Abstract
Several laboratories are pursuing the synthesis of cellular systems from different directions, including those that begin with simple chemicals to those that exploit existing cells. The methods that begin with nonliving components tend to focus on mimicking specific features of life, such as genomic replication, protein synthesis, sensory systems, and compartment formation, growth, and division. Conversely, the more prevalent synthetic biology approaches begin with something that is already alive and seek to impart new behavior on existing cells. Here we discuss advances in building cell-like systems that mimic key features of life with defined components.
Keywords: Cell-like, minimal cell, origin of life, protocell, riboswitch, synthetic biology.
BUILDING CELLULAR SYSTEMS FROM THEIR PARTS
Building life from scratch in the laboratory is an old dream with new tools at its disposal. We can now rapidly and affordably synthesize genes [1], assemble genomes [2], evolve new function [3] and make precise changes throughout an existing genome [4]. Most of these technological advances are applied to the modification of existing cells (Fig. 1), and so the resulting data do not directly address life's beginnings or clearly delineate the required components of cellular function. Any work that either exploits existing cells or cell lysates makes use of a complex, undefined mixture of reaction components that we do not have the tools to fully understand. Here we try to highlight steps forward in building fully defined life-like systems from a minimum number of components.
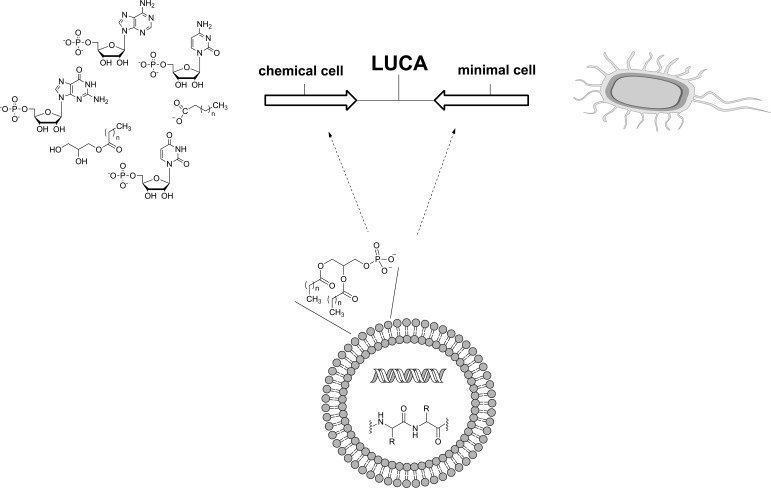
Fig. (1).
Different approaches to building new, artificial cells. Typically, laboratories either begin with chemicals (left) or an existing cell (right). Presumably an approximation of the last universal common ancestor (LUCA) exists in between these two extremes. A third approach is to piece together cellular systems from existing biological components (bottom).
It is hoped that the process of building cell-like systems in the laboratory will give us insight into what is required to endow a system with the properties of life. There is no currently agreed upon threshold that must be crossed in order to label a chemical system living. However, as progress is made in recreating the functions of life in the laboratory, we may reach a point in which a threshold is crossed, even if it is not recognized until afterwards. Further, since the system would have been built with fully defined components, this systematic approach should give us a much better understanding of the necessary components of life. It is worth noting that this approach does not probe how the molecules of life were built, nor does it directly address the historical path taken between the prebiotic chemistry of Earth [5-11] and the emergence of protocellular structures [12-14].
The central dogma [15] of molecular biology offers one perspective on what is needed to build a cell. Typically, information stored in a replicating DNA genome flows through RNA and then finally to proteins. However, this simple description of cellular life is in fact not very simple, which is born out by analyses of microorganisms. E. coli, for example, has well over 4000 genes, and purposeful genetic reductions only reduces the genetic content by 15% [16]. On the smaller end of the spectrum, Mycoplasma genitalium has 482 genes, and genetic knockouts have shown that approximately 100 of these genes are not required under laboratory condi tions [17]. But even with this small genome, which is small enough to be synthesized [2] approximately one third of the genes provide unknown function to the cell. We have reached a point where our technological ability to synthesize genomes has outpaced our understanding of what we are synthesizing.
By comparing sequences of disparate microorganisms, Moya and others suggest that a minimal cell would contain on the order of 200 genes, of which more than half would be necessary for protein synthesis [18]. Interestingly, a natural symbiotic microorganism, Carsonella ruddii, has only 182 genes, a value similar to theoretical predictions of a minimum gene set [19]. Further, over half of the C. ruddii genome is dedicated to protein synthesis. In some ways, the impression is given that living systems are nothing more than just a bag of protein synthesizing machinery. Clearly life is more than just protein synthesis, but at least as far back as the last universal common ancestor, protein synthesis has been a crucial aspect of cellular function [20].
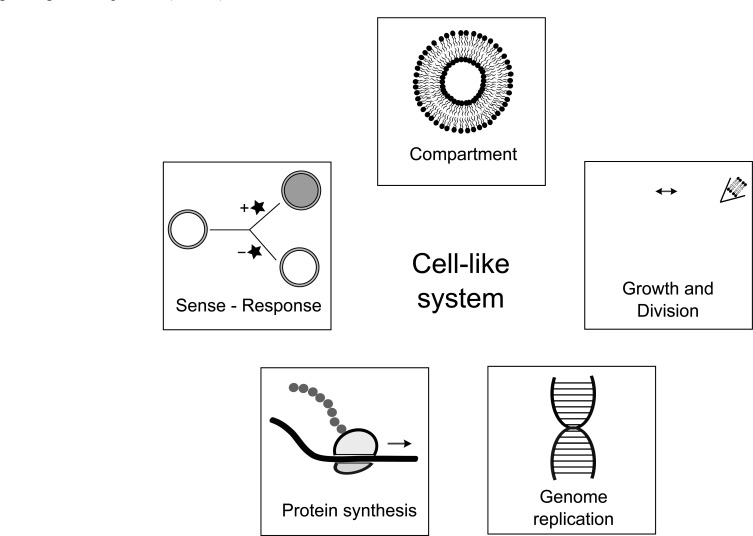
One conception of a simplified, laboratory-made cell consists of a vesicle compartment that contains a replicating DNA genome and transcription-translation machinery that responds to changing environmental conditions (Fig. 2). Much of the needed functions for such a cell-like system appears to depend on protein function. Nevertheless, origins of life research has shown that under specific chemical conditions, several features of life emerge without the participation of proteins. Perhaps future approaches that combine the lessons learned from origins research with those gained from attempts to exploit biological machinery will allow for the synthesis of a simplified cell.
Fig. (2).
Features of cellular life that are mimicked by cell-like systems.
COMPARTMENT TYPES
Compartmentalization is considered to be one of the key steps along the transition from simple chemistry to cellular life [21]. The enclosure of a chemical system within a semipermeable membrane causes several useful features to emerge. For example, encapsulation facilitates evolutionary processes [22, 23, 24], provides for an energy storage mechanism [21-25] and likely influenced accessible chemistry. Although it is possible that prebiotic boundary structures were defined by substances other than lipids, no living systems to date have been identified that are capable of surviving without lipid membranes. Further, several lines of evidence argue for the presence of lipids on prebiotic Earth, including simulated prebiotic syntheses of lipids [5-11] and the identification of lipid molecules within carbonaceous meteorites [8]. Finally, vesicles form easily in aqueous solution, thus suggesting that there were vesicles on Earth even before there was life. The latter point has led some to suggest that there once existed a lipid world in which hereditary was mediated by lipid composition rather than by specific nucleic acid sequences [26].
Prebiotically plausible lipids are generally thought to be saturated, single-chained amphiphiles. In the laboratory, fatty acids and fatty acid derivatives are often used as an approximation of what could have been present. Not only do such lipids form vesicles, they also exhibit many useful features not dependent upon protein function, including the ability to grow and divide, uptake nutrients, and retain macromolecules [23,25,27-34]. The main disadvantages of fatty acid based vesicles is the encountered difficulty in recovering encapsulated enzymatic activity from some enzymes and the vesicle's decreased stability in comparison with phospholipid vesicles. For example, fatty acid vesicles are stable over a narrow pH and salinity range [30,35,36] and difficulty has been encountered in reconstituting DNA polymerase activity within fatty acid vesicles [32-37]. Therefore, fatty acid vesicles are typically used for protocellular research rather than for attempts to build cell-like systems similar to life as we know it.
Contemporary cells exploit membranes of complex composition including monoacyl and diacyl lipids and proteins. Laboratory constructions tend to ignore this complexity and instead rely on the ease in which many lipids alone form vesicles. Of the commonly used vesicle systems, those built with diacylphospholipids are the most robust. However, this robustness comes at a cost. Diacylphospholipids are generally impermeable, thereby posing a difficulty in their use for building cell-like structures. One approach to overcome this limitation is to exploit membrane proteins, such as the bacterial toxin α-hemolysin. This protein expresses as a soluble monomer that then spontaneously oligomerizes into a pore in the presence of a membrane [38]. An alternative approach is to utilize the phase transition temperature of lipid membranes to create packing defects that can be exploited for permeation [39]. Both the α-hemolysin and packing defect mechanisms have been used to feed in substrates for encapsulated RNA [40] and protein synthesis reactions [38]. Interestingly, symbiotic organisms with small genomes, such as C. ruddii and Buchner aphidicola, are thought to be heavily dependent on passive diffusion mechanisms for nutrient uptake [41]. In short, phospholipid membranes need not be viewed as an impenetrable barrier. Simple passive diffusion based mechanisms exist for nutrient uptake and waste release that may not be much different than what is used by some microorganisms with small genomes.
There is another approach to building compartments with dimensions similar to extant cells. Water-in-oil (w/o) emulsions are easy to make and are extremely efficient in encapsulating hydrophilic molecules. They have been used extensively for molecular evolution experiments [42, 43] and further developments in the technology allow for the delivery of reactants directly to the water droplets without breaking the emulsion [44]. However, w/o emulsions have not proved useful for constructing cell-like systems due the lack of solute exchange across the water-oil interface.
COMPARTMENT GROWTH & DIVISION
A compartment must grow and divide to allow for the cell-like system to replicate. For fatty acid vesicle systems, growth and division can be accomplished simply by adding lipids to preexisting vesicles [27-29,33,34]. Similar mechanisms for phospholipids are not available because of the decreased dynamics of diacyl phospholipids [45]. However, if phospholipid synthesis reactions were reconstituted within vesicles, then once the lipids were produced they would naturally partition into the membrane resulting in vesicle growth. The difficulty with reconstituting such enzymatic activities is that many of the lipid synthesis enzymes are membrane proteins. Nevertheless, Luisi and colleagues have built an encapsulated enzyme system that can produce diacyl glycerophospholipids when provided with fatty acids and glycerol [46]. Although this is an important development, more work is needed to move beyond the use of simple lipids (fatty acids) to build more complex lipids (phospholipids). Also, additional enzymes may be required to flip a fraction of the newly synthesized lipids from the inner- to the outer-leaflet.
Phospholipid vesicles can be coerced into dividing through simple chemical-physical forces. For example, Baumgart et al. found that phospholipid vesicles containing liquid ordered (Lo) and liquid disordered (Ld) domains can bud and divide when placed under osmotic stress [47]. However, since the daughter vesicles do not retain the same membrane lipid composition as the parental vesicle, further rounds of division are not possible. Presumably if an additional mechanism were incorporated to restore the original membrane composition then further rounds of division could occur. More recently, Andes-Koback and Keating revealed that osmotic gradients can induce budding and division of phospholipid vesicles that contain an aqueous two-phase system, one enriched in dextran and the other aqueous phase enriched in polyethyleneglycol [11]. Since the resulting daughter vesicles retain the composition of the parental vesicle, they are also capable of dividing. If the Andes-Koback and Keating mechanism were combined with a vesicle growth system, then a growth – division cycle could be built. The system in its current state, however, does require external intervention to adjust the osmolality of the extravesicular space. It is interesting to note that some biological evidence exists suggesting that life can persist in the absence of protein mediated division [48, 49].
Protein based systems have also shown promise in dividing vesicles. Although in vivo cell division mechanisms are complex [50], recent studies have shown that parts of the system can be reconstituted in vitro [51, 52]. For example, FtsZ is a highly conserved division protein that polymerizes into a ring that interacts with other members of the divisome to split the cell into two. Even if FtsZ is not thought to directly interact with the inner membrane in natural systems, FtsZ constructs can be engineered to anchor into the membrane of synthetic vesicles by the addition of a small helix to the C-terminus of FtsZ [52]. Further, in the presence of GTP this engineered version of FtsZ forms constricting rings that cause visible indentations within the membranes of tubular vesicles [52]. While these vesicles do not divide, the data suggest that a proper mix of Fts proteins could be sufficient to reconstitute functional cell division machinery within vesicles.
REPLICATING GENOMES
There are many ways to copy nucleic acids in vitro, but none are currently amenable to the construction of cell-like systems. The use of an RNA genome is attractive for at least two reasons. First, RNA genomes appear to simplify the system by removing the need of a class of molecules, i.e. DNA. Second, RNA polymerases do not require a primer. Although there are no known cells that use an RNA genome, there are viral systems that could be exploited. For example, the bacteriophage phi6 uses a double stranded RNA genome that is fully replicated by a single RNA polymerase [53]. This may seem like a simple system to reconstitute in the laboratory for building cell-like systems, but significant complications may arise when protein synthesis machinery is incorporated. Ribosomes require single stranded RNA as a template. This means that in addition to the double stranded RNA genome, single stranded RNA also must be present for protein synthesis. In other words, transcription of the genome, even an RNA genome, is always required. Although there are several classes of RNA viral systems with different genome organizations, e.g. viruses that use a circular RNA genome, they all share these same difficulties. An exciting alternative RNA system would exploit an RNA replicase, i.e. an RNA enzyme that copies an RNA genome. Despite the impressive advancements made in building better RNA replicases [54] much more progress is needed before they serve as a practical alternative.
The use of a DNA genome has the advantage of better separating replication and transcription, thus avoiding competition between the two processes [55]. Additionally, methods to replicate DNA in vitro have existed for several decades. Some attempts have been made to exploit these technologies for building cell-like systems. PCR based mechanisms for DNA copying inside of phospholipid vesicles have been demonstrated [56]. While useful as a first step, this approach is not practical, because it requires manipulation that is not regulated by the cell-like system (i.e. thermocycling), and it requires the addition of oligonucleotide primers. Another approach uses a more complex mixture of proteins, including a helicase and single strand binding proteins, in addition to a DNA polymerase to replicate DNA inside of vesicles. This system overcomes the thermocycling limitation of PCR, but still requires the addition of oligonucleotides, and it is only capable of replicating short (<100 bp) strands of DNA [37]. An extension of this technique would exploit a primase [57] to remove the need of adding oligonucleotide primers, but this would also add a significant problem. Genomic replication must be complete, which means that there must be a mechanism to ensure that there is no loss of the genomic termini, i.e. telomeres. Since primases add RNA primers that must later be removed, the simplified systems described so far are incapable of fully replicating a genome end-to-end.
The telomere problem is perhaps the biggest challenge to constructing a simple genomic replication mechanism. Viral DNA replication systems could potentially give a simpler solution to this problem than the isothermal bacterial systems described above. For example, the Bacillus subtilis bacteriophage phi29 uses only four proteins to replicate its entire genome, including a highly processive DNA polymerase that possesses strong strand displacement activity, a single strand DNA binding protein, a double strand DNA binding protein, and an additional protein that is required to initiate replication (terminal protein) [58]. The natural phi29 genome is linear, contains covalently attached terminal protein at the 5'-termini, and is replicated fully via a protein priming mechanism. Despite the peculiarities of the phi29 system, Salas and colleagues recently demonstrated that DNA can be fully replicated in vitro with this four protein component system if the template DNA contains appropriate nucleotide sequences at both termini to define the origins of replication [59]. Thus far the phi29 genomic replication system is the simplest and best characterized isothermal system available.
PROTEIN SYNTHESIS
Much of what has been described is dependent upon the activity of proteins. Cell-like systems that require protein function, therefore, require transcription and translation machinery. Transcription is a very straightforward process to reconstitute in vitro, particularly if the commonly used bacteriophage RNA polymerases are exploited. T7 and SP6 RNA polymerases consist of a single protein domain, do not require accessory factors for function, and provide robust activity. Conversely, the synthesis of proteins is a highly complex process requiring over 100 genome encoded components [60]. Nevertheless, this complicated process is understood well enough to be reconstituted in vitro from purified, defined components [61]. Moreover, several laboratories have shown that this minimal transcription-translation system functions in water-in-oil emulsion droplets [62] and in vesicles [63, 64] despite the statistical difficulty associated with encapsulating multiple components within a single compartment [65, 66].
ADAPTING TO CHANGING ENVIRONMENTAL CONDITIONS
Not every process within a cell needs to be monitored and coordinated with other cellular functions. For example, mitochondrial genome replication is not thought to be coordinated with division. Nevertheless, no living system simply repeats the same functions over and over again without regard to its surroundings. Cells must constantly adapt to changing intracellular and extracellular conditions. This is largely due to the fact that life and the environment are intimately linked. Life both feeds off of and shapes the environment. Conversely, the environment dictates which living things can and cannot survive. Therefore, if a cell is to survive for an appreciable length of time, the cell must be able to continually adapt to changing environmental conditions, some of which are induced by the cell's own existence.
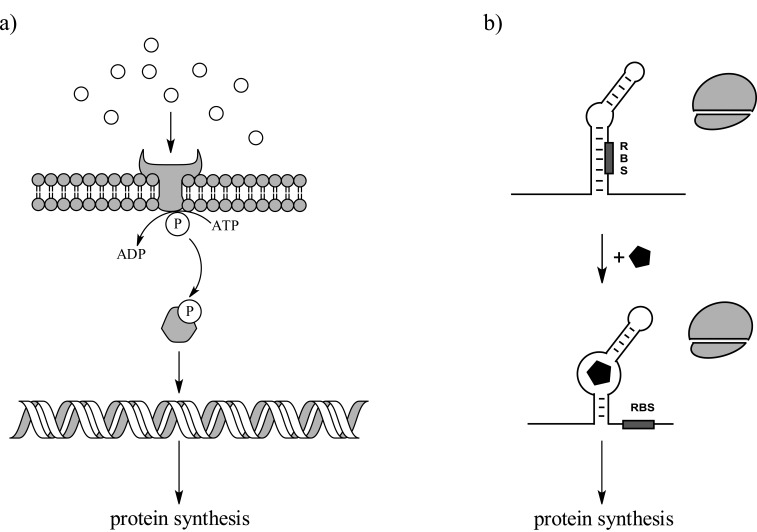
To adapt to changing environmental conditions, a cell needs to be capable of sensing and responding to stimuli. A straightforward solution to the problem would be to encode a sense-response system that exploits environmentally responsive transcription factors. For example, proteins such as FNR, IscR, and CooA control gene expression in response to oxygen, iron, and carbon monoxide levels, respectively [67,68]. Alternatively, bacterial two component systems [69] could be similarly used to alter gene expression profiles in response to environmental changes. Here a sensor-kinase would control the phosphorylation state and thus the activity of a response-regulator protein, which would then result in changes in gene expression (Fig. 3). Further layers of control could be built into sensory pathways by exploiting the sigma factors of bacterial polymerases. While bacterial RNA polymerases are more complicated than T7 or SP6 bacteriophage RNA polymerases, their use of initiation factors to guide promoter recognition provide for an opportunity to build in additional control elements. Although thoroughly studied in vivo, few attempts have been made to reconstitute any of these protein sensory systems in vitro [70].
Fig. (3).
Simple biological sensors could be built from protein or RNA components. (a) A two component protein system consisting of a sensor-kinase and a response regulator. In this example, a membrane bound sensor-kinase phosphorylates the response regulator only in the presence of ligand. The phosphorylated response regulator then activates gene expression. (b) An example of one type of riboswitch system that directly controls protein production in response to the presence or absence of ligand. Upon the binding of ligand, the ribosome binding site (RBS) is unmasked, thereby allowing for ribosome binding and thus protein synthesis. Both systems shown here illustrate an "on" switch; however, analogous "off" switches also exist.
A disadvantage of relying on existing proteins for sensing capabilities is that the use of existing proteins severely limits the types of cell-like systems achievable. In other words, we are forced to build cell-like systems that sense what natural cells are already capable of sensing if we use existing proteins. Although some examples of building new proteins with desired activities have been reported [71, 72], the present methods are not yet sufficiently developed to be widely applicable. Therefore, other sensing mechanisms that are easier to engineer are desirable.
Natural and synthetic RNA sensors that control protein production exist and are an attractive alternative to protein based sensing mechanisms. One class of RNA sensors is known as riboswitches and exist in the 5'-untranslated regions of bacterial genes. Riboswitches either turn on or off transcription or translation in response to ligand binding directly to the mRNA [73] (Fig. 3). The distinct advantage of RNA sensors over protein sensors is that we have years of RNA selection and engineering technologies [74] to build on, and so it is possible to build synthetic riboswitches capable of sensing and responding to molecules beyond those currently sensed by natural cells [75].
The majority of the riboswitch research has focused on measuring sensing activity inside of existing cells, as is the case for the protein based systems described above. However, recent work has shown that a previously selected theophylline riboswitch [76, 77] was capable of turning on gene expression in vitro, in water-in-oil emulsions, and in vesicles [78]. Further, this cell-like system could be built to sense molecules outside of the vesicle compartment, thereby demonstrating that artificial RNA sensors could serve as a foundation for the construction of cellular mimics that go beyond what natural biology gives us.
LIMITATIONS
Many challenges still remain. The progress made in mimicking some of the features of life is tempered by the fact that the majority of the experiments have been performed under different conditions. Therefore, it is unclear whether the integration of different cell-like functions in their present form into a single system is possible. There are also fundamental problems associated with the use of purified translation machinery. Although reconstituted transcription-translation systems are able to drive cascading, genetically encoded networks [61,79], none of the described systems are capable of supporting long-term activity. We are currently able to engineer in vitro systems that turn gene expression on and then off [38,80,81]. However, we are not able to begin another cycle, because we are not able to regenerate the translation machinery in vitro. Currently, the minimal translation system is composed of proteins and RNAs that are isolated from E. coli. Until we are able to produce functioning ribosomes from in vitro transcription-translation reactions, there will be severe limits on what types of cell-like systems can be manufactured in the laboratory.
ACKNOWLEDGEMENTS
We thank the Armenise-Harvard foundation and CIBIO for financial support.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- 2.Gibson D G, Glass J I, Lartigue C, Noskov V N, Chuang R Y, Algire M A, Benders G A, Montague M G, Ma L, Moodie M M, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova E A, Young L, Qi Z Q, Segall-Shapiro T H, Calvey C H, Parmar P P, Hutchison C A 3rd, Smith H O, Venter J C. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- 3.Ellington A D, Szostak J W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Isaacs F J, Carr P A, Wang H H, Lajoie M J, Sterling B, Kraal L, Tolonen A C, Gianoulis T A, Goodman D B, Reppas N B, Emig CJ, Bang D, Hwang S J, Jewett M C, Jacobson J M, Church G M. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deamer D W. Boundary structures are formed by organic components of the Murchison carbonaceous chondrite. Nature. 1985;317:792–794. [Google Scholar]
- 6.Miller S L. A production of amino acids under possible primitive earth conditions. Science. 1953;117:528–529. doi: 10.1126/science.117.3046.528. [DOI] [PubMed] [Google Scholar]
- 7.Oro J. Mechanism of synthesis of adenine from hydrogen cyanide under possible primitive earth conditions. Nature. 1961;191:1193–1194. doi: 10.1038/1911193a0. [DOI] [PubMed] [Google Scholar]
- 8.Pizzarello S. The chemistry of life's origin: a carbonaceous meteorite perspective. Acc. Chem. Res. 2006;39:231–237. doi: 10.1021/ar050049f. [DOI] [PubMed] [Google Scholar]
- 9.Powner M W, Gerland B, Sutherland J D. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 10.Ricardo A, Carrigan M A, Olcott A N, Benner S A. Borate minerale stabilize ribose. Science. 2004;303:196. doi: 10.1126/science.1092464. [DOI] [PubMed] [Google Scholar]
- 11.Rushdi A I, Simoneit B R. Lipid formation by aqueous Fischer-Tropschtype synthesis over a temperature range of 100 to 400 degrees C. Orig. Life Evol. Biosph. 2001;31:103–118. doi: 10.1023/a:1006702503954. [DOI] [PubMed] [Google Scholar]
- 12.Deamer D W, Dworkin J P. Chemistry and physics of primitive membranes. Top. Curr. Chem. 2005;259:1–27. [Google Scholar]
- 13.Mansy S S, Szostak J W. Reconstructing the Emergence of Cellular Life through the Synthesis of Model Protocells. Cold Spring Harb. Symp. Quant. Biol. 2009;74:47–54. doi: 10.1101/sqb.2009.74.014. [DOI] [PubMed] [Google Scholar]
- 14.Stano P, Luisi P L. Achievements and open questions in the selfreproduction of vesicles and synthetic minimal cells. Chem. Commun. (Camb.) 2010;46:3639–3653. doi: 10.1039/b913997d. [DOI] [PubMed] [Google Scholar]
- 15.Crick F. Central dogma of molecular biology. Nature. 1970;227:561–563. doi: 10.1038/227561a0. [DOI] [PubMed] [Google Scholar]
- 16.Posfai G, Plunkett G 3rd, Feher T, Frisch D, Keil G M, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma S S, de Arruda M, Burland V, Harcum S W, Blattner F R. Emergent properties of reduced-genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- 17.Glass J I, Assad-Garcia N, Alperovich N, Yooseph S, Lewis M R, Maruf M, Hutchison C A 3rd, Smith H O, Venter J C. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gil R, Silva F J, Pereto J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar H E, Moran N A, Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 20.Woese C. The universal ancestor. Proc. Natl. Acad. Sci. USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morowitz H J, editor. Yale University Press: New Haven; 1992. Beginnings of cellular life: Metabolism recapitulates biogenesis. [Google Scholar]
- 22.Chen I A, Roberts R W, Szostak J W. The emergence of competition between model protocells. Science. 2004;305:1474–1476. doi: 10.1126/science.1100757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szostak J W, Bartel D P, Luisi P L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- 24.Deamer D W. The first living systems: a bioenergetic perspective. Microbiol. Mol. Biol. Rev. 1997;61:239–261. doi: 10.1128/mmbr.61.2.239-261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen I A, Szostak J W. Membrane growth can generate a transmembrane pH gradient in fatty acid vesicles. Proc. Natl. Acad. Sci. USA. 2004;101:7965–7970. doi: 10.1073/pnas.0308045101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segre D, Ben-Eli D, Deamer D W, Lancet D. The lipid world. Orig. Life Evol. Biosph. 2001;31:119–145. doi: 10.1023/a:1006746807104. [DOI] [PubMed] [Google Scholar]
- 27.Blochliger E, Blocher M, Walde P, Luisi P L. Matrix effect in the size distribution of fatty acid vesicles. J. Phys. Chem. B. 1998;102:10383–10390. [Google Scholar]
- 28.Chen I A, Szostak J W. A kinetic study of the growth of fatty acid vesicles. Biophys. J. 2004;87:988–998. doi: 10.1529/biophysj.104.039875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanczyc M M, Fujikawa S M, Szostak J W. Experimental models of primitative cellular compartments: encapsulation growth and division. Science. 2003;302:618–622. doi: 10.1126/science.1089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargreaves W R, Deamer D W. Liposomes from ionic, single-chain amphiphiles. Biochemistry. 1978;17:3759–3768. doi: 10.1021/bi00611a014. [DOI] [PubMed] [Google Scholar]
- 31.Mansy S S, Schrum J P, Krishnamurthy M, Tobe S, Treco D A, Szostak J W. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansy S S, Szostak J W. Thermostability of model protocell membranes. Proc. Natl. Acad. Sci. USA. 2008;105:13351–13355. doi: 10.1073/pnas.0805086105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walde P, Wick R, Fresta M, Mangone A, Luisi P L. Autopoietic selfreproduction of fatty acid vesicles. J. Am. Chem. Soc. 1994;116:11649–11654. [Google Scholar]
- 34.Zhu T F, Szostak J W. Coupled growth and division of model protocell membranes. J. Am. Chem. Soc. 2009;131:5705–5713. doi: 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen I A, Salehi-Ashtiani K, Szostak J W. RNA catalysis in model protocell vesicles. J. Am. Chem. Soc. 2005;127:13213–13219. doi: 10.1021/ja051784p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monnard P A, Apel C L, Kanavarioti A, Deamer D W. Influence of ionic inorganic solutes on self-assembly and polymerization processes related to early forms of life: implications for a prebiotic aqueous medium. Astrobiology. 2002;2:139–152. doi: 10.1089/15311070260192237. [DOI] [PubMed] [Google Scholar]
- 37.Torino D, Del Bianco C, Ross L A, Ong J L, Mansy S S. Intravesicle isothermal DNA replication. BMC Res. Notes. 2011;4:128. doi: 10.1186/1756-0500-4-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Noireaux V, Libchaber A. A vesicle bioreactor as a step toward an artificial cell assembly. Proc. Natl. Acad. Sci. USA. 2004;101:17669–17674. doi: 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monnard P A, Deamer D W. Nutrient uptake by protocells: a liposome model system. Orig. Life Evol. Biosph. 2001;31:147–155. doi: 10.1023/a:1006769503968. [DOI] [PubMed] [Google Scholar]
- 40.Monnard P A, Luptak A, Deamer D W. Models of primitive cellular life: polymerases and templates in liposomes. Phil. Trans. R. Soc. B. 2007;362:1741–1750. doi: 10.1098/rstb.2007.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCutcheon J P, Moran N A. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc. Natl. Acad. Sci. USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pietrini A V, Luisi P L. Cell-free protein synthesis through solubilisate exchange in water/oil emulsion compartments. Chembiochem. 2004;5:1055–1062. doi: 10.1002/cbic.200400014. [DOI] [PubMed] [Google Scholar]
- 43.Tawfik D S, Griffiths A D. Man-made cell-like compartments for molecular evolution. Nat. Biotechnol. 1998;16:652–656. doi: 10.1038/nbt0798-652. [DOI] [PubMed] [Google Scholar]
- 44.Bernath K, Magdassi S, Tawfik D S. Directed evolution of protein inhibitors of DNA-nucleases by in vitro compartmentalization (IVC) and nanodroplet delivery. J. Mol. Biol. 2005;345:1015–1026. doi: 10.1016/j.jmb.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Mansy S S. Membrane transport in primitive cells.In The Origins of Life Cold Spring Harb. Perspect. Biol. 2010;2:193–206. doi: 10.1101/cshperspect.a002188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurama H, Stano P, Ueda T, Luisi P L. A synthetic biology approach to the construction of membrane proteins in semi-synthetic minimal cells. Biochim. Biophys. Acta. 2009;1788:567–574. doi: 10.1016/j.bbamem.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Baumgart T, Hess S T, Webb W W. Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature. 2003;425:821–824. doi: 10.1038/nature02013. [DOI] [PubMed] [Google Scholar]
- 48.Chen I A. Cell division: breaking up is easy to do. Curr. Biol. 2009;19:R237–R238. doi: 10.1016/j.cub.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 49.Leaver M, Dominguez-Cuevas P, Coxhead J M, Daniel R A, Errington J. Life without a wall or division machine in Bacillus subtilis. Nature. 2009;457:849–853. doi: 10.1038/nature07742. [DOI] [PubMed] [Google Scholar]
- 50.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 51.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 52.Osawa M, Anderson D E, Erickson H P. Reconstitution of contractile FtsZ rings in liposomes. Science. 2008;320:792–794. doi: 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makeyev E V, Grimes J M. RNA-dependent RNA polymerases of dsRNA bacteriophages. Virus Res. 2004;101:45–55. doi: 10.1016/j.virusres.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 54.Wochner A, Attwater J, Coulson A, Holliger P. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011;332:209–12. doi: 10.1126/science.1200752. [DOI] [PubMed] [Google Scholar]
- 55.Ichihashi N, Matsuura T, Kita H, Hosoda K, Sunami T, Tsukada K, Yomo T. Importance of translation-replication balance for efficient replication by the self-encoded replicase. Chembiochem. 2008;9:3023–3028. doi: 10.1002/cbic.200800518. [DOI] [PubMed] [Google Scholar]
- 56.Oberholzer T, Albrizio M, Luisi P L. Polymerase chain reaction in liposomes. Chem. Biol. 1995;2:677–682. doi: 10.1016/1074-5521(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Kim H J, Zheng C, Chow W H, Lim J, Keenan B, Pan X, Lemieux B, Kong H. Primase-based whole genome amplification. Nucleic Acids Res. 2008;36 (13):e79. doi: 10.1093/nar/gkn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salas M. 40 years with bacteriophage phi29. Annu. Rev. Microbiol. 2007;61:1–22. doi: 10.1146/annurev.micro.61.080706.093415. [DOI] [PubMed] [Google Scholar]
- 59.Mencia M, Gella P, Camacho A, de Vega M, Salas M. Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage phi29. Proc. Natl. Acad. Sci. USA. 2011;108:18655–18660. doi: 10.1073/pnas.1114397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Forster A C, Church G M. Towards synthesis of a minimal cell. Mol. Syst. Biol. 2006;2:45. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 62.Zheng Y, Roberts R J. Selection of restriction endonucleases using artificial cells. Nucleic Acids Res. 2007;35 (11):e83. doi: 10.1093/nar/gkm410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stano P, Kuruma Y, Souza T P, Luisi P L. Biosynthesis of proteins inside liposomes. Methods Mol. Biol. 2010;606:127–145. doi: 10.1007/978-1-60761-447-0_11. [DOI] [PubMed] [Google Scholar]
- 64.Sunami T, Matsuura T, Suzuki H, Yomo T. Synthesis of functional proteins within liposomes. Methods Mol. Biol. 2010;607:243–256. doi: 10.1007/978-1-60327-331-2_20. [DOI] [PubMed] [Google Scholar]
- 65.Pereira de Souza T, Stano P, Luisi P L. The minimal size of liposome based model cells brings about a remarkably enhanced entrapment and protein synthesis. Chembiochem. 2009;10:1056–1063. doi: 10.1002/cbic.200800810. [DOI] [PubMed] [Google Scholar]
- 66.Pereira de Souza T, Steiniger F, Stano P, Fahr A, Luisi P L. Spontaneous crowding of ribosomes and proteins inside vesicles: a possible mechanism for the origin of cell metabolism. Chembiochem. 2011;12(15):2325–2330. doi: 10.1002/cbic.201100306. [DOI] [PubMed] [Google Scholar]
- 67.Dufour Y S, Kiley P J, Donohue T J. Reconstruction of the core and extended regulons of global transcription factors. PLoS Genet. 2010;6 (7):e1001027. doi: 10.1371/journal.pgen.1001027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleischhacker A S, Kiley P J. Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 2011;15:335–341. doi: 10.1016/j.cbpa.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stock A M, Robinson V L, Goudreau P N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 70.Moker N, Kramer J, Unden G, Kramer R, Morbach S. In vitro analysis of the two-component system MtrB-MtrA from Corynebacterium glutamicum. J. Bacteriol. 2007;189:3645–3649. doi: 10.1128/JB.01920-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seelig B, Szostak J W. Selection and evolution of enzymes from a partially randomized non-catalytic scaffold. Nature. 2007:448, 828–831. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siegel J B, Zanghellini A, Lovick H M, Kiss G, Lambert A R, St Clair J L, Gallaher J L, Hilvert D, Gelb M H, Stoddard B L, Houk K N, Michael F E, Baker D. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winkler W C, Breaker R R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 74.Lorsch J R, Szostak J W. Chance and necessity in the selection of nucleic acid catalysts. Acc. Chem. Res. 1996;29:103–110. doi: 10.1021/ar9501378. [DOI] [PubMed] [Google Scholar]
- 75.Topp S, Gallivan J P. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lynch S A, Gallivan J P. A flow cytometry-based screen for synthetic riboswitches. Nucleic Acids Res. 2009;37:184–192. doi: 10.1093/nar/gkn924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Topp S, Gallivan J P. Guiding bacteria with small molecules and RNA. J. Am. Chem. Soc. 2007;129:6807–6811. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martini L, Mansy S S. Cell-like systems with riboswitch controlled gene expression. Chem. Commun. (Camb.) 2011;47:10734–10736. doi: 10.1039/c1cc13930d. [DOI] [PubMed] [Google Scholar]
- 79.Ishikawa K, Sato K, Shima Y, Urabe I, Yomo T. Expression of a cascading genetic network within liposomes. FEBS Lett. 2004;576:387–390. doi: 10.1016/j.febslet.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 80.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc. Natl. Acad. Sci. USA. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shin J, Noireaux V. Study of messenger RNA inactivation and protein degradation in an Escherichia coli cell-free expression system. J. Biol. Eng. 2010;4:9. doi: 10.1186/1754-1611-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]