Abstract
Borderline ovarian tumors represent an understudied subset of ovarian tumors. Most studies investigating aberrations in borderline tumors have focused on KRAS/BRAF mutations. In this study we conducted an extensive analysis of mutations and single nucleotide polymorphisms in borderline ovarian tumors.
Using the Sequenom MassARRAY platform we investigated 160 mutations/polymorphisms in 33 genes involved in cell signalling, apoptosis, angiogenesis, cell cycle regulation, and cellular senescence.
Of 52 tumors analysed, 33 were serous, 18 mucinous and 1 endometrioid. KRAS c.35G>A p.Gly12Asp mutations were detected in 8 tumors (6 serous and 2 mucinous), BRAF V600E mutations in 2 serous tumors, and PIK3CA H1047Y and PIK3CA E542K mutations in a serous and an endometrioid BOT respectively. CTNNB1 mutation was detected in a serous tumor. Potentially functional polymorphisms were found in VEGF, ABCB1, FGFR2 and PHLPP2. VEGF polymorphisms were the most common and detected at 4 loci. PHLPP2 polymorphisms were more frequent in mucinous as compared to serous tumors (p=0.04), with allelic imbalance in one case.
This study represents the largest and most comprehensive analysis of mutations and functional single nucleotide polymorphisms in borderline ovarian tumors to date. At least 25% of borderline ovarian tumors harbour somatic mutations associated with potential response to targeted therapeutics.
Keywords: Borderline, Ovarian, Sequenom, Tumours
INTRODUCTION
Borderline ovarian tumors account for 10% of all ovarian neoplasms predominantly affecting women in the reproductive age 1. Borderline ovarian tumors are histologically a heterogeneous group of slow growing, non-invasive tumors, the majority (85%) of which present with stage I disease confined to the ovary. Overall borderline ovarian tumors have a significantly better prognosis with over 95% 5-year overall survival compared to 30–40% 5-year overall survival rates for their invasive counterparts 1, 2. Approximately 1% of borderline ovarian tumors show progression to invasive epithelial cancer. Non-resectable recurrent disease is responsible for the majority of disease related deaths and present similar problems to invasive ovarian cancer such as bowel obstruction and drug resistance 1.
Prognostic factors in patients with borderline ovarian tumors and features potentially associated with recurrent and/or progressive disease include tumor type, patient age, FIGO stage, invasive implants, microinvasion in the primary tumor, and micropapillary architecture 3–6. No single clinical or pathologic feature or combination of features identify all adverse outcomes 7. Treatment options for patients with persisting/progressive disease are limited 8, 9, with limited activity of currently available chemotherapeutic agents used in invasive ovarian cancers in the treatment of borderline ovarian tumors 9,10. New systemic therapeutic agents are therefore urgently required 1.
Although the clinical and pathologic characteristics of borderline ovarian tumors are well described, the molecular aspects are poorly understood 11, 12. Borderline ovarian tumors have not been as extensively studied at a molecular level as invasive ovarian carcinomas, and knowledge of genetic abnormalities associated with borderline ovarian tumors is limited. Studies looking into gene mutations in borderline ovarian tumors have mainly been in comparison to invasive carcinoma and focused on TP53, BRAF and KRAS 13–16. TP53 mutations are not commonly associated with borderline ovarian tumors, in contrast to their high frequency in high grade carcinoma 15. Conversely, KRAS and BRAF mutations are both much more common in borderline ovarian tumors and low grade serous carcinomas 13, 17.
In addition to KRAS, BRAF and TP53, a few studies have also investigated the frequency of PIK3CA 18, BRCA1 19, EGFR 20, CTNNB1 21 and PTEN 21 mutations in borderline ovarian tumors in comparison to invasive ovarian carcinomas. Gene amplifications have also been studied in borderline ovarian tumors including ERBB2 22 and AKT2 23.
In this study we used the high-throughput Sequenom MassArray approach to investigate single nucleotide mutations and polymorphisms in 33 genes in a cohort of borderline ovarian tumors to determine the frequency of genetic changes associated with borderline ovarian tumors.
MATERIALS AND METHODS
Tumor samples
Frozen tissue from 52 borderline ovarian tumors was obtained from the Human Biomedical Resource Centre, Imperial College Healthcare NHS Trust, Hammersmith Hospital. Ethics Committee approval for use of human tissue was obtained. Table 1 summarises patients’ age, tumor types and tumor stage.
Table 1.
Patient and tumor characteristics
| Age (median, years) | 50 |
| Tumor type | |
| Endometroid | 1 |
| Mucinous | 18 |
| Serous | 33 |
| FIGO stage | |
| IA | 22 |
| IC | 16 |
| IIA | 2 |
| IIC | 1 |
| IIIA | 1 |
| IIIB | 3 |
| IIIC | 1 |
| Unstaged | 6 |
DNA extraction
DNA was extracted from fresh snap frozen tissue. An H & E stained section from the frozen tissue used for each specimen was examined to verify the content and the quality of the tissue analysed. Briefly, tissue was homogenised in 180 μL RTL buffer (Qiagen) using a TissueLyser (Tissuelyser I, Retsch, Leeds, UK) at 15 Hz, for 20 seconds. Supernatant containing disrupted tissue was transferred to a 1.5 mL eppendorf, 20 μL Proteinase K was added and the sample incubated at 56 °C overnight. DNA was then extracted using the QIAamp DNA Mini kit (Qiagen) according to manufacturer’s protocol.
Sequenom MassArray
The list of genes, mutations and polymorphisms assessed by Sequenom are presented in Appendix 1. PCR and extension primers were designed using Assay Design (Sequenom). PCR-amplified DNA was cleaned using EXO-SAP (Sequenom), and primer was extended by IPLEX chemistry, desalted using Clean Resin (Sequenom), and spotted onto Spectrochip matrix chips using a nanodispenser (Samsung). Chips were run in duplicate on a Sequenom MassArray MALDI-TOF MassArray system. Sequenom Typer Software and visual inspection were used to interpret mass spectra. Reactions where >15% of the resultant mass ran in the mutant site in both reactions were scored as positive. Mutations and polymorphisms for a subset of samples and targets were confirmed by Sanger sequencing and pyrosequencing respectively.
Pyrosequencing
PHLPP2 polymorphisms and allelic imbalance in tumors was assessed using pyrosequencing of genomic DNA. The primers for PHLPP2 amplification and sequencing were: 5′-AAACAAAGCATTGTGGGAACACT -3′ (forward), 5′-biotin- AAACTACCATCGCCCCTACATT -3′ (reverse) and 5′-CTAAGAAGCTGTGCACAT -3′ (sequencing). Initial PCR was performed using Jumpstart Taq (Sigma), 60°C degree annealing, 2.5 mM MgCl, 200 nM primer and 10 ng genomic DNA. Pyrosequencing of PCR products was performed using PyroGold Reagent kit (Biotage, Uppsala, Sweden) according to the manufacturer’s instructions. The individual genotypes of the rs61733127 (L1016S) polymorphism were estimated manually using the Pyro Q-CpG Software (Qiagen, UK)) with thresholds for TT (<10% C), CT (40–60% C) and CC (>90% C). We used the quantitation of C vs T alleles in heterozygotes to identify tumors displaying loss of heterozygosity with a threshold of (10–40% C).
Sanger sequencing for KRAS
Mutations in KRAS were verified using primers (forward: TTTGATAGTGTATTAACCTTATG, reverse: GAGGTAAATCTTGTTTTAATA), using 10 ng DNA, 200 nM primer, 2.5 mM MgCl and JumpStartTaq (Sigma) at 52°C degrees for 40 cycles. Sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems). Cycle conditions were 94°C for 1 min followed by 30 cycles of 94°C for 10secs, 55°C for 15secs, 60°C for 4mins. PCR products were cleaned by EDTA-Ethanol precipitation, resuspended in HiDi formamide and run on a 3730xl DNA Analyser (Applied Biosystems Ltd). Base calling, quality assessment, and assembly were carried out using the Phred, Phrap, Polyphred, Consed software suite. All potential sequence variants were verified by manual inspection of the chromatograms.
EGFR and PDGFRA mutational analysis
EGFR mutations in exons 19 to 21 and PDGFRA mutations in exons 12 and 18 were analysed by capillary electrophoresis single-strand conformation analysis (CE-SSCA) using a 3130xl genetic analyser (Life Technologies, Warrington, UK) and non-denaturing polymer at three different temperatures. Any conformation changes were subjected to bi-directional Sanger sequencing.
Immunohistochemistry
The expression of beta-catenin was evaluated by immunohistochemistry using the avidin biotin immunodetection complex method. Two micron thick sections from formalin-fixed, paraffin embedded tissue were prepared, deparaffinised and rehydrated. Endogenous peroxidase was blocked by incubation in hydrogen peroxide. Antigen retrieval was performed by microwaving in in 0.01M citrate buffer (pH 6.0) at 750W for 20 minutes. Non-specific binding was blocked with normal goat serum for 10 minutes. Tissue sections were then incubated with primary antibody for beta-catenin (BD Biosciences 1:500 dilution) at room temperature for 60 minutes. The sections were washed, then incubated with goat anti-mouse biotinylated immunoglobulin (Dako, 1:2000 dilution) for 30 minutes, followed by streptavidin peroxidase for 30 minutes. The slides were developed in DAB, followed by a haematoxylin counterstain. For each case a section in which the primary antibody was replaced by phosphate buffered saline was used as a negative control.
Statistical analysis
The chi-square test was used to test for the presence of associations between the different gene mutations and polymorphisms and the histopathological features of tumors. A p-value of less than 0.05 was considered statistically significant. Statistical analysis was performed using SPSS (version 16.0, Chicago, IL, USA).
RESULTS
Fifty-two borderline tumors were studied (Table 1, Appendix 2). The tumors included 33 serous (63%), 18 mucinous (35%) and 1 endometrioid tumor (2%). Six (18%) of the serous tumors had a micropapillary component, 2 (6%) showed microinvasion and 7 (21%) were associated with non-invasive implants. Of the mucinous tumors 2 (11%) showed microinvasion, and 4 (22%) showed intramucosal carcinoma. Patients’ aged from 26–82 years (median 50 years), with a median follow-up period of 3 years (range 2 – 7 years). One patient with a mucinous borderline ovarian tumour treated with unilateral oophorectomy developed a cyst on the other ovary that was detected on a follow-up scan 18 months after initial surgery. The patient did not undergo surgical removal of the cyst, so the histological nature of the cyst is unknown to us. None of the patients with serous and endometrioid tumors or other patients with mucinous tumours developed recurrence or disease progression.
Borderline endometrioid tumours are very rare, including the one case from which frozen tissue was available was an opportunity to study the status of a large set of genes in such a rare tumor.
Gene mutations
Mutations were detected in 6 (KRAS, BRAF, PIK3CA, EGFR, PDGFRA and CTNNB1) of the 33 genes studied using the Sequenom assay (Table 2, Appendix 2). KRAS c.35G>A p.Gly12Asp mutations were detected in in 8/52 tumors (15%), which included 6/33 serous (18%) and 2/18 mucinous (11%) tumors. BRAF mutations were detected in only 2/52 (4%) tumors, both of which were in serous tumors (2/33, 6%). PIK3CA mutations were found in 2/52 tumors (4%). PIK3CA_E542K (heterozygous mutation - het) was present in 1/33 (3%) serous tumor, and PIK3CA_H1047Y (het) was detected in the one borderline endometrioid tumor. PDGFRA_V824L (het) was detected in 3 tumors (6%). These included 2/33 serous (6%) and 1/18 (6%) mucinous tumors. CE-SSCA and Sanger sequencing analysis of these samples failed to confirm the PDGFRA_V824L mutation, instead, 2/3 cases showed a synonymous polymorphism very near to that genomic position (c.2472C>T; p.V834V). The third case showed heavily degraded DNA and a wild type conformation, which can also result in false priming.
Table 2.
Frequency of mutations
| Mutation | Sequence | Genotype | Frequency (n=52) |
|---|---|---|---|
| PIK3CA E542 | 1624G | GA | 1/52 (2%) 1/33 serous (3%) |
| PIK3CA H1047Y | TC | 1/52 (2%); 1/1 endometrioid (100%) | |
| KRAS G12 | 35G | GA GT |
8/52 (15%) 4/33 serous (12%); 1/18 (6%) mucinous 2/33 (6%) serous; 1/18 (6%) mucinous |
| BRAF V600E | 1799T | AT | 2/52 (4%); 2/33 serous (6%) |
| CTNNB1 S37 | 110C | CG | 1/52 (2%); 1/33 serous (3%) |
Sequenom also identified EGFR L858R mutations in 5 cases. However, these mutations were not validated on Sanger sequencing.
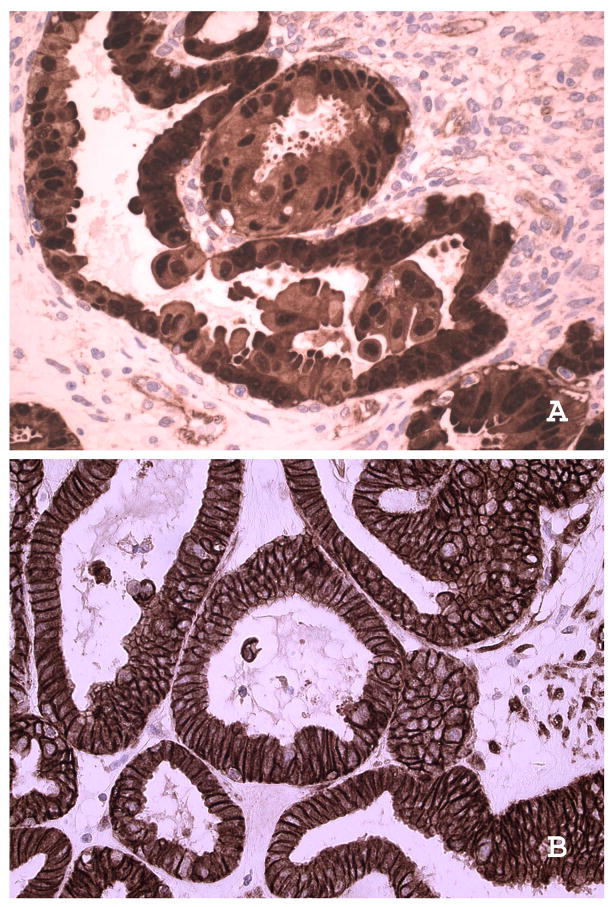
CTNNB1_S37C (het) mutation was detected in one of the 33 serous tumors (3%). Immunostaining of this sample showed nuclear localisation of beta catenin, a feature seen in the presence of beta-catenin mutations (Figure 1). This case was reviewed by 2 gynaecological histopathologists and confirmed to be of the serous type.
Figure 1. Expression of beta catenin in a serous borderline ovarian tumour with beta-catenin mutation.
In the serous borderline tumour with beta-catenin mutation there is cytoplasmic and notable nuclear localisation with absence of membranous staining (A: X400). This is in contrast to distinct membranous localisation and absence of nuclear localisation in a serous borderline ovarian tumor with wild type beta-catenin (B: X100).
Thirteen (25%) of 52 tumors had at least one mutation in the genes tested (Appendix 2). Eleven of the 13 (85%) cases had a single mutation, while 2 cases (15%) had 2 mutations (KRAS and CTNNB1 in one serous tumor and KRAS_G12_35G and PIK3CA_H1047Y in one endometrioid tumor). Table 3 summarises the correlation between the presence of mutations and histopathological features of serous and mucinous tumors.
Table 3.
Correlation between mutations and histopathological features of serous and mucinous tumors
| Non invasive implants (S) | Micropapillary component (S) | Microinvasion (S & M) | Intramucosal carcinoma (M) | |||||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | |
| KRAS | 1/7 (14%) | 5/26 (19%) | 0/6 (0%) | 6/27 (22%) | 0/2 S (0%) 0/2 M (0%) |
6/31 S (19%) 2/16 M (13%) |
0/4 (0%) | 2/14 (14%) |
| BRAF | 0/7 (0%) | 2/26 (7%) | 0/6 (0%) | 2/27 (7%) | 0/2 S (0%) 0/2 M (0%) |
2/31 S (6%) 0/16 M (0%) |
0/4 (0%) | 0/14 (0%) |
| PIK3CA | 0/7 (0%) | 1/26 (4%) | 0/6 (0%) | 1/27 (4%) | 0/2 S (0%) 0/2 M (0%) |
1/31 S (3%) 0/16 M (0%) |
0/4 (0%) | 0/14 (0%) |
| β-catenin | 0/7 (0%) | 1/26 (4%) | 0/6 (0%) | 1/27 (4%) | 0/2 S (0%) 0/2 M (0%) |
1/31 S (3%) 0/16 M (0%) |
0/4 (0%) | 0/14 (0%) |
(S) = serous; (M) = mucinous
Gene polymorphisms
Potential functional single nucleotide gene polymorphisms (SNPs) were found in 4 of the 33 genes studied (Table 4, Appendix 2). Only 4/52 (8%) of tumors featured SNPs in only one of the genes tested, whereas 48/52 (92%) tumors showed polymorphisms in two or more of the genes tested with the majority of genes being heterozygous (Table 4). The most common polymorphism observed was in VEGF in 50/52 borderline ovarian tumors (96%), with SNPs detected at 4 loci. There were 2 or more VEGF SNPs in 24/33 serous tumors, 1/1 endometrioid and 11/18 mucinous tumors. ABCB1 polymorphisms were detected in 37/52 borderline ovarian tumors (71%), followed by FGFR2 polymorphisms in 29/52 borderline ovarian tumors (56%).
Table 4.
Frequency of polymorphisms.
| Polymorphism | PHLPP2_L1016S_T3047C | ABCB1_G2677 TA | FGFR2_rs2981582_CT | VEGF_5_1154_GA_ref | VEGF_5_1498_CT | VEGF_5_2573_CA | VEGF GC_ref_5_634_ |
|---|---|---|---|---|---|---|---|
| Serous | 5/33(15%)* (CT) | 23/33(70%) (T: 8; GT: 15) | 18/33(55%) (T:8; CT: 10) | 13/33(39%) (GA) | 26/33(79%) (T:4; TC: 22) | 7/33(21%) (CA) | 23/33(70%) (C: 4; CG: 19) |
| Mucinous | 8/18(44%) *(CT) | 13/18(72%) (T:4; GT: 9) | 11/18(61%) (T:2; CT: 9) | 9/18 (50%) (A: 1; GA: 8) | 12/18(67%) (T: 3; TC: 9) | 1/18 (6%) (A) | 9/18 (50%) (C:2; CG:7) |
| Endometrioid | 1/1(100%) (CT) | 1/1 (100%) (GT) | 0/1 (0%) | 0/1 (0%) | 1/1 (100%) (TC) | 0/1(0%) | 1/1 (100%) (CG) |
Heterozygote PHLPP2 polymorphisms were detected in 14/52 (27%) tumors. These included 5/33(15%) serous, 8/18 mucinous (44%) and one endometrioid borderline ovarian tumor. Tumors with PHLPP2 polymorphisms included 2/4 (50%) mucinous tumors with intramucosal carcinoma and 1/6 (17%) serous tumor of the micropapillary type, and 1/2 (50%) serous tumors with microinvasion (50%).
Table 5 shows the correlation between polymorphisms and histopathological features of tumors. PHLPP2 polymorphisms were more common in mucinous tumors compared to serous BOTs; 44% (n=8/18) vs 15% (n=5/33), chi-sq p=0.04).
Table 5.
Correlation between polymorphisms and histopathological features of serous and mucinous tumors.
| Non invasive implants (S) | Micropapillary component (S) | Microinvasion (S & M) | Intramucosal carcinoma (M) | |||||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | |
| PHLPP2_L1016S _T3047C | 0/7 (0%) | 5/26 (19%) | 2/6 (33%) | 3/27 (11%) | 1/2 S (50%) 0/2 M (0%) |
4/31 S (13%) 8/16 M (50%) |
2/4 (50%) | 6/14 (43%) |
| ABCB1_G2677T A | 4/7 (57%) | 19/26 (73%) | 5/6 (83%) | 18/27 (67%) | 1/2 S (50%) 1/2 M (50%) |
22/31 S (71%) 12/16 M (75%) |
1/4 (25%) | 12/14 (86%) |
| FGFR2_rs2981582_CT | 3/7 (43%) | 15/26 (58%) | 4/6 (66%) | 13/27 (48%) | 1/2 S (50%) 2/2 M (100%) |
17/31 S (55%) 9/16 M (56%) |
3/4 (75%) | 8/14 (57%) |
| VEGF_5_1154_A_ref | 1/7 (14%) | 12/26 (46%) | 2/6 (33%) | 11/27 (41%) | 1/2 S (50%) 1/2 M (50%) |
12/31 S (39%) 8/16 M (50%) |
2/4 (50%) | 7/14 (50%) |
| VEGF_5_1498_CT | 6/7 (86%) | 20/26 (77%) | 5/6 (83%) | 21/27 (78%) | 2/2 S (100%) 2/2 M (100%) |
24/31 S (77%) 10/16 M (62%) |
3/4 (75%) | 9/14 (64%) |
| VEGF_5_2573_CA | 1/7 (14%) | 6/26 (23%) | 1/6 (17%) | 6/27 (22%) | 2/2 S **(100%) 0/2 M (0%) |
5/31 S **(16%) 1/16 M (6.2%) |
0/4 (0%) | 1/14 (7%) |
| VEGF_5_634_GC_ref | 5/7 (71%) | 18/26 | 4/6 (66%) | 19/27 (70%) | 2/2 S (100%) 0/2 M |
21/31 S 9/16 M |
3/4 (75%) | 6/14 (43%) |
(S) = serous; (M) = mucinous
Validation of gene mutations and polymorphisms in selected genes
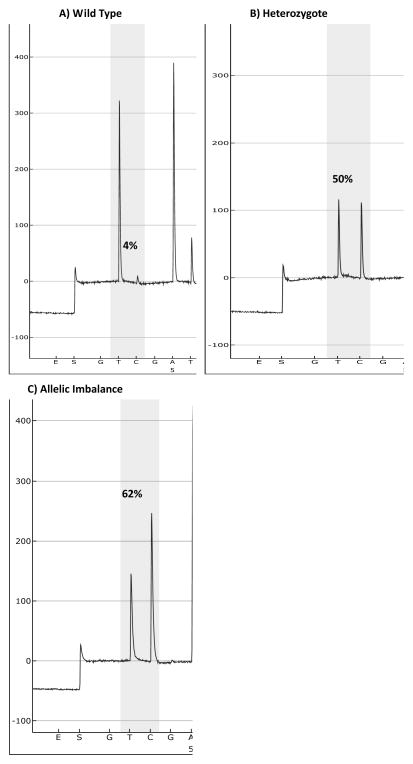
KRAS gene mutation status was assessed by direct Sanger sequencing in 5 cases, and confirmed the results of Sequenom Massarray in all cases. PHLPP2 polymorphisms were confirmed by pyrosequencing in all 52 cases. In addition relative allelic quantification demonstrated evidence of allelic imbalance in one borderline ovarian tumor (Figure 2).
Figure 2. Pyrosequencing results for PHLPP2 polymorphisms.
A) Normal female genomic DNA homozygous for PHLPP2 major allele, B) Borderline ovarian tumor heterozygous for major and minor alleles, C) Borderline ovarian tumor with allelic imbalance for the minor allele.
DISCUSSION
We used the Sequenom Massarray technique to profile gene single nucleotide mutations and polymorphisms in borderline ovarian tumors. We and others have previously demonstrated that the sensitivity of mass spectrometric methods exceeds that of traditional Sanger sequencing where the aberration must be present in approximately 20% of the DNA present and is highly concordant with Sanger sequencing, pyrosequencing, and allele-specific PCR 24,25. Fifty-two tumors, representing the largest set of borderline ovarian tumors analysed for mutational status, were studied for changes in 33 genes known to be involved in tumor pathology with the majority being potential targets, including genes in the RAS-RAF-MEK and RTK-PI3K-AKT pathways.
Our study of borderline ovarian tumors shows that somatic mutations occur predominately in a subset of the genes studied. Overall the frequency and pattern of mutations is consistent with borderline ovarian tumors showing more similarity and being potential precursors of Type I ovarian carcinoma. Jones et al comprehensively analysed somatic mutations in low-grade serous carcinomas, by exome sequencing and showed that the genes showing the most frequent mutations were BRAF and KRAS, occurring in 38% and 19% of low-grade tumours, respectively and a single case showed a PIK3CA mutation. Their mutational analysis demonstrates that point mutations are much less common in low-grade serous tumours of the ovary 17. In agreement with previous reports in our study KRAS and BRAF mutations were the most common mutations detected 13, 26–30. Mutations in decreasing order of frequency were identified in KRAS, BRAF, PDGFRA, PIK3CA, and CTNNB1. Mutations in these genes are likely to perturb several signalling cascades as well as signalling networks involved in cell proliferation, survival and motility. The most frequent polymorphisms were found in the VEGF gene with 4 SNPs, followed by ABCB1, FGFR2 and PHLPP2. Allelic imbalance in favour of the minor allele of PHLPP2 was shown in one tumor suggesting selection of the minor allele may play a role in tumor development and progression.
Although the frequency of RAS/RAF mutations in our cohort is relatively less than that in previous reports 16, 31, the majority of detected mutations and polymorphisms seemed to focus on the RAS/RAF and PI3K/AKT pathway and upstream growth factors and growth factor receptors. The Ras/Raf/MEK/ERK and PI3K/PTEN/AKT signalling cascades interact and play critical roles in the transmission of signals from growth factor receptors to regulate gene expression and prevent apoptosis. These signalling and anti-apoptotic pathways can have different effects on growth, prevention of apoptosis and induction of drug resistance in cells of various lineages. Components of these pathways or upstream receptors are mutated or aberrantly expressed in human cancer and subjects of active drug development programs 32.
One serous tumor had concurrent CTNNB1 and KRAS mutations, and an endometrioid borderline ovarian tumor PIK3CA and KRAS mutations. The finding of concordant mutations, despite the overall low frequency of mutations, suggests that additional genetic abnormalities are selected in the presence of mutations in KRAS and BRAF during the pathogenesis of borderline ovarian tumors. The presence of co-ordinate mutations suggests that alternative functions of the two genes are selected or that an aberration in a single node in the pathway does not engender sufficient pathway activation for tumor initiation or progression. CTNNB1 mutations have been reported previously in borderline ovarian tumors, exclusively in endometrioid but not serous borderline ovarian tumors as in the case here.
Deregulated signalling via the phosphatidylinositol 3-kinase (PI3K) pathway is common in many cancer lineages. For example, PIK3CA is the most commonly mutated oncogene in uterine endometrioid carcinoma (UEC) 33 and breast carcinomas 34. Mutations occur in predominately in exon 20 (kinase domain), and exon 9 (helical domain) in breast cancer 35–37 but in other sites in different lineages such as UEC. The frequency of PIK3CA hotspot mutations throughout the coding region of 3% is similar to the previously reported rate of 5% in borderline ovarian tumors 38. In endometrial carcinoma, PIK3CA mutations occur more frequently in KRAS-mutant samples (7/18, 39%; p = 0.06) than in KRAS wild type (17/90, 19%) tumors 39. Consistent with these results, the only case of endometrioid borderline ovarian tumor harboured both KRAS and PIK3CA mutations. In contrast, and in keeping with published studies AKT mutations were not identified in borderline ovarian tumors 40.
The value of SNPs, the most common form of genetic variation, as biomarkers in cancer for risk and prognosis is well established. There may be quantitative variation of transcript levels associated with distinct alleles or haplotypes found in promoters and coding regions of genes. These changes in expression due to allelic variation are often associated with additional genomic or transcript modifications such as DNA methylation or RNA editing. The Sequenom MassArray platform is a rapid, high throughput platform that has been extensively used for SNP detection 41, 42.
The physiological role of vascular endothelial growth factor (VEGF) in angiogenesis, and the activity of anti-VEGF agents such as bevacizumab in ovarian cancer, makes it an important target for evaluation in genetic association studies 43. Ovarian cancer patients with the VEGF C+936T polymorphism C/T genotype have a longer median PFS of 11.8 months, compared with those with the C/C and T/T genotype, with median PFS of 5.5 and 3.2 months, respectively 44. In our study VEGF polymorphisms were the most common genetic variation detected, being present in 96 % of tumors. Polymoprhisms were found in 4 loci of the gene and 69% of the tumors had polymorphisms in 2 or more loci.
In this study we detected polymorphisms and allelic imbalance in one of the members of the PHLPP gene family. The two members of this recently discovered family, PHLPP1 and PHLPP2, control the degree of agonist-evoked signalling by Akt and the cellular levels of PKC 45,46. Brognard et al identified a T-C SNP at position 3047 of PHLPP2, with a population frequency of 30%, which results in an amino acid change from Leucine to Serine at codon 1016 in the PP2C phosphatase domain with a reduction in phosphatase activity, thus driving constitutive phosphorylation of Akt 47.
In this study we detected this PHLPP2 polymorphism in 27% of tumors with a significant correlation with mucinous tumors as compared to serous. Pyrosequencing technology provides qualitative sequencing data simultaneously with quantitative allele dose information (allele quantification). Using pyrosequencing we confirmed the presence of the PHLPP2 polymorphisms detected on the Sequenom platform, with loss of heterozygosity in one case. The association between PHLPP2 polymorphism and mucinous borderline ovarian tumors to our knowledge has not previously been reported, and may imply a role for the activation of AKT/PKC pathways in the genesis of this phenotype.
PDGFRA_V824L (het) was detected in 3 tumors (6%). CE-SSCA and Sanger sequencing analysis of these samples failed to confirm the PDGFRA_V824L mutation, instead, 2/3 cases showed a synonymous polymorphism very near to that genomic position (c.2472C>T; p.V834V). In most primer designs, care in taken to avoid annealing positions that overlap known polymorphic sites; however, if secondary structure interferes with primer design on the opposite strand, this is not always possible. Sequenom also identified EGFR L858R mutations in 5 cases. However, these mutations were not validated on Sanger sequencing. The discordance can be due to the increased sensitivity of the mass-spec based sequencing; these mutations were detected at an allelic frequency of 15% which is below the detection level of Sanger sequencing.
Highly accurate MALDI-TOF-based detection provides unparalleled specificity and sensitivity for studies of genetic variation including somatic mutation detection in heterogeneous samples. The limitations of this platform is that only “hotspot” mutations are detectable. Although the Sequenom MassArray system is highly sensitive and accurate, prior to use in clinical practice, all mutations should be confirmed using an approved CLIA or equivalent assay in a laboratory medicine facility. Indeed, the EGFR and PDGFRA mutations detected by MALDI-TOF could not be confirmed by a validated clinical method based on CE-SSCA and Sanger sequencing. In the case of PDGFRA mutations, this discordance is likely due to the presence of a known polymorphism in the area adjacent to the mutation hotspot for which the test is designed in 2 out of 3 cases. Although our study was carried out on frozen tissue, these mutations and polymorphisms can in principle be detected on analysing DNA extracted from formalin fixed paraffin embedded tissue, and hence can be used in routine practice. However, the degraded nature of some DNA samples derived from formalin fixed paraffin embedded tissue, can cause artefacts in the analysis.
In summary, this study expands the repertoire of mutations and polymorphisms implicated in the pathogenesis of borderline ovarian tumors. The genes and pathways associated with these mutations and polymorphisms are clinically important with active drug development programmes, as the trials using MEK inhibitors in low grade serous carcinoma, offering the opportunity for the implementation of similar targeted therapy in borderline ovarian tumors, an area of unmet clinical need 48. A larger sample size linked to clinical trials is required to establish that a gene mutation has a significant impact for prediction of response to therapy or prognosis. Future studies to determine if the presence of specific mutations predicts anti-tumor activity of targeted biologic agents in borderline ovarian tumors are warranted.
Supplementary Material
Acknowledgments
The authors are grateful for support from the Imperial College Biomedical Research Centre and Experimental Cancer Medicine Centre grant from Cancer Research UK and the Dept of Health. RA is supported by a Clinician Scientist Fellowship from Cancer Research UK.
Contributor Information
Katherine Stemke-Hale, Department of Systems Biology, MD Anderson, Texas, USA.
Kristy Shipman, Department of Oncology, Hammersmith Hospital, Imperial College London, UK.
Isidora Kitsou-Mylona, Department of Histopathology, Hammersmith Hospital, Imperial College London, UK.
David Gonzalez de Castro, The Institute of Cancer Research and Royal Marsden NHS Foundation Trust, Sutton, UK.
Vicky Hird, Department of Obstetrics and Gynaecology, Queen Charlottes Hospital, Imperial College Healthcare NHS Trust, London, UK.
Robert Brown, Department of Oncology, Hammersmith Hospital, Imperial College London, UK.
James Flanagan, Department of Oncology, Hammersmith Hospital, Imperial College London, UK.
H Hani Gabra, Department of Oncology, Hammersmith Hospital, Imperial College London, UK.
Gordon B. Mills, Department of Systems Biology, MD Anderson, Texas, USA
R Agarwal, Department of Oncology, Hammersmith Hospital, Imperial College London, UK.
Mona El-Bahrawy, Department of Histopathology, Hammersmith Hospital, Imperial College London, UK and Faculty of Medicine, University of Alexandria, Egypt.
References
- 1.Morice P, Uzan C, Fauvet R, et al. Borderline ovarian tumour: pathological diagnostic dilemma and risk factors for invasive or lethal recurrence. Lancet Oncol. 2012;13:e103–15. doi: 10.1016/S1470-2045(11)70288-1. [DOI] [PubMed] [Google Scholar]
- 2.Sherman ME, Berman J, Birrer MJ, et al. Current challenges and opportunities for research on borderline ovarian tumors. Hum Pathol. 2004;35:961–70. doi: 10.1016/j.humpath.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Bell DA, Scully RE. Ovarian serous borderline tumors with stromal microinvasion: a report of 21 cases. Hum Pathol. 1990;21:397–403. doi: 10.1016/0046-8177(90)90201-f. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy AW, Hart WR. Ovarian papillary serous tumors of low malignant potential (serous borderline tumors). A long-term follow-up study, including patients with microinvasion, lymph node metastasis, and transformation to invasive serous carcinoma. Cancer. 1996;78:278–86. doi: 10.1002/(SICI)1097-0142(19960715)78:2<278::AID-CNCR14>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 5.McKenney JK, Balzer BL, Longacre TA. Patterns of stromal invasion in ovarian serous tumors of low malignant potential (borderline tumors): a reevaluation of the concept of stromal microinvasion. Am J Surg Pathol. 2006;30:1209–21. doi: 10.1097/01.pas.0000213299.11649.fa. [DOI] [PubMed] [Google Scholar]
- 6.Seidman JD, Kurman RJ. Ovarian serous borderline tumors: a critical review of the literature with emphasis on prognostic indicators. Hum Pathol. 2000;31:539–57. doi: 10.1053/hp.2000.8048. [DOI] [PubMed] [Google Scholar]
- 7.Longacre TA, McKenney JK, Tazelaar HD, Kempson RL, Hendrickson MR. Ovarian serous tumors of low malignant potential (borderline tumors): outcome-based study of 276 patients with long-term (> or =5-year) follow-up. Am J Surg Pathol. 2005;29:707–23. doi: 10.1097/01.pas.0000164030.82810.db. [DOI] [PubMed] [Google Scholar]
- 8.Romagnolo C, Gadducci A, Sartori E, Zola P, Maggino T. Management of borderline ovarian tumors: results of an Italian multicenter study. Gynecol Oncol. 2006;101:255–60. doi: 10.1016/j.ygyno.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Wong HF, Low JJ, Chua Y, et al. Ovarian tumors of borderline malignancy: a review of 247 patients from 1991 to 2004. Int J Gynecol Cancer. 2007;17:342–9. doi: 10.1111/j.1525-1438.2007.00864.x. [DOI] [PubMed] [Google Scholar]
- 10.Faluyi O, Mackean M, Gourley C, Bryant A, Dickinson HO. Interventions for the treatment of borderline ovarian tumours. Cochrane Database Syst Rev. 2010;9:CD007696. doi: 10.1002/14651858.CD007696.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Micci F, Haugom L, Ahlquist T, et al. Genomic aberrations in borderline ovarian tumors. J Transl Med. 2010;8:21. doi: 10.1186/1479-5876-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shih Ie M, Kurman RJ. Molecular pathogenesis of ovarian borderline tumors: new insights and old challenges. Clin Cancer Res. 2005;11:7273–9. doi: 10.1158/1078-0432.CCR-05-0755. [DOI] [PubMed] [Google Scholar]
- 13.Caduff RF, Svoboda-Newman SM, Ferguson AW, Johnston CM, Frank TS. Comparison of mutations of Ki-RAS and p53 immunoreactivity in borderline and malignant epithelial ovarian tumors. Am J Surg Pathol. 1999;23:323–8. doi: 10.1097/00000478-199903000-00012. [DOI] [PubMed] [Google Scholar]
- 14.Ho CL, Kurman RJ, Dehari R, Wang TL, Shih Ie M. Mutations of BRAF and KRAS precede the development of ovarian serous borderline tumors. Cancer Res. 2004;64:6915–8. doi: 10.1158/0008-5472.CAN-04-2067. [DOI] [PubMed] [Google Scholar]
- 15.Kupryjanczyk J, Bell DA, Dimeo D, et al. p53 gene analysis of ovarian borderline tumors and stage I carcinomas. Hum Pathol. 1995;26:387–92. doi: 10.1016/0046-8177(95)90138-8. [DOI] [PubMed] [Google Scholar]
- 16.Mayr D, Hirschmann A, Lohrs U, Diebold J. KRAS and BRAF mutations in ovarian tumors: a comprehensive study of invasive carcinomas, borderline tumors and extraovarian implants. Gynecol Oncol. 2006;103:883–7. doi: 10.1016/j.ygyno.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 17.Jones S, Wang TL, Kurman RJ, et al. Low-grade serous carcinomas of the ovary contain very few point mutations. The Journal of pathology. 2012;226:413–20. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama K, Nakayama N, Kurman RJ, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–85. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 19.Lakhani SR, Manek S, Penault-Llorca F, et al. Pathology of ovarian cancers in BRCA1 and BRCA2 carriers. Clin Cancer Res. 2004;10:2473–81. doi: 10.1158/1078-0432.ccr-1029-3. [DOI] [PubMed] [Google Scholar]
- 20.Steffensen KD, Waldstrom M, Olsen DA, et al. Mutant epidermal growth factor receptor in benign, borderline, and malignant ovarian tumors. Clin Cancer Res. 2008;14:3278–82. doi: 10.1158/1078-0432.CCR-07-4171. [DOI] [PubMed] [Google Scholar]
- 21.Oliva E, Sarrio D, Brachtel EF, et al. High frequency of beta-catenin mutations in borderline endometrioid tumours of the ovary. J Pathol. 2006;208:708–13. doi: 10.1002/path.1923. [DOI] [PubMed] [Google Scholar]
- 22.Mayr D, Kanitz V, Amann G, et al. HER-2/neu gene amplification in ovarian tumours: a comprehensive immunohistochemical and FISH analysis on tissue microarrays. Histopathology. 2006;48:149–56. doi: 10.1111/j.1365-2559.2005.02306.x. [DOI] [PubMed] [Google Scholar]
- 23.Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–5. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 24.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68:6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli D, Gavin PG, Taniyama Y, et al. A rapid, sensitive, reproducible and cost-effective method for mutation profiling of colon cancer and metastatic lymph nodes. BMC Cancer. 2010;10:101. doi: 10.1186/1471-2407-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cuatrecasas M, Erill N, Musulen E, et al. K-ras mutations in nonmucinous ovarian epithelial tumors: a molecular analysis and clinicopathologic study of 144 patients. Cancer. 1998;82:1088–95. [PubMed] [Google Scholar]
- 27.Diebold J, Seemuller F, Lohrs U. K-RAS mutations in ovarian and extraovarian lesions of serous tumors of borderline malignancy. Lab Invest. 2003;83:251–8. doi: 10.1097/01.lab.0000056994.81259.32. [DOI] [PubMed] [Google Scholar]
- 28.Giordano G, Azzoni C, D’Adda T, et al. Human papilloma virus (HPV) status, p16INK4a, and p53 overexpression in epithelial malignant and borderline ovarian neoplasms. Pathol Res Pract. 2008;204:163–74. doi: 10.1016/j.prp.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Mok SC, Bell DA, Knapp RC, et al. Mutation of K-ras protooncogene in human ovarian epithelial tumors of borderline malignancy. Cancer Res. 1993;53:1489–92. [PubMed] [Google Scholar]
- 30.Sieben NL, Macropoulos P, Roemen GM, et al. In ovarian neoplasms, BRAF, but not KRAS, mutations are restricted to low-grade serous tumours. J Pathol. 2004;202:336–40. doi: 10.1002/path.1521. [DOI] [PubMed] [Google Scholar]
- 31.Singer G, Oldt R, 3rd, Cohen Y, et al. Mutations in BRAF and KRAS characterize the development of low-grade ovarian serous carcinoma. J Natl Cancer Inst. 2003;95:484–6. doi: 10.1093/jnci/95.6.484. [DOI] [PubMed] [Google Scholar]
- 32.McCubrey JA, Steelman LS, Abrams SL, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzyme Regul. 2006;46:249–79. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 33.Hayes MP, Wang H, Espinal-Witter R, et al. PIK3CA and PTEN mutations in uterine endometrioid carcinoma and complex atypical hyperplasia. Clin Cancer Res. 2006;12:5932–5. doi: 10.1158/1078-0432.CCR-06-1375. [DOI] [PubMed] [Google Scholar]
- 34.Campbell IG, Russell SE, Choong DY, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 35.Catasus L, Gallardo A, Cuatrecasas M, Prat J. PIK3CA mutations in the kinase domain (exon 20) of uterine endometrial adenocarcinomas are associated with adverse prognostic parameters. Modern Pathol. 2008;21:131–9. doi: 10.1038/modpathol.3800992. [DOI] [PubMed] [Google Scholar]
- 36.Catasus L, Gallardo A, Cuatrecasas M, Prat J. Concomitant PI3K-AKT and p53 alterations in endometrial carcinomas are associated with poor prognosis. Modern Pathol. 2009;22:522–9. doi: 10.1038/modpathol.2009.5. [DOI] [PubMed] [Google Scholar]
- 37.Mori N, Kyo S, Sakaguchi J, et al. Concomitant activation of AKT with extracellular-regulated kinase 1/2 occurs independently of PTEN or PIK3CA mutations in endometrial cancer and may be associated with favorable prognosiss. Cancer science. 2007;98:1881–8. doi: 10.1111/j.1349-7006.2007.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakayama K, Nakayama N, Kurman RJ, et al. Sequence mutations and amplification of PIK3CA and AKT2 genes in purified ovarian serous neoplasms. Cancer Biol Ther. 2006;5:779–85. doi: 10.4161/cbt.5.7.2751. [DOI] [PubMed] [Google Scholar]
- 39.Konstantinova D, Kaneva R, Dimitrov R, et al. Rare mutations in the PIK3CA gene contribute to aggressive endometrial cancer. DNA Cell Biol. 2010;29:65–70. doi: 10.1089/dna.2009.0939. [DOI] [PubMed] [Google Scholar]
- 40.Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer. 1995;64:280–5. doi: 10.1002/ijc.2910640412. [DOI] [PubMed] [Google Scholar]
- 41.Jurinke C, Denissenko MF, Oeth P, et al. A single nucleotide polymorphism based approach for the identification and characterization of gene expression modulation using MassARRAY. Mut Res. 2005;573:83–95. doi: 10.1016/j.mrfmmm.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Tang K, Oeth P, Kammerer S, et al. Mining disease susceptibility genes through SNP analyses and expression profiling using MALDI-TOF mass spectrometry. J Proteome Res. 2004;3:218–27. doi: 10.1021/pr034080s. [DOI] [PubMed] [Google Scholar]
- 43.Perren TJ, Swart AM, Pfisterer J, et al. A phase 3 trial of bevacizumab in ovarian cancer. NEJM. 2011;365:2484–96. doi: 10.1056/NEJMoa1103799. [DOI] [PubMed] [Google Scholar]
- 44.Schultheis AM, Lurje G, Rhodes KE, et al. Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clin Cancer Res. 2008;14:7554–63. doi: 10.1158/1078-0432.CCR-08-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brognard J, Newton AC. PHLiPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab. 2008;19:223–30. doi: 10.1016/j.tem.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem. 2008;283:6300–11. doi: 10.1074/jbc.M707319200. [DOI] [PubMed] [Google Scholar]
- 47.Brognard J, Niederst M, Reyes G, Warfel N, Newton AC. Common polymorphism in the phosphatase PHLPP2 results in reduced regulation of Akt and protein kinase C. J Biol Chem. 2009;284:15215–23. doi: 10.1074/jbc.M901468200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matulonis UA, Hirsch M, Palescandolo E, et al. High throughput interrogation of somatic mutations in high grade serous cancer of the ovary. PLoS One. 2011;6:e24433. doi: 10.1371/journal.pone.0024433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.