Abstract
Aim
To simultaneously determine the localization of histones and protamines within human sperm nuclei.
Methods
Immunofluorescence of the core histones and protamines and fluorescence in situ hybridization of the telomere region of chromosome 16 was assessed in decondensed human sperm nuclei.
Results
Immunofluorescent localization of histones, protamine 1 (PRM1) and protamine 2 (PRM2) along with fluorescence in situ hybridization localization of chromosome 16 telomeric sequences revealed a discrete distribution in sperm nuclei. Histones localized to the posterior ring region (i.e. the sperm nuclear annulus), whereas PRM1 and PRM2 appeared to be dispersed throughout the entire nucleus.
Conclusion
The co-localization of the human core sperm histones with the telomeric regions of chromosome 16 is consistent with the reorganization of specific non-protamine regions into a less compacted state.
Keywords: human sperm nucleus, histone, protamine, telomere
1 Introduction
Spermatozoa are the product of a complex cellular program of differentiation initiating from the spermatogonia [1, 2]. This is characterized by a myriad of changes including chromosomal recombination and segregation culminating in the postmeiotic reorganization of the genome into a highly condensed transcriptionally inactive form [3]. These dynamic global changes are keyed through a variety of mechanisms including DNA methylation, phosphorylation, acetylation, methylation and ubiquitination of the histone component [4, 5]. This sets the stage for the ordered substitution of histones with transition proteins then subsequently with sperm-specific protamines yielding a condensed state that is a hallmark of this transition.
Although most of the histones are replaced with protamines, the human male gamete retains approximately 15% of its genome in a histone-bound state [6]. This is in direct contrast to other species such as the mouse where replacement exceeds 98% [7]. Association of histone and protamines with the spermatozoon’s genome is not random [6]. Structural sequences, such as Alu repeats and telomeres, associate with protamines and histones, respectively, whereas centromeric regions are partitioned between the histone and protamine compartments [8] and are part of the loop DNA [9].
Some gene-rich regions are specifically distributed between the two nucleoprotein components. In human spermatozoa, the embryo-specific ε-globin and γ-globin and the paternally imprinted IGF2 genes are histone-associated, like the promoter and nuclear matrix attachment regions of the PRM1→PRM2→TNP2 locus. In contrast, the postnatally activated β-globin and δ-globin genes are protamine-associated [8, 10]. However, the majority of the PRM1→PRM2→TNP2 locus shows no enrichment for histone or protamine association [8]. Even though specific examples have been delineated, the sub-cellular distribution of histones and protamines in the sperm nucleus has yet to be defined. To begin to address this issue the distribution of histones and protamines within the mature human sperm nucleus was examined by immunofluorescence. The core histones and chromosome 16 telomeric regions were localized at the base of the sperm nuclei. In contrast, both protamines were distributed more evenly throughout the nucleus. The significance of this distribution as a means to partition the genome is discussed.
2 Materials and methods
2.1 Semen samples, preparation of nuclei and slides
Human semen samples were randomly collected from the donor pool in accordance with the Wayne State University Human Investigation Committee protocol 095701MP2F, processed then frozen in FSB, Frozen Storage Buffer at −80°C as described [11].
Briefly, frozen human spermatozoa were quickly thawed at room temperature, then washed once with cold 50 mmol/L Tris-HCl buffer, pH 7.4 containing a complete protease inhibitor cocktail (CPI, Complete Mini tablets; Roche Applied Science, Indianapolis, IN, USA). After centrifugation at 2 000 × g for 5 min at 4°C, the cells were resuspended to a concentration of 1 × 107 cells/mL in 50 mmol/L Tris-HCl buffer, pH 7.4, containing 0.5% sodium lauryl sulfate (SDS) and nuclei isolated as described [12]. Briefly, resuspended spermatozoa were transferred in a type A glass dounce homogeneizer to dislodge the tails from the heads. Dissociation was verified under bright field microscopy. The heads were then pelleted by centrifugation at 2 000 × g for 10 min at 4°C. The supernatant was discarded and the heads were resuspended in a sucrose solution (2.2 mol/L sucrose, 50 mmol/L Tris pH 7.4, 5 mmol/L MgOAc) to a final concentration of 5 × 106 cells/mL and transferred to a type C glass dounce homogeneizer. After homogenizing with three strokes, the suspension of nuclei was layered onto sucrose/CsCl cushions (2.2 mol/L sucrose, 10 mmol/L Tris pH 7.4, 5 mmol/L MgCl2, 0.89 mol/L CsCl) and centrifuged at 113 000 × g for 90 min at 4°C in a Beckman XL-90 Ultracentrifuge using a SW41 Ti rotor (Beckman Coulter Inc., Fullerton, CA, USA). The nuclei recovered from centrifugation were then resuspended in FSB to a final concentration of 2 × 107/mL and stored at −80°C.
2.2 Decondensation of sperm nuclei and cells
Sperm nuclei were mixed with 0.5 mL of cold phosphate buffered saline (PBS), pH 7.4, containing CPI. An aliquot containing 500 nuclei was then subjected to centrifugation onto a slide at room temperature for 5 min at 2 000 × g using a cytospin 2 cytofuge (Thermo Shandon, Pittsburgh, PA, USA). The attached nuclei were then swollen by gently overlaying 50 μL of a solution containing 0.05 mg/mL heparin and 10 mmol/L dithiothreitol followed by incubation on ice for 30 min, as described [13]. Alternatively, the samples were treated with 2.5 mmol/L DTT, 0.2% Triton X-100 and 100 U of heparin in PBS, for 30 min [14]. The slides were then washed for 5 min in PBS and the cells fixed in 4% formaldehyde. Samples were then dehydrated through a 1 min series of graded cold ethanol washes of 50%, 70%, 95% and 100%, then fixed in −20°C methanol for 20 min.
2.3 Immunolocalization
Anti-core histone sheep polyclonal (1.15 mg/mL) antibody was purchased from Abcam (Cambridge, UK). Mouse monoclonal anti-human protamine 1 (PRM1) and protamine 2 (PRM2) antibodies were a generous gift from Dr Rod Balhorn, Lawrence Livermore National Laboratory (Livermore, CA, USA).
Non-specific epitopes to mouse or goat antibodies revealed by nuclear swelling were blocked with a solution of PBS containing 5% BSA or 5 % rabbit serum for 1 h at room temperature. Before use, the anti-core histone antibody was diluted 1:300; anti-PRM1 and PRM2, 1:200, in their blocking buffer. The slides were then overlayed with their respective primary antibody solution then hybridized overnight at 4°C. The following day the hybridized samples were washed using a series of 10 min PBS washes to remove unreacted antibody. PRM1 and PRM2 immunoreactive species were revealed following a 1 h, room temperature incubation with 1:500 diluted CY5-conjugated goat anti-mouse antibody (Biomeda, Foster City, CA, USA). Histone immunoreactive species were first incubated for 1 h at room temperature, with biotin conjugated rabbit anti-sheep antibody (PIERCE, Rockford, IL, USA), followed by a 30 min incubation with Alexa Fluor 488-conjugated streptavidin (Molecular Probes, Carlsbad, CA, USA). The nuclei were then counterstained by overlaying 10 μL of 0.5 μg/mL 4′-6-Diamidino-2-phenylindole (DAPI) containing Vectashield (Vector Labs Inc, Burlingame, CA, USA).
2.4 Fluorescence in situ hybridization (FISH)
FISH was essentially as described by Zalensky et al. [13]. Briefly, following immunolocalization, nuclei were fixed in 4% formaldehyde for 10 min at room temperature. The samples were then dehydrated through a series of 50%, 70%, 95% and 100% ethanol washes then denaturated in 70% deionized formamide buffered with 2 × SSC that contained 300 mmol/L NaCl, 30 mmol/L sodium citrate buffer, pH 7.0, at 70°C for 3 min. The chromosome 16 telomeric probe was labeled with dUTP-biotin by polymerase chain reaction (PCR) using specific forward and reverse primers 5′-AAAGCTCTCAGAACCTCCCC-3′, 5′-AGAGGTTCCCATGTAGTTCC-3′ respectively. The PCR reaction was carried out for 35 cycles of 30 s at 94°C, 45 s at 55°C and 40 s at 72°C using HotStarTaq DNA polymerase (Qiagen, Valentia, CA, USA). After synthesis, the probe was purified by FlexiPrep, as suggested by the manufacturer (Amersham Bioscience, Piscataway, NJ, USA), then denatured in hybridization solution containing 50 % deionized formamide buffered with 2 × SSC at 75°C for 5 min. Hybridization was then carried out at 37°C in a humidified chamber for 16 h. The post-hybridized sample was then washed at room temperature in 2 × SSPE (300 mmol/L NaCl, 20 mmol/L NaH2PO4, 2 mmol/L EDTA, pH 7.0) for 10 min, followed by 1 × SSPE for 10 min, then 2 × SSPE at 45°C for 10 min and, finally, 2 × SSPE at room temperature for 10 min. The signal was then visualized with Alexa Fluor 594-conjugated streptavidin by fluorescence microscopy.
2.5 Image capture and processing
Images were captured using a Leica DMRA2 fluorescence microscope (100 × oil immersion objective) then processed using Image Pro-express 5.1 (MediaCybernetics, Bethesda, MD, USA). Images were pseudocolored then merged. DNA stained with DAPI was colored blue whereas immunofluorescence signals from the various antibodies were colored yellow, green or red. A random set of 10 fields that contained at least 50 nuclei were selected for immunofluorescence signal frequency analysis. The number of spermatozoa or nuclei showing the observed pattern in each field examined was assessed.
3 Results
3.1 Distribution of histones and protamines in the sperm nucleus
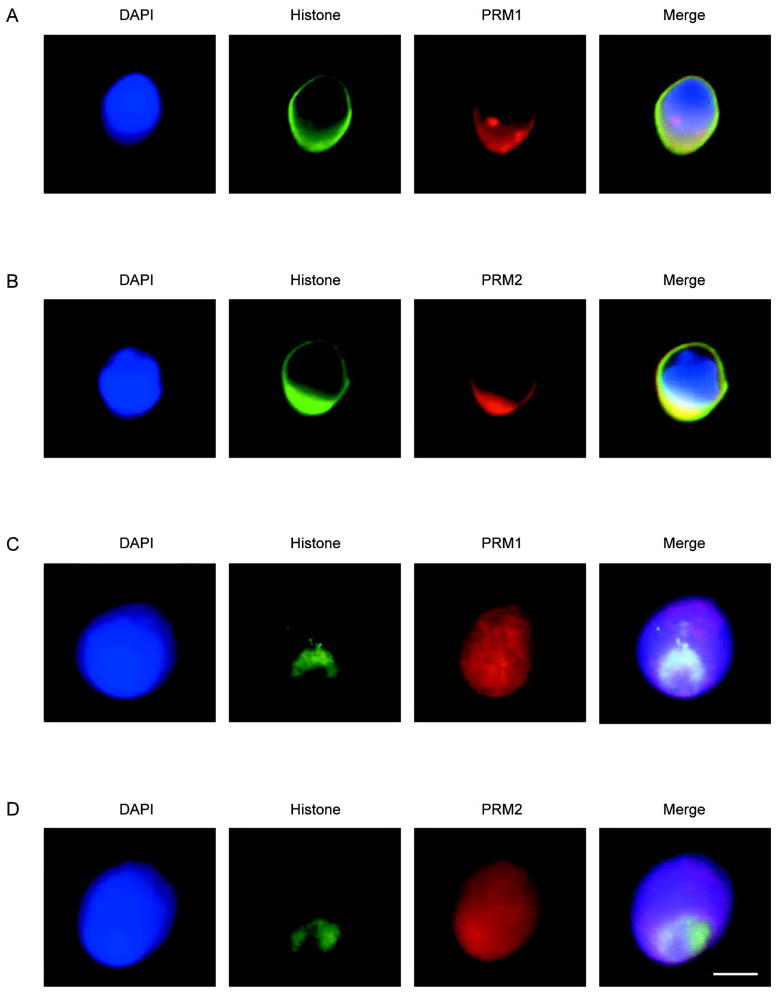
An immunofluorescence assay using an anti-core histone antibody and antibodies prepared against PRM1 and PRM2 was used to immunolabel histones and protamines respectively in human sperm nuclei. Epitopes were revealed by decondensation prior to the application of the antibodies [19]. Two decondensing protocols essentially differing in the concentration of dithiothreitol (DTT) and heparin, were assessed [13, 14]. As shown in Figure 1A and 1B, both histones and protamines were distributed towards the surface of the nuclei when the nuclei were treated with 2.5 mmol/L DTT plus 100 U of heparin. In contrast, when the nuclei were first treated with 10 mmol/L DTT plus 0.05 mg/mL heparin prior to the application of the anti-histone antibody, the distribution of the core histones appeared altered. Surprisingly, as shown in Figure 1C and 1D, 95% of the images analyzed showed that the histones were radically distributed from the posterior in what appeared as a ring-like structure to the postacrosomal region. As summarized in Table 1, this pattern of the histone antibody was observed in the majority of the fields examined. In contrast, at least 98% of images showed that the PRM1 and PRM2 protamines were distributed throughout the nucleus.
Figure 1.
Immunofluorescent localization of the core histones, protamine 1 (PRM1) and protamine 2 (PRM2) in human sperm nuclei. (A): Dual histone and PRM1 immunofluorescence in decondensed nuclei. (B): Dual histone and PRM2 immunofluorescence in decondensed nuclei. (C): Immunofluorescent colocalization of core histones with PRM1 in decondensed and permeablized nuclei. (D) Immunofluorescent colocalization of histones with PRM2 in decondensed and permeablized nuclei. DNA stained with blue 4′-6-diamidino-2-phenylindole (DAPI), histones green, with Alexa Fluor 488-conjugated streptavidin and protamines red with CY5-conjugated anti-mouse antibody. The merged pseudo-color image is shown in the right most panel. Scale bar=5 μm. of a histone-enriched territory well apart from the PRM1 and PRM2 regions distributed throughout the nucleus. The question arises, what does this unique packaging reflect?
Table 1.
The frequency of immunofluorescence observed. PRM1, protamine 1; PRM2, protamine 2.
| Antibody | Signal frequency (f) | Total (n) | Ratio (%) (f/n) |
|---|---|---|---|
| Core histone | 95 | 100 | 95 |
| PRM1 | 98 | 100 | 98 |
| PRM2 | 99 | 100 | 99 |
To examine whether this distribution resulted from the physical preparation of nuclei, dual immunolabeling and differential interference contrast (DIC) microscopy of intact spermatozoa using the histone antibody together with either the PRM1 antibody or the PRM2 antibody was undertaken. The results are summarized in Figure 2A and 2B and confirm that the histones are distributed from the posterior ring to the postacrosomal region. Perhaps this reflects the sequence-specific compartmentalization
Figure 2.
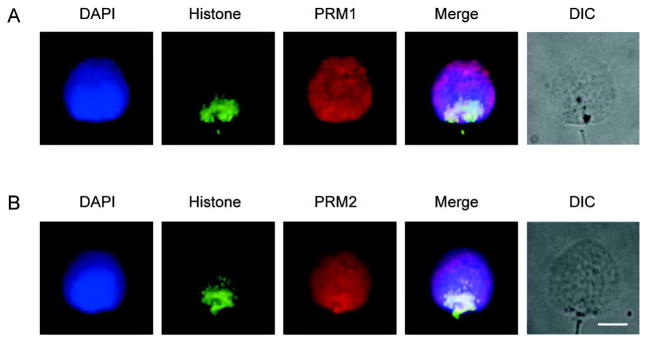
Immunofluorescent colocalization of the core histones with protamine 1 (PRM1) and protamine 2 (PRM2) in permeablized and decondensed human sperm cells. (A): Immunofluorescent colocalization of histones with anti-core histones antibody plus PRM1 with anti-protamine 1. (B): Immunofluorescent colocalization of histones with anti-core histones antibody plus PRM2 with anti-protamine 2. The corresponding grey-scale DIC image of the intact permeablized cells is shown. DNA stained with blue 4′-6-diamidino-2-phenylindole (DAPI), histones green, with Alexa Fluor 488-conjugated streptavidin and protamines red with CY5-conjugated goat anti-mouse antibody. The merged pseudo-color image is shown in the right most panel. Scale bar=5 μm. Note that core the histones localize towards the post acrosomal posterior ring region end while PRM1 and PRM2 occupy the entire nucleus.
3.2 Histone-Telomere
Previous studies report the association of telomere regions with sperm histones, and those of chromosome 16 using PCR analysis [8, 15, 16]. Co-localization of the chromosome 16 telomere together with the core histones within the human sperm nuclei was determined. This employed fluorescence in situ hybridization analysis of the chromosome 16 telomere along with the immunofluorescent detection of the core histones. The results of this analysis in human sperm nuclei are shown in Figure 3. As summarized in Table 2 the chromosome16 telomere is histone-bound.
Figure 3.
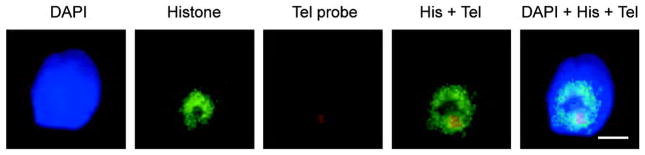
Distribution of the core histones and the telomeric region of human chromosome 16 within the permeablized and decondensed sperm nucleus. The fluorescence in situ hybridization signal (FISH) shows that the human chromosome 16 telomeric region is located within an immunofluorescent core histone rich region. The relative location of the biotin-labeled human chromosome 16 telomeric region is indicated in red with streptavidin Alexa Fluor 594 whereas the core histone region marked by green with Alexa Fluor 488. The merged pseudo-color image is shown in the right most panel. Scale bar=5 μm. DAPI, 4′-6-diamidino-2-phenylindole; Tel, telomere; His, histones.
Table 2.
The frequency of dual immunofluorescence observed. PRM1, protamine 1; PRM2, protamine 2.
| Antibody | Signal frequency (f) | Total (n) | Ratio (%) (f/n) |
|---|---|---|---|
| Core histone + PRM1 | 45 | 50 | 90 |
| Core histone + PRM2 | 42 | 50 | 84 |
| Core histone + telomere | 35 | 50 | 70 |
4 Discussion
Unlike spermatozoa from other species, those from man are characterized by their somewhat incomplete replacement of histones with protamines. Approximately 15% of the human sperm genome remains histone-associated [7], in comparison with 1%–2% of the mouse sperm genome. Immunofluorescence analysis has previously shown that the histones are peripherally distributed around the mouse spermatozoon nucleus [14]. Their corresponding distribution in human spermatozoa is less well-characterized, although histone H2B has been shown to localize specifically to the telomeres and is observed as a punctuate structure [17]. This punctuate distribution is reiterated in specific regions of human gene clusters enriched in the histone complement [8, 10]. We have yet to garner a global understanding of the unique arrangement of these nuclear proteins in human spermatozoa.
The physical organization of the nucleoprotamine and nucleohistone components of human sperm nuclei was assessed by immunolocalization and in situ hybridization. Sperm nuclei present a highly compact structure that is only accessible to probing when relaxed and the membrane has been compromised. This is typically facilitated with the use of DTT and heparin [18]. As demonstrated from the above, the concentration of the reducing agent, i.e., the use of 2.5 mmol/L or 10 mmol/L DTT can markedly affect the outcome. It is clear that independent of this factor, the histones localize towards the posterior region of the nucleus and can be observed at the periphery when mild reducing conditions are employed. Interestingly, when higher concentrations of reducing agent are used, the protamines are distributed throughout the nucleus, appearing well-separated from the dominant histone fraction. A uniform distribution of protamines within human sperm nuclei is also reported by Zalensky and collegues [19]. The results obtained using mild decondensation conditions are comparable to those previously observed with intact mouse sperm nuclei [14]. Together, these studies emphasize the importance of sperm nuclei decondensation to permit the antibodies to recognize their epitopes.
It is clear that human sperm chromatin is distributed between histone and protamines in a non-random manner [8], and that these two components localize to distinct regions of the sperm nucleus. These reiterate the independent uniform distribution of PRM1 [19] and postacrosomal localization of histones [17, 20] that were previously shown in human spermatozoa.
The unique packaging of the sperm nucleus might be essential for proper unpacking of the male genome during fertilization [8, 21]. For example, the nucleohistone component of the nucleus could serve as a template for the replacement of protamines by histones after fertilization [22]. This could be initiated by the binding of chromatin remodelers at acetylated histones, such as H4K8ac and H4K12ac, which are present in the nuclei prior to fertilization [22]. Others have suggested that the histones in the sperm nuclei could influence which genes are to be transcribed after fertilization [7, 10].
Further characterization of the human sperm chromatin components revealed the co-localization of chromosome 16 telomere with the core histones. Previous studies report that histones associate with specific regions that include the telomeres, promoter and repeating regions and certain small gene-rich regions [6, 8, 23]. The nucleohistone components localized in the posterior region of the nuclei, and extend to the region occupied by the sperm nuclear annulus, likely provide an anchor [24, 25].
Taken together, the data presented in this study show the physical organization of the nucleoprotamine and nucleohistone components within human sperm nuclei. These two components are not randomly distributed, but are generally organized into specific regions, with the protamine-compacted chromatin splayed throughout the nucleus and equatorial regions of sperm nuclei, and the histone-compacted chromatin localized in the postacrosomal region, extending to the sperm nuclear annulus. It is not known to what extent the histone replacement with protamines is precise or stochastic. However, studies of the human protamine locus show that in human spermatozoa the promoter region remains histone-bound, whereas the coding region of PRM1 is protamine-bound [8]. Perhaps this unique packaging is essential for appropriate decondensation and reorganization of the paternal genome following fertilization.
Acknowledgments
This work was supported by National Institute of Child Health and Development Grant HD36512 to Stephen A. Krawetz, and a Biotechnology and Biological Sciences Research Council (BBSRC), UK, grant to David Miller. Claudia Lalancette is supported inpart by a Wayne State University postdoctoral recruiting award.
References
- 1.Holstein AF, Schulze W, Davidoff M. Understanding spermatogenesis is a prerequisite for treatment. Reprod Biol Endocrinol. 2003;1:107. doi: 10.1186/1477-7827-1-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krawetz SA. Paternal contribution: new insights and future challenges. Nat Rev Genet. 2005;6:633–42. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- 3.Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, Khochbin S. Establishment of male-specific epigenetic information. Gene. 2005;345:139–53. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–8. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 5.Govin J, Caron C, Lestrat C, Rousseaux S, Khochbin S. The role of histones in chromatin remodelling during mammalian spermiogenesis. Eur J Biochem. 2004;271:3459–69. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 6.Gatewood JM, Cook GR, Balhorn R, Bradbury EM, Schmid CW. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–4. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- 7.Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6. [PubMed] [Google Scholar]
- 8.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 9.Yaron Y, Kramer JA, Gyi K, Ebrahim SA, Evans MI, Johnson MP, et al. Centromere sequences localize to the nuclear halo of human spermatozoa. Int J Androl. 1998;21:13–8. doi: 10.1046/j.1365-2605.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- 10.Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. Histone- and protamine-DNA association: conservation of different patterns within the beta-globin domain in human sperm. Mol Cell Biol. 1998;18:3350–6. doi: 10.1128/mcb.18.6.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodrich R, Johnson G, Krawetz SA. The preparation of human spermatozoal RNA for clinical analysis. Arch Androl. 2007;53:161–7. doi: 10.1080/01485010701216526. [DOI] [PubMed] [Google Scholar]
- 12.Kramer JA, Krawetz SA. Nuclear matrix interactions within the sperm genome. J Biol Chem. 1996;271:11619–22. doi: 10.1074/jbc.271.20.11619. [DOI] [PubMed] [Google Scholar]
- 13.Zalensky AO, Tomilin NV, Zalenskaya IA, Teplitz RL, Bradbury EM. Telomere-telomere interactions and candidate telomere binding protein(s) in mammalian sperm cells. Exp Cell Res. 1997;232:29–41. doi: 10.1006/excr.1997.3482. [DOI] [PubMed] [Google Scholar]
- 14.Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, et al. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci. 1999;112 (Pt 20):3537–48. doi: 10.1242/jcs.112.20.3537. [DOI] [PubMed] [Google Scholar]
- 15.Gineitis AA, Zalenskaya IA, Yau PM, Bradbury EM, Zalensky AO. Human sperm telomere-binding complex involves histone H2B and secures telomere membrane attachment. J Cell Biol. 2000;151:1591–8. doi: 10.1083/jcb.151.7.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–8. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- 17.Zalensky AO, Siino JS, Gineitis AA, Zalenskaya IA, Tomilin NV, Yau P, et al. Human testis/sperm-specific histone H2B (hTSH2B). Molecular cloning and characterization. J Biol Chem. 2002;277:43474–80. doi: 10.1074/jbc.M206065200. [DOI] [PubMed] [Google Scholar]
- 18.Mudrak O, Tomilin N, Zalensky A. Chromosome architecture in the decondensing human sperm nucleus. J Cell Sci. 2005;118:4541–50. doi: 10.1242/jcs.02581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zalensky AO, Breneman JW, Zalenskaya IA, Brinkley BR, Bradbury EM. Organization of centromeres in the decondensed nuclei of mature human sperm. Chromosoma. 1993;102:509–18. doi: 10.1007/BF00368344. [DOI] [PubMed] [Google Scholar]
- 20.van Roijen HJ, Ooms MP, Spaargaren MC, Baarends WM, Weber RF, Grootegoed JA, et al. Immunoexpression of testis-specific histone 2B in human spermatozoa and testis tissue. Hum Reprod. 1998;13:1559–66. doi: 10.1093/humrep/13.6.1559. [DOI] [PubMed] [Google Scholar]
- 21.Zalensky A, Zalenskaya I. Organization of chromosomes in spermatozoa: an additional layer of epigenetic information? Biochem Soc Trans. 2007;35:609–11. doi: 10.1042/BST0350609. [DOI] [PubMed] [Google Scholar]
- 22.van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 2006;298:458–69. doi: 10.1016/j.ydbio.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 23.Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, et al. Postmeiotic sex chromatin in the male germline of mice. Curr Biol. 2006;16:660–7. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- 24.Barone JG, De Lara J, Cummings KB, Ward WS. DNA organization in human spermatozoa. J Androl. 1994;15:139–44. [PubMed] [Google Scholar]
- 25.Ward WS, Coffey DS. Identification of a sperm nuclear annulus: a sperm DNA anchor. Biol Reprod. 1989;41:361–70. doi: 10.1095/biolreprod41.2.361. [DOI] [PubMed] [Google Scholar]