, 4.6 × 250mm, Waters, Elstree, England). Elution was performed by a gradient of acetonitril/water (Solvent A: 80:20, and Solvent B: 20:80) at a flow rate of 0.7 ml/min and detection by UV absorption at 195 nm.
Reduction of oligosaccharides
NaBH4 reagent (20 μl, 0.05 M NaBH4 in 0.01 M NaOH) was added to the freeze-dried oligosaccharide (typically 2-5 μg) and the reduction was carried out at 4 °C overnight as described25. The reaction solution was then neutralized with a solution of AcOH/H2O (1:1) to destroy borohydride before passing through a mini-column of cation exchange (AG50W-X8, H-form, Bio-Rad, Hemel Hempstead, UK). Boric acid was removed by repeated co-evaporation with MeOH.
Electrospray ionization mass spectrometry
ESI-MS and CID-MS/MS were carried out on a Waters (Manchester, UK) Q-TOF-type mass spectrometer SYNAPT. Cone voltage was varied between 30-45 eV for different oligosaccharides while capillary voltage was maintained at 3 kV for conventional scale electrospray. Source temperature was at 80°C and the desolvation temperature at 150°C. Product-ion spectra were obtained using argon as the collision gas at a pressure of 1.7 bar. The collision energy was adjusted between 12-20 eV for optimal fragmentation of reducing oligosaccharides and 30 eV for the reduced LNFP-II and LNFP-III alditols in the negative-ion mode, and 6 eV for the reducing LNFP-II and LNFP-III in the positive-ion mode. A scan rate of 1.5 sec/scan was used for both ESI-MS and MS/MS experiments and the acquired spectra were summed for presentation.
For positive-ion detection, to minimize the sodium adduction, potential contaminating salts were removed by a mini mixed-bed column of AG50W-X8 (H-form, 50 μl gel, lower bed) and AG1-X8 (OH-form, 50 μl gel, upper bed).
For analysis, oligosaccharides were dissolved in ACN/H2O 1:1, typically at a concentration of 20-40 pmol/μl, of which 3 μl was injected via HPLC injector. Solvent (ACN/ H2O 1:1) was delivered by a HPLC pump (Waters) at a flow rate of 300 μl/min.
NMR Spectroscopy
The minor and major components of A-Hexa-T1 (50 μg) were taken up in D2O 99.9% (Apollo Scientific, Stockport, UK), exchanged by lyophilization, redissolved in 0.6 ml D2O, and transferred to a 5 mm NMR tube. 1D 1H-NMR spectra were recorded at 500 MHz, 30°C, on a Bruker spectrometer, using pulse sequences supplied by the manufacturer.
RESULTS AND DISCUSSION
The nomenclature to define the fragmentation is based on that proposed by Domon and Costello38. For convenience, the numbering of monosaccharide residues to indicate the cleavage site is based on the oligosaccharide backbone without the fucose, and the glycosidic bond of fucose on the oligosaccharide backbone is denoted by the residue number of the oligosaccharide with a α suffix, e.g. 3α indicating the bond between the fucose and the residue 3 of the oligosaccharide backbone.
As mass spectral data alone can normally only define the monosaccharides in terms of hexose (Hex), N-acetylhexosamine (HexNAc) and deoxyhexose (dHex), their identities rely on information obtained from monosaccharide composition, methylation analysis, and NMR spectroscopy. However, other information, for example the source of the materials and biochemical analysis, can also provide important information on the possible identities of the monosaccharides present in the oligosaccharide chains. In the present work, to establish the principles of the method, standard oligosaccharides were used, and therefore, the monosaccharide identities are known and are indicated.
Unique fragmentation of reducing oligosaccharides in negative-ion ESI-CIDMS/MS
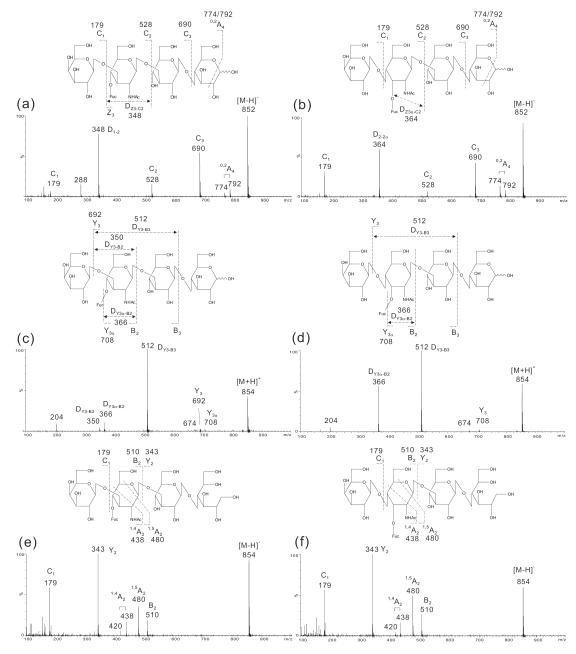
Lea- and Lex-pentasaccharides (LNFP-II and LNFP-III, respectively, Table 1) were used to investigate whether the fragmentation patterns obtained previously28 for typing of blood-group Lea and Lex are unique to negative-ion ESI-CID-MS/MS of reducing sugars. The product-ion spectra of LNFP-II and -III were compared using the deprotonated molecule [M−H]− (Fig. 1a and 1b) and the protonated molecule [M+H]+ (Fig. 1c and 1d) of the reducing sugars, and also [M−H]− of the reduced alditols (Fig. 1e and 1f) as the precursors.
Figure 1.
ESI-CID-MS/MS product-ion spectra of Lea and Lex pentasaccharides LNFP-II and LNFP-III.: (a) [M−H]− of LNFP-II, (b) [M−H]− of LNFP-III, (c) [M+H]+of LNFP-II, (d) [M+H]+of LNFP-III, (e) [M−H]− of the reduced alditol LNFP-II-ol, and (f) [M−H]− of the reduced alditol LNFP-III-ol.
In the product-ion spectra of [M−H]− of the reducing sugars (Fig. 1a and 1b), a full set of glycosidic C-type ions (C1, C2, and C3 at m/z 179, 528, and 690, respectively) was obtained in both spectra to indicate the sequences, while the fragment ion pair of 0,2A4 and its dehydrated form at m/z 792/774 to demonstrate a 4-linked Glc at the reducing terminus. More importantly the double glycosidic cleavage D-ions at m/z 348 (Fig. 1a) and m/z 364 (Fig. 1b) were unique and can be used to differentiate a blood-group Lea and a Lex determinant in LNFP-II and LNFP-III, respectively28.
These important features were not observed in the positive-ion spectra using [M+H]+ as the precursors. Glycosidic cleavages were very limited and only weak Y3 (m/z 692) and Y3α (m/z 708) were present in the spectrum of LNFP-II (Fig. 1c) and Y3α (m/z 708) in the spectrum of LNFP-III (Fig. 1d). The incomplete Y-ions and the lack of any other glycosidic (e.g. the B-, C- and Z-type) cleavages made it impossible to define the sequences of the oligosaccharides. Positive-ion spectra were dominated by double glycosidic cleavage: DY3-B3 (m/z 512) and DY3α-B2 (m/z 366) in both spectra while DY3-B2 in the spectrum of LNFP-II only. The ion at m/z 204 is likely to be from a GlcNAc residue from a triple cleavage.
Negative-ion ESI-CID-MS/MS of the reduced LNFP-II and-III alditols (LNFP-II-ol and LNFP-III-ol, respectively) was also carried out. As shown in Fig. 1e and 1f, the spectra of the two isomeric alditols were almost identical. Only limited glycosidic cleavages were obtained and these were insufficient to define the sequence. The unique D- and 0,2A-type cleavages observed in the spectra of reducing oligosaccharides used for defining the Lea/Lex epitopes and the 4-linked GlcNAc, respectively, were absent. These indicated the importance of the reducing termini for obtaining the characteristic fragmentations.
Clearly from the three different types of product-ion spectra, the [M−H]− of the reducing sugars gave the most informative fragmentation and can be used for typing of blood-group Lea and Lex. Negative-ion ESI-CID-MS/MS was then used to assess its utility in typing of other blood-group determinants on reducing oligosaccharide chains.
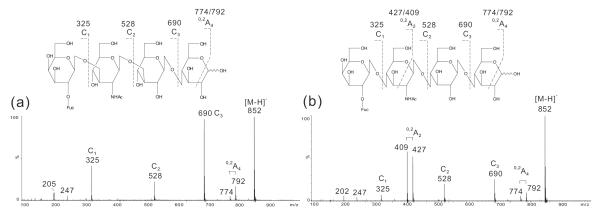
Blood-group H on type 1 and type 2 chains
In the negative-ion spectrum of LNFP-I (Fig. 2a) three C-type ions at m/z 325 (C1), 528 (C2) and 690 (C3) clearly identified the linear sequence of Fuc-Gal-GlcNAc-Gal-Glc (Table 1). The presence of a 0,2A4 doublet at m/z 792/774 indicated a reducing terminal 4-linked Glc. In the spectrum of LnNFP-I (Fig. 2b), the C-type ions (C1 to C3) were similarly obtained together with a 0,2A4 doublet at m/z 792/774. The apparent difference is the presence of an intensive 0,2A2 doublet at m/z 427/409, indicating that the glycosidic bond between Gal and GlcNAc is a 4-linkage but not 3-linkage and therefore defined a type 2 chain.
Figure 2.
Negative-ion ESI-CID-MS/MS product-ion spectra of H pentasaccharides LNFP-I (a) and LnNFP-I (b).
Although not all the linkage information is obtainable from the product-ion spectra under negative-ion ESI-CID-MS/MS condition, the important linear sequence information unambiguously identifies a blood-group H determinant on either a type 1 or type 2 chains.
Blood-group A on type 1 and type 2 chains
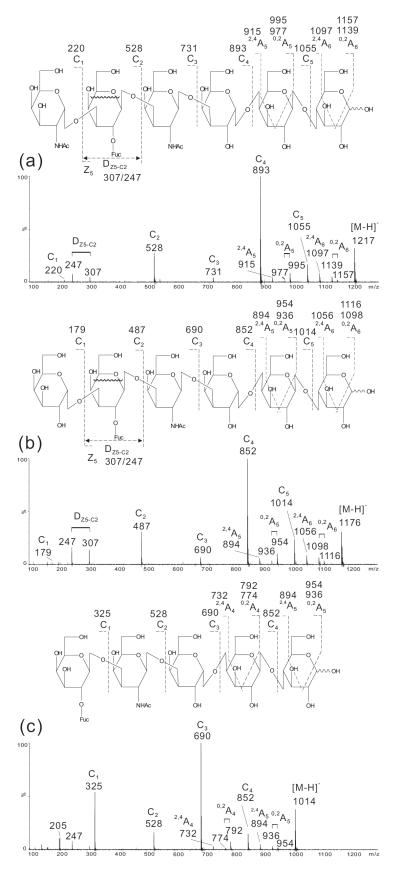
In the spectrum of the hexasaccharide containing blood-group A antigen on type 1 chain (A-Hexa-T1, Fig. 3a) a full set of C-type fragment ions were obtained and the gap between C1 (m/z 220) and C2 (m/z 528) with a mass difference of 308 clearly indicated a branched Fuc at the Gal (162+146). A 0,2A5 doublet at m/z 995/977, together with a 2,4A5 at m/z 935, suggested a reducing terminal 4-linked Glc. A sequence of GlcNAc-Gal(Fuc)-GlcNAc-Gal1-4Glc (Table 1) can be deduced. A unique and prominent DZ4-C2 ion at m/z 307, together with the associated fragment ion at m/z 247 produced by loss of further 2CH2O through multiple cleavages (indicated by the wavy line in the structure of Fig. 3a), further identified the Fuc linked to the sub-terminal Gal, and moreover the Gal linked to a 3-position of the GlcNAc as a double glycosidic cleavage is favored for a 3-linked GlcNAc residue28. The latter was also supported by the lack of a typical 0,2A doublet for a 4-linked GlcNAc. Therefore, a blood-group A determinant on a type 1 chain was identified.
Figure 3.
Negative-ion ESI-CID-MS/MS product-ion spectra of blood group A hexasaccharides A-Hexa-T1 (a) and A-Hexa-T2 (b).
The spectrum of A-hexasaccharide with a type 2 chain (A-Hexa-T2) exhibited similar ions (Fig. 3b), e.g. C1 (m/z 220) C2 (m/z 528), C3 (m/z 731) and C4 (m/z 893), although with lower abundances, and also exhibited the reducing terminal 0,2A5 doublet (m/z 995/977) and 2,4A5 (m/z 935). The unique ion doublet at m/z 307/247 for a Fuc branch at the sub-terminal Gal was also present but with much lower intensity, as the Gal is linked to a 4-position of the GlcNAc and a double glycosidic D-cleavage is less favored as compared with A-Hexa-T1, in which the Gal linked to the 3-position of the GlcNAc (Fig. 3a). The main feature of the product-ion spectrum of A-Hexa-T2 is the 0,2A3 doublet at m/z 630/612 to indicate the Gal 4-linked to a GlcNAc, therefore a type 2 chain.
In both spectra the presence of C1 at m/z 220 and C2 at m/z 528 together with a D-ion doublet at m/z 307/247 defined a GalNAc at the non-reducing end which is linked to a Fuc-branched Gal, indicating a blood-group A determinant. The type 1 or 2 chains can be readily assigned by the absence or presence of 0,2A doublet specific for a 4-linked GlcNAc.
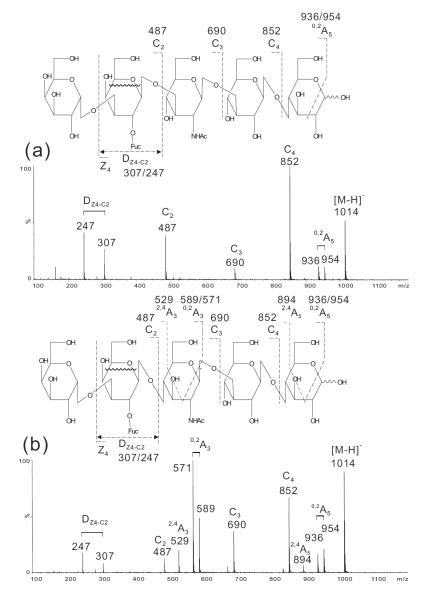
Blood-group B on type 1 and type 2 chains
The spectra of blood-group B hexasaccharides (Fig. 4), B-Hexa-T1 and B-Hexa-T2, are similar to those of hexasaccharides carrying the blood-group A determinants (Fig. 3) apart from mass shifts of 41 due to the difference between the residue Gal and GalNAc at the non-reducing termini for a blood-group B and A, respectively. For example, the C2, C3, C4 and 0,2A5 ions at m/z 487, 690, 852 and 954/936, respectively,were 41 mass units lower than those obtained from the blood-group A hexasaccharides (Fig. 3).
Figure 4.
Negative-ion ESI-CID-MS/MS product-ion spectra of blood group B hexasaccharides B-Hexa-T1 (a) and B-Hexa-T2 (b).
Although the DZ4-C2 ion doublet at m/z 307/247 was similarly produced as those of the blood-group A hexasaccharides, this can be used in conjunction with the C2 ion at m/z 487 to define a blood-group B determinant. Again, the intense 0,2A3 doublet at m/z 571/589 (Fig. 4b) can be used to differentiate a type 2 chain from the type 1 chain (Fig.4a).
Blood-group A, B and H on globoside type 4 chains
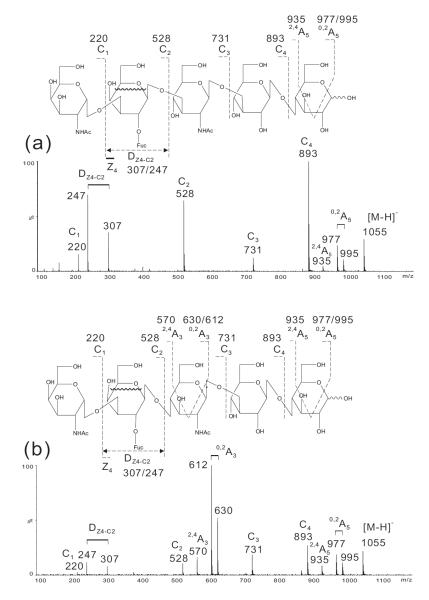
The main features obtained from the negative-ion CID-MS/MS experiments of blood-group A, B and H on type 1 and 2 chains of lacto-N-tetraose (LNT) and lacto-N-neotetraose (LNnT) backbones were further assessed using oligosaccharides with type-4 chains on globoside backbones (Table 1).
Using three oligosaccharides we demonstrate that the assignment of blood-group determinants can be similarly made. In the spectrum of Globo-A-Hepta (Fig. 5a) the DZ5-C2 doublet at m/z 307/247 together with C2 at m/z 528 clearly identified the blood-group A determinant while in the spectrum of Globo-B-Hepta (Fig. 5b) the DZ5-C2 doublet m/z 307/247 and C2 m/z 487 indicated a blood group B. The complete sets of C-type ions, C1, C2, C3, C4 and C5 at m/z 220, 528, 731, 893 and 1055, and at m/z 179, 487, 690, 852 and 1014, for Globo-A-Hepta (Fig. 5a) and Globo-B-Hepta (Fig. 5b), respectively, can be used to define the globo-backbone. For Globo-H-Hexa, the backbone can be similarly defined by the C1 to C4 ions while C1 at m/z 325 indicated a Fuc linked to the terminal Gal (Fig. 5c) and therefore a blood-group H determinant.
Figure 5.
Negative-ion ESI-CID-MS/MS product-ion spectra of Globo-A-Hepta (a), Globo-B-Hepta (b), and Globo-H-Hexa (c).
For all the three globo-oligosaccharides with type 4 chains, the sub-terminal Gal on the reducing side is 4-linked (Table 1). This is different from that of the oligosaccharides with LNT and LNnT backbones, in which the sub-terminal Gal 3-linked. Therefore, 0,2A- and 2,4A-type of fragmentation also occurred for this Gal residue, similar to those obtained with the reducing terminal 4-linked Glc (Fig. 5). These additional fragmentations (e.g. 0,2A5 and 2,4A5 at m/z 995/977 and 915 in Fig. 5a, and at m/z 954/936 and 894 in Fig. 5b, respectively,) are apparently different from the spectra of A-, B- and H-containing oligosaccharides with type 1 and type 2 chains and the LNT/LNnT backbones (see Figs 2, 3 and 4).
Blood-group typing of minor oligosaccharide components of bacterial fermentation products
As shown in the HPLC chromatograms (Fig. S1 in the Supporting Information), several minor components were observed in each of the bacterial fermentation products. Among these, peak 2 of B-Hexa-T1 and peak 3 of A-Hexa-T1 were of good purities and were characterized by the ESI-CID-MS/MS method.
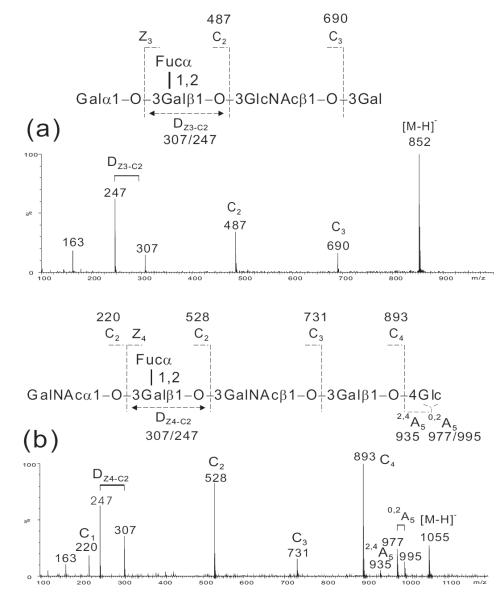
The molecular mass 853 Da obtained from the primary mass spectrum (not shown) of the minor component, peak 2, of B-Hexa-T1 clearly indicated a pentasaccharide with one Hex less than the intended hexasaccharide product (molecular mass 1015 Da). The missing hexose is likely to be either a Glc at the reducing or a Gal at the non-reducing end. The product-ion spectrum (Fig. 6a) of this minor component clearly indicated the absence of a reducing terminal Glc and the presence of a non-reducing terminal blood-group B antigen by the characteristic ion pair at m/z 307/247. A reducing terminal 3-linked Gal rather than a 4-linked Glc can be deduced from the lacking of A-type fragmentations typical for a reducing terminal 4-linked Glc. Therefore, a pentasaccharide Galα1-3(Fucα1-2)Galβ1-3GlcNAcβ1-3Gal can be concluded.
Figure 6.
Negative-ion ESI-CID-MS/MS product-ion spectra of HPLC fraction 1 of B-Hexa-T1 (a) and HPLC fraction 3 of A-Hexa-T1 (b).
The assignment of the sequence of the minor component, peak 3, of A-Hexa-T1 (Fig. S1a in the Supporting Information) was not as straightforward. The molecular mass and the product-ion spectrum (Fig. 6b) were identical to those of the main component, peak 2, of A-Hexa-T1 (Fig. 3a), indicating a hexasaccharide with a non-reducing terminal blood-group A antigen and similar linkages within the hexasaccharide sequence. The possibility of peaks 2 and 3 as α/β isomers can be ruled out as re-injection of either the collected peak 2 or peak 3 gave a single peak at identical retention time without splitting.
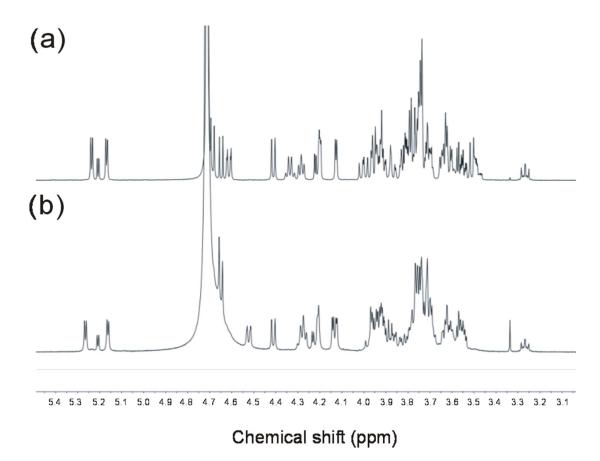
1H-NMR was then carried out to determine the structure of this blood-group A-containing isomeric hexasaccharide. The proton NMR spectra of the major (peak 2; Fig. 7a) and minor (peak 3; Fig. 7b) components of A-Hexa-T1 were acquired and it is clear by simple inspection that the minor component is related to, but not identical with, the major component. Assignments for the anomeric signals were made by comparison with literature data (Table S1 in the Supporting Information). The signals for the non-reducing terminal blood-group A trisaccharide epitope in both major and minor components could be derived by comparison with published values for blood-group A-containing tetra41- and pentasaccharide42 alditols. The reducing end Glc gave rise to signals for H1α at 5.21 ppm and H1β at 4.65 ppm43; the reducing end βGlc H2 signal (3.27 ppm41) is also clearly visible in both spectra (Fig. 7 a and b). The H1 resonance for GlcNAc (4.61 ppm) in the major component (Fig. 7a) was assigned by comparison with the corresponding value for the pentasaccharide alditol R9 reported by Dua et al42. By elimination the doublet at 4.41 ppm can be assigned to βGal at the reducing end. Major differences between the spectra of the major component (Fig. 7a) and the minor component (Fig. 7b) can be seen in the β-anomeric region between 4.7 and 4.4 ppm. The anomeric signal for GlcNAc is not present in the spectrum of the minor component (Fig. 7b), and a new doublet at 4.53 ppm has appeared. In addition, the anomeric signal of the 2,3-linked βGal in the spectrum of the minor component (Fig. 7b) coincides with the reducing end βGlc H1 at 4.65 ppm. Multiplets at 3.53 and 3.51 ppm, in the spectrum of the major component (Fig. 7a), which may be assigned to H4 and H5 of GlcNAc42, are not present in the spectrum of the minor component (Fig. 7b). The NAc methyl signals for the major component coincide at 2.03 ppm (not shown) and those for the minor component are separated at 2.03 and 2.05 ppm (not shown). All the above evidence is consistent with the absence in the minor component of -3GlcNAcβ1- and the presence of an extra -3GalNAcβ1-. Taken together a hexasaccharide carrying a blood-group A antigen with a type 4 chain can tentatively be deduced: GalNAcα1-3(Fucα1-2)Galβ1-3GalNAcβ1-3Galβ1-4Glc.
Figure 7.
1H-NMR at 500 MHz of the major component HPLC-2 (a) and the minor component HPLC-3 (b) in A-Hexa-T1.
In the preparation of these oligosaccharides, the substrate for the enzymatic synthesis was lactose Galβ1-4Glc. The reasons for these minor by-products by removal of a -4Glc from the reducing end (peak 2, B-Hexa-T1) and addition of a -3GalNAcβ1- to the lactose instead of -3GlcNAβ1- (peak 3, A-Hexa-T1) during the fermentation process were not clear and require further investigation. However, the identification of these minor components would certainly contribute to a better understanding of the bacterial fermentation process and assist the optimization of conditions to minimize by-products.
CONCLUSIONS
It has now been recognized that fragmentation in negative-ion mode is much more informative than the positive-ion fragmentation, but unfortunately, negative-ions by deprotonation of neutral oligosaccharides in MALDI are not always straightforward to obtain44 and most negative-ion mass spectrometry has been carried out with ESI. Using Lea and Lex pentasaccharides we clearly demonstrated that the characteristic fragmentation observed for typing of blood-group antigens on either type 1 and type 2 chains are unique to the negative-ion ESI-CID-MS/MS of reducing sugars. These fragmentations did not occur in either the negative-ion ESI-CID-MS/MS of reduced alditols or the positive-ion ESI-CID-MS/MS of reducing sugars. The importance of the reducing terminal hemiacetal functionality for the structure-informative fragmentation we showed here may explain the reason for the lack of the important structural information in the product-ion spectra of oligosaccharides modified by reduction, permethylation or reducing-terminal derivatization. As demonstrated again in the present study, for the internal 4-linked GlcNAc residue of a type 2 chain the prominent 0,2A-doublet is diagnostic. However, the reason for this unique fragmentation is not yet clear and requires further investigation.
The method established is not only applicable to the reducing free oligosaccharides obtained from mammalian milk5 and other secretions14 but also to those released from e.g. polysaccharides by acid hydrolysis45, glycolipids by endoglycoceramidase46,47 or by ozonolysis48, and N-glycans by different peptide-N-glycanases or by hydrazinolysis49. For O-glycans, although some considerable effort has been made towards non-reductive alkaline release11,50, to prevent ‘peeling’ (oligosaccharide chain degradation and/or saccharide destruction) the classical reductive-alkaline hydrolysis, Carlson degradation51, is still the method of choice. Unfortunately, the released sugars are in reduced alditol forms and the structural information obtained here are not available in the negative-ion tandem mass spectra.
Oligosaccharide chains with blood-group antigens on the LNT/LNnT and globoside backbones and the type 1 to type 4 chains are widely distributed in human and mammalian tissues52,53. Identification of the blood-group and chain types is important for defining the roles of carbohydrate involved in various biological events. We have in the past identified a number of novel neutral and acidic oligosaccharide sequences bearing the Lea/x and Leb/y determinant based on the strategy of negative-ion ESI-CID-MS/MS complemented by NMR. Here using the minor unknown oligosaccharide components isolated from the enzymatic synthesis by bacterial fermentation as examples, we again demonstrated the utility of this method for characterization of blood-group H, A and B antigens.
Supplementary Material
Supporting Information Figure S1. HPLC profiles of A-Hexa-T1 (a) and B-Hexa-T1 (b).
Table S1. Assignments for anomeric signals in the 1H spectrum of the minor component (HPLC-3) in A-Hexa-T1.
ACKNOWLEDGMENTS
This work was supported, in part, by a Chinese NSFC grant (31171640), a Wellcome Trust grant (WT093378), and a NCI grant ‘Alliance of Glycobiologists for Detection of Cancer and Cancer Risk’ (U01 CA128416). We thank Dr Colin Herbert for HPLC analysis, and Dr Chris Jones and Ms Caroline Swann, of NIBSC, for recording and interpretation of NMR spectra.
Reference List
- 1.Fredriksson S-A, Podbielska M, Nilsson B, Krotkiewska B, Lisowska E, Krotkiewski H. Archives of Biochemistry and Biophysics. 2010;498:127–135. doi: 10.1016/j.abb.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Podbielska M, Fredriksson S-A, Nilsson B, Lisowska E, Krotkiewski H. Archives of Biochemistry and Biophysics. 2004;429:145–153. doi: 10.1016/j.abb.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 3.Letts JA, Rose NL, Fang YR, Barry CH, Borisova SN, Seto NOL, Palcic MM, Evans SV. Journal of Biological Chemistry. 2006;281:3625–3632. doi: 10.1074/jbc.M507620200. [DOI] [PubMed] [Google Scholar]
- 4.Zivkovic AM, German JB, Lebrilla CB, Mills DA. Proceedings of the National Academy of Sciences. 2011;108:4653–4658. doi: 10.1073/pnas.1000083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode L. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakhour M, Ruvoen-Clouet N, Charpilienne A, Langpap B, Poncet D, Peters T, Bovin N, Le Pendu J. PLoS Pathog. 2009;5:e1000504. doi: 10.1371/journal.ppat.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nystrom K, Le Gall-Recule G, Grassi P, Abrantes J, Ruvoen-Clouet N, Le Moullac-Vaidye B, Lopes AM, Esteves PJ, Strive T, Marchandeau S, Dell A, Haslam SM, Le Pendu J. PLoS Pathog. 2011;7:e1002188. doi: 10.1371/journal.ppat.1002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aspholm-Hurtig M, Dailide G, Lahmann M, Kalia A, Ilver D, Roche N, Vikstrom S, Sjostrom R, Linden S, Backstrom A, Lundberg C, Arnqvist A, Mahdavi J, Nilsson UJ, Velapatino B, Gilman RH, Gerhard M, Alarcon T, Lopez-Brea M, Nakazawa T, Fox JG, Correa P, Dominguez-Bello MG, Perez-Perez GI, Blaser MJ, Normark S, Carlstedt I, Oscarson S, Teneberg S, Berg DE, Bordn T. Science. 2004;305:519–522. doi: 10.1126/science.1098801. [DOI] [PubMed] [Google Scholar]
- 9.Feizi T. Nature. 1985;314:53–57. doi: 10.1038/314053a0. [DOI] [PubMed] [Google Scholar]
- 10.Wang CC, Huang YL, Ren CT, Lin CW, Hung JT, Yu JC, Yu AL, Wu CY, Wong CH. Proceedings of the National Academy of Sciences. 2008;105:11661–11666. doi: 10.1073/pnas.0804923105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chai W, Feizi T, Yuen C-T, Lawson AM. Glycobiology. 1997;7:861–872. doi: 10.1093/glycob/7.6.861. [DOI] [PubMed] [Google Scholar]
- 12.Yuen C-T, Bezouska K, O’Brien J, Stoll MS, Lemoine R, Lubineau A, Kiso M, Hasegawa A, Bockovich NJ, Nicolaou KC, Feizi T. J. Biol. Chem. 1994;269:1595–1598. [PubMed] [Google Scholar]
- 13.Bevilacqua MP, Nelson RM. J. Clin. Invest. 1993;91:379–387. doi: 10.1172/JCI116210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalabi S, Easton RL, Patankar MS, Lattanzio FA, Morrison JC, Panico M, Morris HR, Dell A, Clark GF. Journal of Biological Chemistry. 2002;277:32562–32570. doi: 10.1074/jbc.M205152200. [DOI] [PubMed] [Google Scholar]
- 15.Hong P, Ninonuevo MR, Lee B, Lebrilla C, Bode L. British Journal of Nutrition. 2009;101:482–486. doi: 10.1017/s0007114508025804. [DOI] [PubMed] [Google Scholar]
- 16.Lesman-Movshovich E, Lerrer B, Gilboa-Garber N. Canadian Journal of Microbiology. 2003;49:230–235. doi: 10.1139/w03-027. [DOI] [PubMed] [Google Scholar]
- 17.Idota T, Kawakami H, Yuji M, Makihiro S. Bioscience, Biotechnology, and Biochemistry. 1995;59:417–419. doi: 10.1271/bbb.59.417. [DOI] [PubMed] [Google Scholar]
- 18.Yolken RH, Peterson JA, Vonderfecht SL, Fouts ET, Midthun K, Newburg DS. J Clin Invest. 1992;90:1984–1991. doi: 10.1172/JCI116078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sodetz JM, Paulson JC, McKee PA. Journal of Biological Chemistry. 1979;254:10754–10760. [PubMed] [Google Scholar]
- 20.Watkins WM. Glycoproteins: Their composition, structure and function. In: Gottschalk A, editor. Blood-Group Specific Substances. Elsevier; Amsterdam: 1972. pp. 830–899. [Google Scholar]
- 21.Podbielska M, Fredriksson SA, Nilsson B, Lisowska E, Krotkiewski H. Arch. Biochem. Biophys. 2004;429:145–153. doi: 10.1016/j.abb.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Savage AV, Donohue JJ, Koeleman CA, Van den Eijnden DH. Eur. J. Biochem. 1990;193:837–843. doi: 10.1111/j.1432-1033.1990.tb19407.x. [DOI] [PubMed] [Google Scholar]
- 23.Dell A, Morris HR. Science. 2001;291:2351–2356. doi: 10.1126/science.1058890. [DOI] [PubMed] [Google Scholar]
- 24.Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, Backstrom M, Costello CE, Hansson GC, Hiki Y, Ishihara M, Ito H, Kakehi K, Karlsson N, Hayes CE, Kato K, Kawasaki N, Khoo KH, Kobayashi K, Kolarich D, Kondo A, Lebrilla C, Nakano M, Narimatsu H, Novak J, Novotny MV, Ohno E, Packer NH, Palaima E, Renfrow MB, Tajiri M, Thomsson KA, Yagi H, Yu SY, Taniguchi N. Molecular & Cellular Proteomics. 2010;9:719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kogelberg H, Piskarev VE, Zhang Y, Lawson AM, Chai W. Eur. J Biochem. 2004;271:1172–1186. doi: 10.1111/j.1432-1033.2004.04021.x. [DOI] [PubMed] [Google Scholar]
- 26.Wheeler SF, Harvey DJ. Analytical Chemistry. 2000;72:5027–5039. doi: 10.1021/ac000436x. [DOI] [PubMed] [Google Scholar]
- 27.Zaia J, McClellan JE, Costello CE. Anal. Chem. 2001;73:6030–6039. doi: 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- 28.Chai W, Piskarev V, Lawson AM. Anal. Chem. 2001;73:651–657. doi: 10.1021/ac0010126. [DOI] [PubMed] [Google Scholar]
- 29.Chai W, Piskarev V, Lawson AM. J. Am. Soc. Mass Spectrom. 2002;13:670–679. doi: 10.1016/S1044-0305(02)00363-X. [DOI] [PubMed] [Google Scholar]
- 30.Chai W, Piskarev VE, Zhang Y, Lawson AM, Kogelberg H. Arch. Biochem. Biophys. 2005;434:116–127. doi: 10.1016/j.abb.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 31.Martin MJ, Feizi T, Leteux C, Pavlovic D, Piskarev VE, Chai W. Glycobiology. 2002;12:829–835. doi: 10.1093/glycob/cwf094. [DOI] [PubMed] [Google Scholar]
- 32.Chai W, Piskarev VE, Mulloy B, Liu Y, Evans P, Osborn HMI, Lawson AM. Anal. Chem. 2006;78:1581–1592. doi: 10.1021/ac051606e. [DOI] [PubMed] [Google Scholar]
- 33.Yu G, Zhao X, Yang B, Ren S, Guan H, Zhang Y, Lawson AM, Chai W. Anal. Chem. 2006;78:8499–8505. doi: 10.1021/ac061416j. [DOI] [PubMed] [Google Scholar]
- 34.Leteux C, Chai W, Nagai K, Herbert CG, Lawson AM, Feizi TJ. Biol. Chem. 2001;276:12539–12545. doi: 10.1074/jbc.M010291200. [DOI] [PubMed] [Google Scholar]
- 35.Chai W, Leteux C, Westling C, Lindahl U, Feizi T. Biochemistry. 2004;43:8590–8599. doi: 10.1021/bi036250k. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Yu G, Zhao X, Liu H, Guan H, Lawson AM, Chai W. J Am. Soc. Mass Spectrom. 2006;17:621–630. doi: 10.1016/j.jasms.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Xu J, Xue C, Dong P, Sheng W, Yu G, Chai W. Glycoconjugate Journal. 2008;25:481–492. doi: 10.1007/s10719-007-9096-2. [DOI] [PubMed] [Google Scholar]
- 38.Domon B, Costello CE. Glycoconjugate J. 1988;5:397–409. [Google Scholar]
- 39.Tseng K, Hedrick JL, Lebrilla CB. Anal. Chem. 1999;71:3747–3754. doi: 10.1021/ac990095r. [DOI] [PubMed] [Google Scholar]
- 40.Drouillard S, Driguez H, Samain E. Angewandte Chemie International Edition. 2006;45:1778–1780. doi: 10.1002/anie.200503427. [DOI] [PubMed] [Google Scholar]
- 41.Azurmendi HF, Bush CA. Carbohydrate Research. 2002;337:905–915. doi: 10.1016/s0008-6215(02)00070-8. [DOI] [PubMed] [Google Scholar]
- 42.Dua VK, Rao BN, Wu SS, Dube VE, Bush CA. Journal of Biological Chemistry. 1986;261:1599–1608. [PubMed] [Google Scholar]
- 43.Roslund MU, T+ñhtinen P, Niemitz M, Sj+Âholm R. Carbohydrate Research. 2008;343:101–112. doi: 10.1016/j.carres.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 44.Domann P, Spencer DIR, Harvey DJ. Rapid Commun. Mass Spectrom. 2012;26:469–479. doi: 10.1002/rcm.5322. [DOI] [PubMed] [Google Scholar]
- 45.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodríguez E, Campanero-Rhodes AS, Costa J, Brown GD, Chai W. J Biol. Chem. 2006;281:5771–5779. doi: 10.1074/jbc.M511461200. [DOI] [PubMed] [Google Scholar]
- 46.Karlsson H, Halim A, Teneberg S. Glycobiology. 2010;20:1103–1116. doi: 10.1093/glycob/cwq070. [DOI] [PubMed] [Google Scholar]
- 47.Benktander J, +àngstr+Âm J, Breimer ME, Teneberg S. Journal of Biological Chemistry. 2012;287:31712–31724. doi: 10.1074/jbc.M112.387654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pacuszka T, Fishman PH. J. Biol. Chem. 1990;265:7673–7678. [PubMed] [Google Scholar]
- 49.Takasaki S, Mizuochi T, Kobata A. Methods Enzymol. 1982;83:263–268. doi: 10.1016/0076-6879(82)83019-x. [DOI] [PubMed] [Google Scholar]
- 50.Yu G, Zhang Y, Zhang Z, Song L, Wang P, Chai W. Anal. Chem. 2010;82:9534–9542. doi: 10.1021/ac102300r. [DOI] [PubMed] [Google Scholar]
- 51.Iyer RN, Carlson DM. Arch. Biochem. Biophys. 1971;142:101–105. doi: 10.1016/0003-9861(71)90263-3. [DOI] [PubMed] [Google Scholar]
- 52.Okajima T, Nakamura Y, Uchikawa M, Haslam DB, Numata S. i., Furukawa K, Urano T, Furukawa K. Journal of Biological Chemistry. 2000;275:40498–40503. doi: 10.1074/jbc.M006902200. [DOI] [PubMed] [Google Scholar]
- 53.Fujitani N, Liu Y, Okamura T, Kimura H. Journal of Histochemistry & Cytochemistry. 2000;48:1649–1655. doi: 10.1177/002215540004801208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1. HPLC profiles of A-Hexa-T1 (a) and B-Hexa-T1 (b).
Table S1. Assignments for anomeric signals in the 1H spectrum of the minor component (HPLC-3) in A-Hexa-T1.