Abstract
The hepatitis B virus (HBV) is a small enveloped DNA virus that belongs to the Hepadnaviridae family. HBV can cause acute and persistent infection which can lead to hepatocellular carcinoma (HCC). MicroRNAs (miRNAs) play a crucial role in the main cellular events. The dysregulation of their expression has been linked to the development of the cancer as well as to viral interference. This chapter will describe the involvement of miRNAs in the case of HBV infection and their implication in the development of the HBV-related diseases.
Keywords: hepatitis B virus, microRNA, hepatocellular carcinoma
1. Introduction
The microRNAs (miRNAs or miRs) are small non-coding RNAs of 19–23 nucleotides that play key roles in the regulation of almost every cellular process in all multicellular eukaryotes [1]. As intracellular pathogens, viruses are affected by these post-transcriptional modulators and have evolved to subvert them. Several viruses, especially the herpesviruses, encode for their own miRNAs that increase their replication potential and/or allow the evasion from the innate immune system [2]. Other viruses, such as the hepatitis B virus (HBV), modulate the cellular miRNAs in order to achieve the same effects.
HBV is a small enveloped DNA virus that belongs to the Hepadnaviridae family. It primarily infects hepatocytes and causes acute and chronic liver disease. Among the 2,000 million people worldwide infected with HBV, more than 350 million remain chronically infected and become carriers of the virus [3]. Epidemiological studies have uncovered chronic HBV infection as the major etiological factor in the development of hepatocellular carcinoma (HCC) [4]. Despite the availability of an efficient vaccine, persistent HBV infection remains a challenging global health issue. The recent discovery of miRNAs involvement in HBV infection provides new insights into the virus biology and pathogenesis [5,6].
This chapter will outline the roles of miRNAs in the HBV biology and associated pathogenesis. We will also outline present and future miRNA-based strategies for the diagnosis, prognosis and treatment of the HBV-related diseases.
2. Biogenesis and Functions of miRNAs
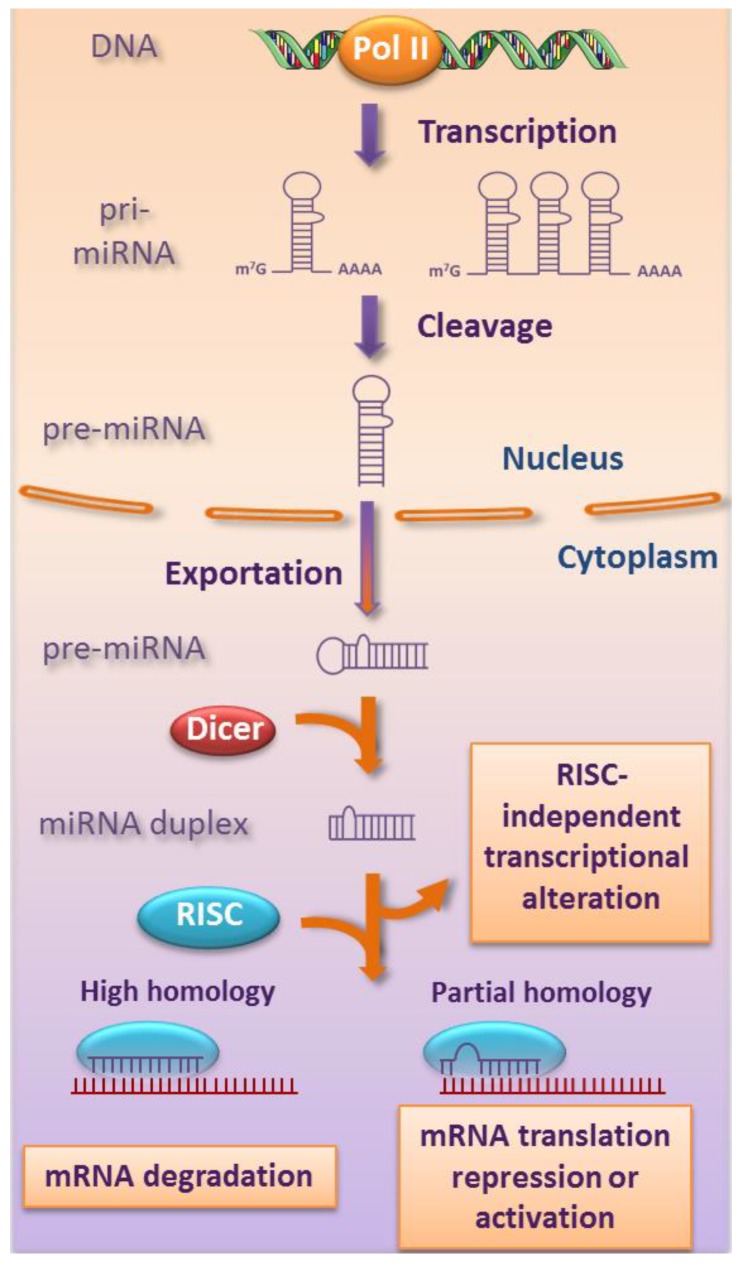
miRNAs are most commonly transcribed in the nucleus by the RNA polymerase II (Pol II), as monocistronic or polycistronic pri-miRNAs that are further processed in pre-miRNAs (Figure 1). These pre-miRNAs are exported to the cytoplasm where they undergo cleavage by the RNAse III enzyme called Dicer that produces a miRNA duplex. This duplex splits to generate the single-stranded mature miRNA that incorporates the RNA-induced silencing complex (RISC). Based on the complementarity with its target gene sequence, the mature miRNA induce either translational repression (partial complementarity) or mRNA degradation (perfect complementarity) [7]. Besides, the mature miRNA can increase the expression of the target gene under growth arrest condition [8]. Finally, it has been recently reported that miRNA can also act in a RISC-independent manner on the transcriptional level by interaction with ribonucleoprotein or direct binding to DNA [9,10,11].
Figure 1.
Schematic representation of miRNA biogenesis. The mature miRNAs originate from successive different steps. An initial DNA transcription generates pri-miRNAs that are cleaved in pre-miRNAs before their exportation to the cytoplasm. There, an RNAse III enzyme, Dicer, cleaves it to generate a miRNA duplex that subsequently joins an RNA‑induced silencing complex (RISC) to produce the mature miRNA. The sequence complementarity to the target will decide its fate. Some miRNAs can act on the transcriptional level independently from the RISC.
One single miRNA has the ability to regulate multiple targets and thereby to affect a broad network of genes (up to 100 genes) [12]. This specific characteristic makes the miRNAs key mediators of most of the cellular events. In animal, miRNAs mainly regulate mRNAs by interacting with their 5' end (5p) to the 3'-untranslated region (3'-UTR) of their target [13]. However, recent studies have revealed miRNAs target sites in the 5'-UTR, which interacts with the 3' end (3p) of miRNAs, and even simultaneous 5'-UTR and 3'-UTR interaction sites [14,15].
The abnormal expression levels of miRNAs have been revealed in various diseases such as cancer [16,17], inflammation [18,19], Alzheimer [20], cardiovascular disease [21] and viral infection including HBV [2,22].
3. Role of miRNAs in HBV Infection
Despite the fact that HBV is a nuclear DNA virus, none viral-encoded miRNA has been so far identified. Only one putative HBV miRNA, with hypothetical regulation role on its own genome, was deduced by computational approach [23]. However, HBV can modulate the expression of several cellular miRNAs in order to promote a favorable environment for its replication and survival. They are presented in this section and summarized in Table 1.
Table 1.
Cellular miRNAs and their effects on HBV infection or HBV related-diseases. HBV (↑): Promotes HBV replication; HBV (↓): Inhibits HBV replication; HCC (↑): Development and/or growth of HCC; Fibrosis (↑): Promotes liver fibrosis.
| miRNAs | miRNA expression | Target genes | HBV or disease status | Ref. |
|---|---|---|---|---|
| Cellular targets | ||||
| miR-1 | up | HDAC4 (histone deacetylase 4) | HBV (↑) | [28] |
| miR-17-92 cluster | up | E2F1 (c-myc repressor) | HBV (↑?), HCC (↑) | [29] |
| miR-155 | up | C/EBPβ (CCAAT/enhancer binding protein) | HCC (↑) | [30] |
| SOCS1 (JAK/STAT signaling) | HBV (↓) | [31] | ||
| miR-181a | up | HLA-A? (MHC class I) | HBV (↑) | [5] |
| miR-372 | up | NFIB (nuclear factor I/B) | HBV (↑) | [32] |
| miR-373 | up | NFIB (nuclear factor I/B) | HBV (↑) | [32] |
| miR-501 | up | HBIP (HBx inhibitor) | HBV (↑) | [33] |
| mir-29 family | down | collagen | Fibrosis (↑) | [22,34] |
| miR-122 | down | cyclin G1 (p53 modulator) | HBV (↑), HCC (↑) | [6] |
| miR-152 | down | DNMT1 (DNA methyltransferase 1) | HBV (↓) | [35] |
| let-7 family | down | STAT3 (transcription factor) | HBV (↑?), HCC (↑) | [36] |
| Viral targets | ||||
| miR-122 | up | HBV DNA polymerase | HBV (↓) | [37] |
| miR-125a-5p | up | HBsAg (HBV surface antigen) | HBV (↓) | [38] |
| miR-199-3p | up | HBsAg | HBV (↓) | [39] |
| miR-210 | up | HBV pre-S1 (pre-surface 1) | HBV (↓) | [39] |
3.1. Brief Description of HBV Infection
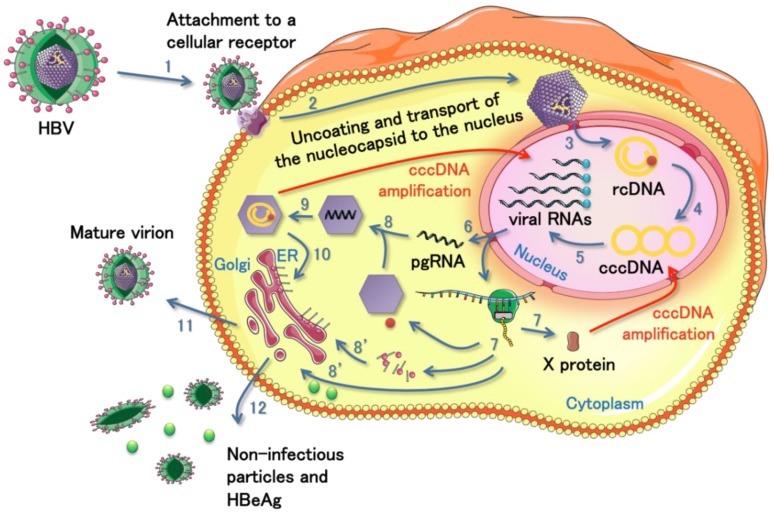
The HBV infection is characterized by two phases; the acute and the chronic infection [24]. The initial stages of the acute infection include virion attachment [25], uncoating and nucleocapsid transport to the cell nucleus (Figure 2, steps 1 and 2). The 3.2 kb relaxed circular DNA genome is released into the nucleus and converted into a covalently closed circular DNA (cccDNA) from which all the viral RNAs are transcribed (Figure 2, steps 3 to 5). The pregenomic RNA (pgRNA) serves as template for reverse transcription (Figure 2, steps 8 and 9). The subgenomic mRNAs comprise the pre-surface (S) and S genes, the pre-core (C) and C genes, the polymerase gene, and the X gene. The newly formed nucleocapsids can either assemble with envelope proteins in the endoplasmic reticulum and form mature virions that will be secreted (Figure 2, steps 10 and 11), or return to the nucleus to maintain the cccDNA amplification. When the immune system fails to clear the virus, the HBV infection becomes chronic and it remains under a dormant state into the cell. Eventually, the viral genetic material or sequences can integrate into the host cellular DNA. The integration has been frequently observed and is associated with HCC [26,27].
Figure 2.
Schematic representation of HBV life cycle. The virus infects a cell by an initial attachment to a cellular receptor that allows its internalization (step 1). In the cytoplasm, the virus is uncoated and the nucleocapsid is transported to the nuclear membrane (step 2). The viral genome is released into the nucleus under its relaxed circular form (rcDNA) and converted into a covalently closed circular DNA (cccDNA) from which all the viral RNAs are produced (steps 3 to 5). The viral RNAs transfer to the cytoplasm for traduction of the different viral proteins (steps 6 and 7) or for subsequent reverse transcription of the pregenomic RNA (pgRNA, steps 6, 8 and 9). All the viral components move to the proper place and assemble together to form new mature virions (steps 8, 10 and 11). The virus also produces non-infectious particles and extracellular antigen (HBeAg) as a decoy for the immune system of the host (step 12). The nucleocapsid containing the rcDNA and the HBV X protein (HBx) can go back to the nucleus in order to amplify the cccDNA and maintain the viral production.
3.2. Role of miRNAs in the HBV Replication
The role of miRNAs in HBV replication is therefore dependent on the phase of HBV infection. During the acute phase, the virus must activate its replication while avoiding destruction by the immune system. During the chronic phase, the virus reaches a “dormant” state where the viral replication must be restricted and viral evasion maintained. This leads to a time-dependent intricate interaction network in which miRNAs play an important role.
One of the best studied miRNAs in HBV infection and other liver-related diseases is miR-122. This liver-specific miRNA is expressed at high levels in normal hepatocytes (about 70% of the total miRNA population in the adult liver) [40] and is pivotal in numerous aspects of the liver function such as lipid metabolism, liver development, differentiation, growth and neoplastic transformation [41]. While the loss of miR-122 expression impedes hepatitis C virus (HCV) replication [42], it enhances the replication in the circumstance of HBV infection [6]. However, miR-122 can negatively regulate the viral gene expression and replication by direct binding to a highly conserved sequence of HBV [37]. This repression effect can apparently be impeded by a negative feedback loop involving the Heme oxygenase-1 [43]. A recent study has reported the indirect implication of the HBV X protein (HBx) in miR-122 dysregulation [44] that could, at least partially, explain the difference observed between the two viruses.
On the other hand, miR-1 can enhance the HBV core promoter transcription by down-regulating the expression of the histone deacetylase 4 (HDAC4) [45]. This miRNA might act complementary to the nuclear HBx in order to induce epigenetic modifications on the cccDNA and amplify the viral genome [45,46].
miR-372, together with miR-373, also supports HBV gene expression by targeting the nuclear factor I/B [47]. This cellular protein is known to be an important regulator of several viruses [48].
The let-7 family of miRNAs has been demonstrated to be negatively regulated by HBx [36]. The consequence of this down-regulation is the increase activity of the signal transducer and activator of transcription 3 (STAT3) that supports cell proliferation, and potentially viral replication and hepatocarcinogenesis.
Finally, miR-501 has been suggested to work with HBx for the benefit of viral replication [33]. HBx itself has also the ability to dysregulate the cellular miRNAs expression. This small protein is a key regulator of HBV infection. It is usually overexpressed in HCC and is involved in hepatocarcinogenesis [48].
3.3. Role of miRNAs in the Immune Evasion of HBV
miRNAs are important in the development and function of immune system [49]. In particular, miR-155 has multi-roles during innate immune response such as regulation of the acute inflammatory response after recognition of pathogens by the toll-like receptors [50,51]. Su and collaborators demonstrated that ectopic expression of miR-155 in human hepatoma cells could enhance the innate immunity through promotion of the janus kinase (JAK)/STAT pathway and down-regulate HBx expression [31].
On the other hand, a study analyzing the modified expression profiles of miRNAs in a stable HBV-expressing cell line revealed the upregulation of miR-181a [5]. The dysregulation of this miRNA in liver cell might participate to HBV replication through inhibition of the human leukocyte antigen A (HLA-A)-dependent HBV antigen presentation.
It is now unclear if the miRNAs altered in the infected hepatocytes, such as miR-181a and miR-146 [5], that have specific regulatory functions in the immune cells as well [49], could affect directly these cells to support viral evasion. The presence of circulating miRNAs and the existence of intercellular nanovesicle-mediated miRNA transfer that modulates the environment, could potentially support that hypothesis [52,53,54,55,56,57].
3.4. Role of miRNAs in the Establishment of HBV Chronic Infection
The natural history of HBV infection shows often a transition from acute to chronic infection, especially in young children. The virus reaches a “dormant” state into the infected hepatocytes, under the cccDNA form, and survive until its eventual life cycle reactivation [3,35,45,58]. One study reported the CpG islands methylation of the cccDNA by DNA methyltransferase 1 (DNMT1) to prevent the viral gene expression and therefore the viral antigen presentation. The DNMT1 overexpression is induced by a decrease of miR-152, under the effect of HBx [35].
miR-1 illustrates the duality of actions that can be observed in the course of HBV infection. As said previously, this miRNA can promote viral replication but it can also inhibit the cell proliferation and even induce a reverse cancer cell phenotype [28]. The effect on HCC was confirmed in another study [59].
Also, miR-122 can bind directly to the polymerase region in order to repress its expression [37]. Similar observations were made for miR-125a-5p, miR-199a-3p that can affect the S region and miR-210 that can affect the pre-S1 region [38,39]. Since the RNA intermediates of HBV (pgRNA and transcripts) are good targets of miRNA action, it is not surprising to observe several cellular miRNAs targeting them. However, it remains to be determined whether the targeting of HBV transcripts represents an active anti-viral mechanism of the host or if the virus has evolved to hijack these cellular miRNAs in order to reach its “dormant” state.
4. Role of miRNAs in HBV-Related Diseases
The modifications induced as a result of HBV infection profoundly alter the cellular and overall organism homeostasis. They are usually associated with diseases, including liver cirrhosis with fibrosis and HCC. The liver cirrhosis turns most of the time into HCC.
4.1. Role of miRNAs in HBV-Related Cirrhosis
Numerous studies have tried to identify and characterize the miRNAs involved in liver cirrhosis and therefore differently expressed during this intermediate phase. Roderburg et al. investigated the role of miRNAs in liver fibrosis on a carbon tetrachloride-induced hepatic fibrogenesis and bile-duct ligation mouse models [34]. They observed a significant down-regulation of all the members of the miR-29 family in the two models. The decreased expression was induced by the transforming growth factor beta (TGF-β), inflammatory signals and the nuclear factor kappa B (NFκB) pathways. miR-29c was also identified in another report focusing on the miRNA expression profile in patients with HCC-positive or HCC-negative chronic hepatitis B and hepatitis C virus [22] (Table 1).
Nevertheless, the global miRNA expression profile analysis of human liver tissues from different inflammation, infection and cancer states are not always consistent. It sometimes revealed a particular profile due to the association of both viral hepatitis and cirrhosis [60] or regarding to the type of viral hepatitis [22] and sometimes showed no difference [61]. Further experiments are therefore required to identify the exact molecular mechanisms implicating miRNAs and viral components in the development of cirrhosis and in the transition from cirrhosis to HCC in the patients with chronic viral hepatitis.
4.2. Role of miRNAs in HBV-Related HCC
When the cellular modifications and inflammation are too high and maintained for too long, the liver cirrhosis usually evolves into HCC.
The miR-17-92 cluster is important in the HBV infection and associated HCC. This polycistron includes six miRNAs (mir-17-5p, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a-1) and its upregulated expression is associated with malignancies [62]. By using human HBV-positive human HCC tissues, hepatoma cell lines and woodchuck hepatitis virus-induced HCC animal model [63], Connolly and colleagues were able to demonstrate the elevated expression of miR-17-92 cluster and its implication in the malignant phenotype [29] (Table 1). The expression could be amplified by c-myc activation [64], under HBx control [65], to contribute to HBV latency state [66]. The consequence is the induction of liver oncogenesis.
Because of its role in immune response, miR-155 is also implicated in hepatocarcinogenesis. Indeed, its upregulation can lead to prolonged exposure to inflammation, a well-known causal agent to cancers like HCC [67]. Using HCC-induced mouse model, Wang and collaborators have demonstrated the oncogenic role of miR-155 at the early stages of the tumorigenesis [30] (Table 1).
To conclude, the liver-specific miR-122 has been extensively studied in the liver-associated diseases. Its expression is low in HCC tissues, including those with viral chronic hepatitis [6,68] (Table 1). As described in point 3.2, the regulation of miR-122 is very complex and helps either promotion or inhibition of the HBV replication. In HCC cells, the “dormant” state of HBV implicates a replication rate very low or inexistent [69]. The recent data accumulate evidence of miR-122 as a highly potential linker between HBV infection and liver carcinogenesis [6,70]. Because of its characteristics, miR-122 is therefore a target of choice for future clinical applications.
5. miRNAs as Molecular Tools Against HBV Infection and HBV-Related Diseases
The significance of miRNAs in viral replication, antiviral immunity and liver carcinogenesis emphasizes their values as diagnostic, prognostic and therapeutic targets for HBV infection and HBV-induced diseases.
miR-122 and miR-18a are of particular interest for diagnostic and/or prognostic applications. They are both released in the blood and could be used as potential non-invasive biomarkers for HBV-related HCC screening [5,53,54]. Some other reports suggest the use of a miRNA panel in order to improve the specificity of the test [55,56]. In addition with the current routinely used markers such as HBV surface antigen (HBsAg), HBV extracellular antigen (HbeAg) and alanine aminotransferase (ALT), the circulating miRNAs represent a significant clinical value for better evaluation of the HBV-infection status, liver injury and early diagnosis of HCC.
In the therapeutic perspective, the liver cirrhosis is an event prior to HCC development and being able to interfere with this process would prevent carcinogenesis. For example, a strategy based on administration of miR-29 mimic might prevent liver fibrosis (Section 4.1) [34]. However, the disease is often discovered when hepatocarcinogenesis has already developed and HCC does not always show underlying cirrhosis [71]. Finding therapeutic targets involved in HCC is thus a major issue.
For this purpose, the work of Ura’s group is valuable [22]. They analyzed the livers of HBV and HCV positive patients with HCC to identify the miRNAs that are differentially expressed. Nineteen miRNAs were clearly differentiated between HBV and HCV groups, six specific for HBV and thirteen specific for HCV. Based on the miRNAs profile, they made a pathway analysis of candidate targeted genes and were also able to distinguish the cellular mechanisms altered in HBV or HCV-infected livers. The HBV infection alters mostly the pathways related to signal transduction, inflammation and natural killer toxicity, DNA damage, recombination, and cell death, while HCV infection modifies those involved in immune response involving antigen presentation, cell cycle and cell adhesion. Although very interesting, their results are not consistent with those presented in other reports [60,61] and confirmation of the targets needs to be done before considering their clinical application.
Finally, technological advances in the delivery of miRNA and RNA interference enable safe and efficient in vivo miRNA gene therapy, as exemplify by the recent study from Kota and colleagues on the liver cancer [72]. They used an adeno-associated virus to deliver miR-26a in a mouse model of HCC. This resulted in the successful inhibition of the cancer cell proliferation, induction of the tumor‑specific apoptosis, and protection from disease progression without toxicity.
6. Conclusions
miRNAs have emerged as new key players in the control of gene expression in cells. Investigations of their profiling have unveiled specific miRNA dysregulations in tumors and during viral infection.
The HBV is a widespread pathogen that is implicated in HCC development. Numerous cellular miRNAs interacting with HBV have been identified. They reflect the cellular pathways that are altered as a result of the viral infection, viral infection that triggers the liver cirrhosis and carcinogenesis as side effects. On the viral point of view, the dysregulated pathways mirror the strategies of the virus to allow its replication and evade the host defense mechanisms to survive. On the cellular point of view, they mirror the immune response that tries to get rid of the intruder and that becomes dysregulated. The present and future knowledge about the interaction between miRNA, HBV infection and HCC development and progress will probably allow developing strategies and tools to cope, efficiently and at various steps, with the liver carcinogenesis induced by HBV infection.
Acknowledgments
This work was supported in part by a grant-in-aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control of Japan; Project for Development of Innovative Research on Cancer Therapeutics (P-Direct); Scientific Research on Priority Areas Cancer from the Japanese Ministry of Education, Culture, Sports, Science, and Technology; and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation of Japan. The authors would like to thank Servier Medical Art for their image bank used to create the illustrations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bartel D.P. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem D., Prince A.M. Hepatitis B virus infection—Natural history and clinical consequences. N. Engl. J. Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 4.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Zhao J.J., Wang C.M., Li M.Y., Han P., Wang L., Cheng Y.Q., Zoulim F., Ma X., Xu D.P. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin. Med. J. 2009;122:10–14. doi: 10.3901/jme.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Wang S., Qiu L., Yan X., Jin W., Wang Y., Chen L., Wu E., Ye X., Gao G.F., Wang F., et al. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1)-modulated P53 activity. Hepatology. 2012;55:730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 7.He L., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Vasudevan S., Tong Y., Steitz J.A. Switching from repression to activation: Micrornas can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 9.Eiring A.M., Harb J.G., Neviani P., Garton C., Oaks J.J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C.J., et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez S., Pisano D.G., Serrano M. Mechanistic principles of chromatin remodeling guided by siRNAs and miRNAs. Cell Cycle. 2008;7:2601–2608. doi: 10.4161/cc.7.16.6541. [DOI] [PubMed] [Google Scholar]
- 11.Kim D.H., Saetrom P., Snove O., Jr., Rossi J.J. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc. Natl. Acad. Sci. USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 13.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 14.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee I., Ajay S.S., Yook J.I., Kim H.S., Hong S.H., Kim N.H., Dhanasekaran S.M., Chinnaiyan A.M., Athey B.D. New class of microRNA targets containing simultaneous 5'-UTR and 3'-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calin G.A., Sevignani C., Dumitru C.D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C.M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calin G.A., Ferracin M., Cimmino A., di Leva G., Shimizu M., Wojcik S.E., Iorio M.V., Visone R., Sever N.I., Fabbri M., et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 18.Bi Y., Liu G., Yang R. MicroRNAs: Novel regulators during the immune response. J. Cell Physiol. 2009;218:467–472. doi: 10.1002/jcp.21639. [DOI] [PubMed] [Google Scholar]
- 19.Fasseu M., Treton X., Guichard C., Pedruzzi E., Cazals-Hatem D., Richard C., Aparicio T., Daniel F., Soule J.C., Moreau R., et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sethi P., Lukiw W.J. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- 21.Latronico M.V., Catalucci D., Condorelli G. Emerging role of microRNAs in cardiovascular biology. Circ. Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- 22.Ura S., Honda M., Yamashita T., Ueda T., Takatori H., Nishino R., Sunakozaka H., Sakai Y., Horimoto K., Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 23.Jin W.B., Wu F.L., Kong D., Guo A.G. HBV-encoded microRNA candidate and its target. Comput. Biol. Chem. 2007;31:124–126. doi: 10.1016/j.compbiolchem.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Seeger C., Mason W.S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64:51–68. doi: 10.1128/MMBR.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H., et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brechot C., Pourcel C., Louise A., Rain B., Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533–535. doi: 10.1038/286533a0. [DOI] [PubMed] [Google Scholar]
- 27.Paterlini-Brechot P., Saigo K., Murakami Y., Chami M., Gozuacik D., Mugnier C., Lagorce D., Brechot C. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Zhang E., Ma Z., Pei R., Jiang M., Schlaak J.F., Roggendorf M., Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 29.Connolly E., Melegari M., Landgraf P., Tchaikovskaya T., Tennant B.C., Slagle B.L., Rogler L.E., Zavolan M., Tuschl T., Rogler C.E. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am. J. Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang B., Majumder S., Nuovo G., Kutay H., Volinia S., Patel T., Schmittgen T.D., Croce C., Ghoshal K., Jacob S.T. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su C., Hou Z., Zhang C., Tian Z., Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol. J. 2011;8 doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H., Liu H., Mitchelson K., Rao H., Luo M., Xie L., Sun Y., Zhang L., Lu Y., Liu R., et al. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808–819. doi: 10.1002/hep.24441. [DOI] [PubMed] [Google Scholar]
- 33.Jin J., Tang S., Xia L., Du R., Xie H., Song J., Fan R., Bi Q., Chen Z., Yang G., et al. MicroRNA-501 promotes HBV replication by targeting HBXIP. Biochem. Biophys. Res. Commun. 2013;430:1228–1233. doi: 10.1016/j.bbrc.2012.12.071. [DOI] [PubMed] [Google Scholar]
- 34.Roderburg C., Urban G.W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M., et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2010;53:209–218. doi: 10.1016/j.jhep.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Huang J., Wang Y., Guo Y., Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y., Lu Y., Toh S.T., Sung W.K., Tan P., Chow P., Chung A.Y., Jooi L.L., Lee C.G. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y., Shen A., Rider P.J., Yu Y., Wu K., Mu Y., Hao Q., Liu Y., Gong H., Zhu Y., et al. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511–4521. doi: 10.1096/fj.11-187781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potenza N., Papa U., Mosca N., Zerbini F., Nobile V., Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157–5163. doi: 10.1093/nar/gkr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang G.L., Li Y.X., Zheng S.Q., Liu M., Li X., Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 40.Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 41.Girard M., Jacquemin E., Munnich A., Lyonnet S., Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 2008;48:648–656. doi: 10.1016/j.jhep.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 42.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 43.Qiu L., Fan H., Jin W., Zhao B., Wang Y., Ju Y., Chen L., Chen Y., Duan Z., Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 44.Song K., Han C., Zhang J., Lu D., Dash S., Feitelson M., Lim K., Wu T. Epigenetic regulation of miR-122 by PPARgamma and hepatitis B virus X protein in hepatocellular carcinoma cells. Hepatology. 2013 doi: 10.1002/hep.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belloni L., Pollicino T., de Nicola F., Guerrieri F., Raffa G., Fanciulli M., Raimondo G., Levrero M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA. 2009;106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pollicino T., Belloni L., Raffa G., Pediconi N., Squadrito G., Raimondo G., Levrero M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Nagata K., Guggenheimer R.A., Hurwitz J. Specific binding of a cellular DNA replication protein to the origin of replication of adenovirus DNA. Proc. Natl. Acad. Sci. USA. 1983;80:6177–6181. doi: 10.1073/pnas.80.20.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian Y., Yang W., Song J., Wu Y., Ni B. HBV X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 2013;33:2810–2816. doi: 10.1128/MCB.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baltimore D., Boldin M.P., O’Connell R.M., Rao D.S., Taganov K.D. MicroRNAs: New regulators of immune cell development and function. Nat. Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 50.O’Connell R.M., Taganov K.D., Boldin M.P., Cheng G., Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tili E., Michaille J.J., Cimino A., Costinean S., Dumitru C.D., Adair B., Fabbri M., Alder H., Liu C.G., Calin G.A., Croce C.M. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007;179:5082–5089. doi: 10.4049/jimmunol.179.8.5082. [DOI] [PubMed] [Google Scholar]
- 52.Arataki K., Hayes C.N., Akamatsu S., Akiyama R., Abe H., Tsuge M., Miki D., Ochi H., Hiraga N., Imamura M., et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med. Virol. 2013;85:789–798. doi: 10.1002/jmv.23540. [DOI] [PubMed] [Google Scholar]
- 53.Waidmann O., Bihrer V., Pleli T., Farnik H., Berger A., Zeuzem S., Kronenberger B., Piiper A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J. Viral Hepat. 2012;19:e58–e65. doi: 10.1111/j.1365-2893.2011.01536.x. [DOI] [PubMed] [Google Scholar]
- 54.Li L., Guo Z., Wang J., Mao Y., Gao Q. Serum miR-18a: A potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig. Dis. Sci. 2012;57:2910–2916. doi: 10.1007/s10620-012-2317-y. [DOI] [PubMed] [Google Scholar]
- 55.Li L.M., Hu Z.B., Zhou Z.X., Chen X., Liu F.Y., Zhang J.F., Shen H.B., Zhang C.Y., Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 56.Zhou J., Yu L., Gao X., Hu J., Wang J., Dai Z., Wang J.F., Zhang Z., Lu S., Huang X., et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J. Clin. Oncol. 2011;29:4781–4788. doi: 10.1200/JCO.2011.38.2697. [DOI] [PubMed] [Google Scholar]
- 57.Kogure T., Lin W.L., Yan I.K., Braconi C., Patel T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belloni L., Allweiss L., Guerrieri F., Pediconi N., Volz T., Pollicino T., Petersen J., Raimondo G., Dandri M., Levrero M. IFN-alpha inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 2012;122:529–537. doi: 10.1172/JCI58847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Datta J., Kutay H., Nasser M.W., Nuovo G.J., Wang B., Majumder S., Liu C.G., Volinia S., Croce C.M., Schmittgen T.D., et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Jiang J., Gusev Y., Aderca I., Mettler T.A., Nagorney D.M., Brackett D.J., Roberts L.R., Schmittgen T.D. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murakami Y., Yasuda T., Saigo K., Urashima T., Toyoda H., Okanoue T., Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 62.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 63.Popper H., Roth L., Purcell R.H., Tennant B.C., Gerin J.L. Hepatocarcinogenicity of the woodchuck hepatitis virus. Proc. Natl. Acad. Sci. USA. 1987;84:866–870. doi: 10.1073/pnas.84.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terradillos O., Billet O., Renard C.A., Levy R., Molina T., Briand P., Buendia M.A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 66.Jung Y.J., Kim J.W., Park S.J., Min B.Y., Jang E.S., Kim N.Y., Jeong S.H., Shin C.M., Lee S.H., Park Y.S., et al. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J. Med. Virol. 2013;85:969–978. doi: 10.1002/jmv.23534. [DOI] [PubMed] [Google Scholar]
- 67.Berasain C., Castillo J., Perugorria M.J., Latasa M.U., Prieto J., Avila M.A. Inflammation and liver cancer: new molecular links. Ann. NY Acad. Sci. 2009;1155:206–221. doi: 10.1111/j.1749-6632.2009.03704.x. [DOI] [PubMed] [Google Scholar]
- 68.Kutay H., Bai S., Datta J., Motiwala T., Pogribny I., Frankel W., Jacob S.T., Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69.Wong D.K., Yuen M.F., Poon R.T., Yuen J.C., Fung J., Lai C.L. Quantification of hepatitis B virus covalently closed circular DNA in patients with hepatocellular carcinoma. J. Hepatol. 2006;45:553–559. doi: 10.1016/j.jhep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 70.Fan C.G., Wang C.M., Tian C., Wang Y., Li L., Sun W.S., Li R.F., Liu Y.G. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol. Rep. 2011;26:1281–1286. doi: 10.3892/or.2011.1375. [DOI] [PubMed] [Google Scholar]
- 71.La Vecchia C., Lucchini F., Franceschi S., Negri E., Levi F. Trends in mortality from primary liver cancer in Europe. Eur. J. Cancer. 2000;36:909–915. doi: 10.1016/S0959-8049(00)00052-6. [DOI] [PubMed] [Google Scholar]
- 72.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H.W., Chang T.C., Vivekanandan P., Torbenson M., Clark K.R., et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]