Abstract
TRAF4 is an adapter protein overexpressed in certain cancers but its contributions to tumorigenesis are unclear. In lung cancer cells and primary lung tumors, we found that TRAF4 is overexpressed. RNAi-mediated attenuation of TRAF4 expression blunted the malignant phenotype in this setting, exerting inhibitory effects on cell proliferation, anchorage-independent growth and tumor development in a xenograft mouse model. Unexpectedly, we discovered that TRAF4, but not Skp2, was required for activation of the pivotal cell survival kinase Akt through ubiquitination. Furthermore, TRAF4 attenuation impaired glucose metabolism by inhibiting expression of Glut1 and HK2 mediated by the Akt pathway. Overall, our work suggests that TRAF4 offers a candidate molecular target for lung cancer prevention and therapy.
Keywords: lung cancer, TRAF4, Akt, ubiquitination
Introduction
Lung cancer is the leading cause of cancer-related death in the world (1). Although advanced molecular biology techniques have greatly accelerated the understanding of the biological mechanisms that underlie lung cancer development, the 5-year survival rate is 15%, which has hardly improved from the 13% reported 35 years ago (2, 3). A major challenge in treating lung cancer is to identify novel therapeutic targets that can complement current chemotherapy (4).
Akt signaling plays a major role in tumorigenesis by regulating cell proliferation, cell cycle, cell survival and metabolism (5–7). Akt is constitutively activated in lung cancer, where it can regulate cancer cell growth and survival and confer resistance to chemotherapy or radiation treatment (8, 9). It is activated by various stimuli, including insulin and EGF, and Akt phosphorylation at Thr308 by PDK1 and at Ser473 by mTORC2 is required for full activation (10). Studies detailing Akt activation indicate that Lys63-mediated ubiquitination is essential for Akt activity (11, 12). Notably, different stimuli utilize distinct E3 ligases for Akt ubiquitination. For example, TRAF6 or Skp2 mediates Akt activation through ubiquitination in IGF-1 or ErbB receptor signaling, respectively (13, 14). Additionally, Akt hyperactivation as a consequence of Skp2 overexpression drives abnormal glycolysis in breast cancer, and Skp2 is a marker for poor prognosis in HER2-positive patients (14).
TRAF4 is an atypical TRAF family member. Unlike other TRAFs, which are not relevant to the genetic phenotype, in vivo evidence strongly suggests that TRAF4 is involved in embryogenesis (15) and central nervous system myelin homeostasis (16). TRAF4 is widely and highly expressed during development and TRAF4 deficiency is embryonic lethal in approximately one third of the homozygote mutants in pure 129/SvJ genetic background mice (17). Furthermore, TRAF4 deficiency results in severe developmental changes in the respiratory system, axial skeleton and nervous system (17, 18).
TRAF4 was first identified by differential screening of human metastatic lymph nodes from a breast cancer cDNA library (19). The amplification and overexpression of TRAF4 suggested that it was involved in the initiation and progression of primary breast cancers and metastases (19–21). Although TRAF4 is overexpressed in various human malignancies (22), the mechanism regarding TRAF4’s role in tumorigenesis remains unclear.
In the present study, we found that TRAF4 plays an important role in lung tumorigenesis. Knockdown of TRAF4 dramatically attenuated the malignant phenotype in lung cancer, including proliferation, anchorage-independent growth and tumor formation ability in nude mice. Furthermore, we demonstrated that TRAF4, but not Skp2, is required for EGF-induced Akt activation through its ubiquitination in lung cancer. TRAF4 deficiency markedly impaired the activity of Akt signaling and Akt-mediated lung cancer glycolysis.
Materials and Methods
Reagents and Antibodies
Tris, NaCl, and SDS for molecular biology and buffer preparation were purchased from Sigma (St. Louis, MO). Cell culture media and supplements were from Invitrogen (Grand Island, NY). Antibodies against phosphorylated Akt (Ser473 or Thr308), Akt1, panAkt, phosphorylated S6 (Ser235/236), S6, HK2 and ubiquitin (P4D1) were obtained from Cell Signaling Technology, Inc. (Beverly, MA). Antibodies against TRAF4, TRAF6, β-actin, α-tubulin, Flag-tag, Myc-tag and HA-tag were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-Flag-HRP was from Sigma. The Glut1 antibody was obtained from Millipore (Billerica, MA) and anti-E-cadherin was from BD Biosciences (San Jose, CA).
Construction of Expression Vectors
Expression constructs, including HA-Akt1, Myc-Akt1, and HA-Lys-63-ubiquitin, pCDNA3.0- Flag-TRAF6-WT, pCDNA3.0-TRAF4-WT, pCDNA3.0-TRAF4-DM-Ring and pCDNA3.0-TRAF4-DM-TRAF were from Addgene (Cambridge, MA)(23). Flag-TRAF4-WT, Flag-TRAF4-DM-Ring and Flag-TRAF4-DM-TRAF were subcloned into the pBabe vector by Xba1 and BamH1 from pCDNA3.0-TRAF4-WT, pCDNA3.0-TRAF4-DM-Ring and pCDNA3.0-TRAF4-DM-TRAF. Lentivirus plasmids containing pLKO.1-shTRAF4 (#1, TRCN0000034242; #2, TRCN0000034243), pLKO.1-shTRAF6 (#1 TRCN0000007348, #2 TRCN0000007352), pLKO.1-shAkt1 (#1, TRCN0000010162, #2, TRCN0000010174) and pLKO.1-shskp2 (#1, TRCN0000007530; #2, TRCN0000007531) were from Thermo Scientific (Huntsville, AL).
Cell Culture and Transfection
Cells from American Type Culture Collection (ATCC, Manassas, VA) were cultured at 37 °C in a humidified incubator with 5% CO2 according to ATCC protocols. Cells were cytogenetically tested and authenticated before being frozen. Each vial of frozen cells was thawed and maintained for 2 months (10 passages). 293T and HaCaT cells were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% antibiotics. Human lung cancer cells, including H460, H1650, H1975, H1299, H520, HCC827, and H358, were grown in RPMI-1640 medium supplemented with 10% FBS and antibiotics. A549 human lung cancer cells were cultured with F-12K medium containing 10% FBS and antibiotics. MRC5 human normal lung fibroblasts were cultured with Eagle’s Minimum Essential Medium (MEM) supplemented with 10% FBS and antibiotics. NIH-3T3 mouse embryo fibroblasts were cultured with DMEM containing 10% bovine calf serum and antibiotics. For transfection experiments, the jetPEI (Qbiogene, Inc., Montreal, Canada) transfection reagent was used following the manufacturer’s instructions. The cells were cultured for 36–48 h and proteins extracted for analysis.
Lentiviral Infection
To generate knockdown TRAF4, TRAF6, Akt1 and Skp2 cells, pLKO.1-shTRAF, pLKO.1-shTRAF6, pLKO.1-shAkt1 and pLKO.1-shskp2 lentivirus plasmids, were co-transfected into 293T cells with PSPAX2 and PMD2-G. Viral supernatant fractions were collected at 48 h after transfection and filtered through a 0.45-μm filter followed by infection into cells together with 10 μg/ml polybrene. At 16 h after infection, the medium was replaced with medium containing puromycin. To generate stable HaCaT cells expressing TRAF4-WT (Flag-TRAF4), TRAF4-Ring domain deletion (Flag-TRAF4-DM-Ring) and TRAF4-TRAF domain deletion (Flag-TRAF4-DM-TRAF), the pBabe-mock, pBabe-TRAF4-WT, pBabe-TRAF4-DM-Ring, or pBabe-TRAF4-DM-TRAF plasmid was co-transfected into 293T cells together with pCI-VSVG and pCI-GPZ. Viral supernatant fractions were collected at 48 h after transfection and filtered through a 0.45-μm filter followed by infection into HaCaT cells together with 10 μg/ml polybrene. At 16 h after infection, the medium was replaced with medium containing 1 μg/ml puromycin and cells incubated for another 6 days.
Immunoblotting and Immunoprecipitation
Protein samples were extracted with Nonidet P-40 buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 0.5% Nonidet P-40, and protease inhibitor mixture). For immunoblotting, proteins (30 μg) were detected with specific antibodies and an alkaline phosphatase (AP)-conjugated secondary antibody. Proteins were visualized by chemiluminescence (Amersham Biosciences, Piscataway, NJ). For immunoprecipitation (IP), extracts were precleared with 30 μl (50% slurry) agarose A/G beads by rocking for 2 h at 4 °C. Beads were removed and 30 μl (50% slurry) of fresh agarose A/G beads and appropriate antibodies were added to the precleared lysates overnight at 4 °C. The beads were washed, mixed with 6×SDS sample buffer, boiled, and then resolved by 8% SDS-PAGE. Proteins were detected with specific antibodies and an AP-conjugated secondary antibody. For immunoprecipitation under denaturing conditions, proteins were extracted using regular IP buffer plus 1% SDS and heated at 95 °C for 5 min. The samples were diluted 1:10 in regular IP buffer before IP. Beads were washed, mixed with 6×SDS sample buffer, boiled, and then resolved by 8% SDS-PAGE. Proteins were visualized by immunoblotting.
Membrane Fractionation
A549-shMock and A549-shTRAF4 cells were starved 24 h in 0.1% FBS/DMEM. After EGF (50 ng/ml) treatment, membrane and cytosolic fractions were prepared (ProteoExtract kit; Calbiochem, San Diego, CA) according to the manufacturer’s instructions.
MTS Assay
Stable HaCaT and lung cancer cells were seeded (1×103 /well/100 μl) into 96-well plates, and proliferation was assessed by MTS assay (Promega, Madison, WI) according to instructions provided.
Immunofluorescence Staining
Cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100 for 30 min. Fixed cells were incubated with an Akt rabbit monoclonal antibody (Cell Signaling Biotechnology, Inc., Danvers, MA) overnight at 4 °C followed by incubation with green fluorescent Alexa Fluor 488 dye-labeled anti-rabbit IgG (Invitrogen, Grand Island, NY). Nuclei were stained with DAPI. Samples were viewed with a confocal fluorescence microscope system (NIKON C1si, NIKON Instruments Co., Melville, NY).
Anchorage-independent Growth
For EGF-induced transformation, cells (8×103/ml/well) were exposed to EGF in 1 ml of 0.3% basal medium Eagle agar containing 10% FBS. The cultures were maintained in a 37 °C, 5% CO2 incubator. For anchorage-independent growth, cells (8×103/ml/well) were seeded into 6-well plates with 0.3% basal medium Eagle agar containing 10 or 20% (H1975) FBS and cultured. Colonies were scored using a microscope and Image-Pro PLUS (v.6) software (Media Cybernetics).
Immunohistochemical Analysis of Tissue Array
A human lung tissue array (LC1005) was purchased from US Biomax, Inc. (Rockville, MD) and included 15 cases of adenocarcinoma, 15 cases of squamous cell carcinoma, 8 cases of large cell carcinoma, 8 cases of lymph node metastasis carcinoma and 38 cases of matched or unmatched adjacent normal tissue, and 8 cases of normal lung tissue from necroscopy. A Vectastain Elite ABC Kit (Vector Laboratories; Burlingame, CA) was used for immunohistochemical staining according to the recommended protocol. Briefly, the slide was baked at 60 °C for 2 h, deparaffinized, and rehydrated. To expose antigens, the slide was unmasked by submersion into boiling sodium citrate buffer (10 mM, pH 6.0) for 10 min, and then treated with 3% H2O2 for 10 min. The slide was blocked with 50% goat serum albumin in 1×PBS in a humidified chamber for 1 h at room temperature and then with a TRAF4 antibody (1:100 dilution in 50% goat serum with PBS) at 4 °C in a humidified chamber overnight. The slide was washed and hybridized with the secondary antibody from Vector Laboratories (Burlingame, CA) (anti-rabbit 1:200) for 1 h at room temperature. Slides were stained using the Vectastain Elite ABC kit. The intensity was estimated by Image-Pro PLUS (v.6) and Image J (NIH) software programs. Statistical analyses were performed using Prism 5.0.
In Vivo Tumor Growth
Athymic nude mice (The Jackson Laboratory; Bar Harbor, ME) were divided into two groups (n = 15) and injected in the right flank with sh-Mock or sh-TRAF4 A549 lung cancer cells (3×106). Tumors were measured by caliper 2x week. All studies were performed following guidelines approved by the University of Minnesota Institutional Animal Care and Use Committee.
Glucose Uptake and Lactate Production Measurement
Cells (5×105) were seeded in 6-well plates and after incubation for 4 h, medium was discarded and cells were incubated in fresh medium for 8 h. Glucose and lactate levels were measured (Automatic Biochemical Analyzer; 7170A, HITACHI, Tokyo, Japan) at the Clinical Biochemical Laboratory of Xiangya Hospital (Changsha, China).
Ubiquitination Assays
In vitro ubiquitination assays were performed as described (14). Briefly, Flag-TRAF4, Flag-TRAF4-DM-Ring, Flag-TRAF4-DM-TRAF and Flag-TRAF6 were expressed in 293T cells, immunoprecipitated with anti-Flag, and eluted from Protein A/G beads using Flag peptides according to manufacturers’ standard procedures. Flag-TRAF4 or TRAF4 mutants, Flag-TRAF4-DM-Ring and Flag-TRAF4-DM-TRAF or Flag-TRAF6 protein along with Akt1 (1 μg) were incubated for 3 h at 37 °C in reaction buffer (20 mM HEPES [pH 7.4], 10 mM MgCl2, 1 mM DTT, 59 mM ubiquitin, 50 nM E1, 850 nM of Ubc13 or UbcH5A, 1 mM ATP, 30 mM creatine phosphate, and 1 U of creatine kinase). After incubation, protein mixtures were diluted in RIPA buffer and supernatant fractions were pre-cleared with Protein A/G beads for 2 h, and immunoprecipitated overnight with anti-Flag or anti-Akt, after which Protein A/G beads were added for an additional 2 h. Beads were centrifuged and washed 4x with E1A Buffer. Proteins were eluted in 6×SDS sample buffer and subjected to immunoblotting.
Statistical analysis
All quantitative data are expressed as mean values ± S.D of at least 3 independent experiments. Significant differences were determined by Student’s t test or by one-way ANOVA. A probability value of p < 0.05 was used as the criterion for statistical significance.
Results
TRAF4 is overexpressed in human lung cancer
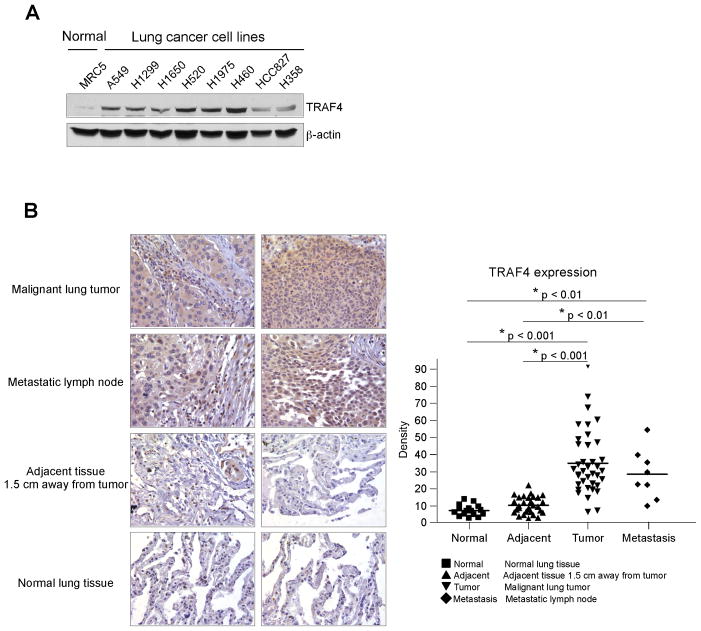
Clinical RNA database sequence analysis of 40 lung cancer patients revealed overexpression of TRAF4 (data not shown). To verify the high expression of TRAF4 in lung cancer, we determined the protein level of TRAF4 in different lung cancer cell lines and in a human lung cancer tissue array. Results showed that, compared with normal MRC5 lung cells or normal tissue, TRAF4 is overexpressed in lung cancer cells (Fig. 1A) and in lung cancer tissues (Fig. 1B). These results indicated that TRAF4 might be a critical molecule in lung cancer development.
Figure 1.
TRAF4 is overexpressed in lung cancer cells. A, TRAF4 is highly expressed in lung cancer cells. Western blot analysis was performed to examine TRAF4 expression in several lung cancer cell lines and normal MRC5 lung cells. B, TRAF4 is highly expressed in lung cancer tissues. Immunohistochemical staining was performed on a lung cancer tissue array using a TRAF4 antibody. The intensity was evaluated using Image-Pro PLUS (v.6) and Image J (NIH) computer software. Statistical analyses were performed using Prism 5.0. The density of each individual sample is shown (right). The asterisk (*) indicates a significant difference between groups as indicated.
TRAF4 has a critical role in cell transformation
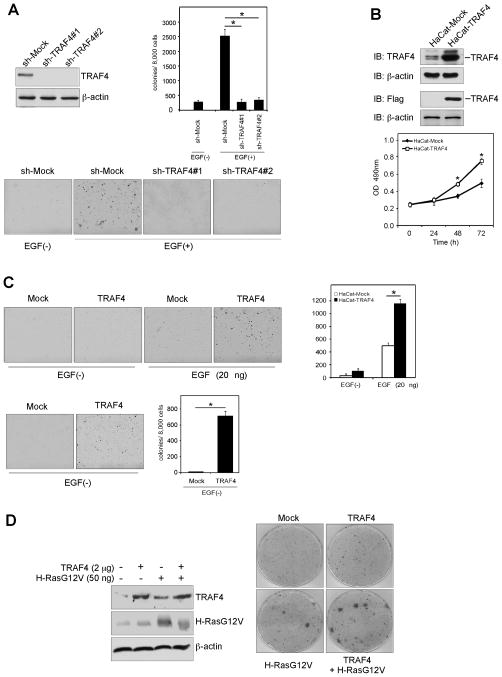
To investigate TRAF4 function in lung cancer, we generated TRAF4 knockdown stable HaCaT cells using two different sh-TRAF4 sequences (Fig. 2A, left top). An anchorage- independent growth assay was performed to assess the effect of TRAF4 on cell transformation. The result indicated that EGF-induced colony formation was attenuated in TRAF4 knockdown cells (Fig. 2A, right, lower). Furthermore, overexpression of TRAF4 promoted anchorage-dependent growth (Fig. 2B, lower) and increased anchorage-independent growth with EGF treatment (Fig. 2C, upper). Usually, without EGF, HaCaT cells cannot form colonies but, unexpectedly, colony formation was observed in HaCaT cells overexpressing TRAF4 even without EGF treatment (Fig. 2C, lower). Furthermore, although TRAF4 alone could not transform 3T3 cells, it dramatically promoted the transformation ability of H-RasG12V (Fig. 2D). These results suggested that TRAF4 is associated with cell transformation.
Figure 2.
TRAF4 plays a critical role in cell transformation. A, knockdown of TRAF4 blocks EGF-induced transformation. Generation of stable knockdown TRAF4 in HaCaT cells (left) was performed and cells exposed to EGF (0, 20 ng/ml) in 0.3% basal medium Eagle agar containing 10% FBS. Cultures were maintained in a 37 °C, 5% CO2 incubator for 15 days, and colonies counted using a microscope and Image-Pro PLUS (v.6). Representative photographs (lower) are shown and the graph (right) shows data from at least 3 independent experiments expressed as means ± S.D. The asterisk (*) indicates a significant (p < 0.05) decrease in colony formation by knockdown cells. B, overexpression of TRAF4 promotes proliferation. An MTS assay was conducted to assess the effect of TRAF4 on cell growth. C, overexpression of TRAF4 increases anchorage-independent growth. Stable overexpression of TRAF4 in HaCaT cells was performed and cells treated with EGF (0, 20 ng/ml) in 3% basal medium Eagle agar containing 10% FBS. The cultures were maintained in a 37 °C, 5% CO2 incubator for 10 (upper) or 30 days (lower), and colonies counted. Data are shown as means ± S.D of at least 3 independent experiments. The asterisk (*) indicates a significant (p < 0.05) increase in colony formation compared to Mock-transfected cells. D, TRAF4 facilitates H-RasG12V-induced cell transformation. NIH-3T3 cells were seeded in 10-cm dishes and the constructs were transfected as indicated. At 100% confluence, cells were cultured in medium containing 3% bovine calf serum. The medium was changed every 2–3 days for 3 weeks and foci were stained with Geimsa stain.
Knockdown TRAF4 in human lung cancer cells reduces tumorigenic properties
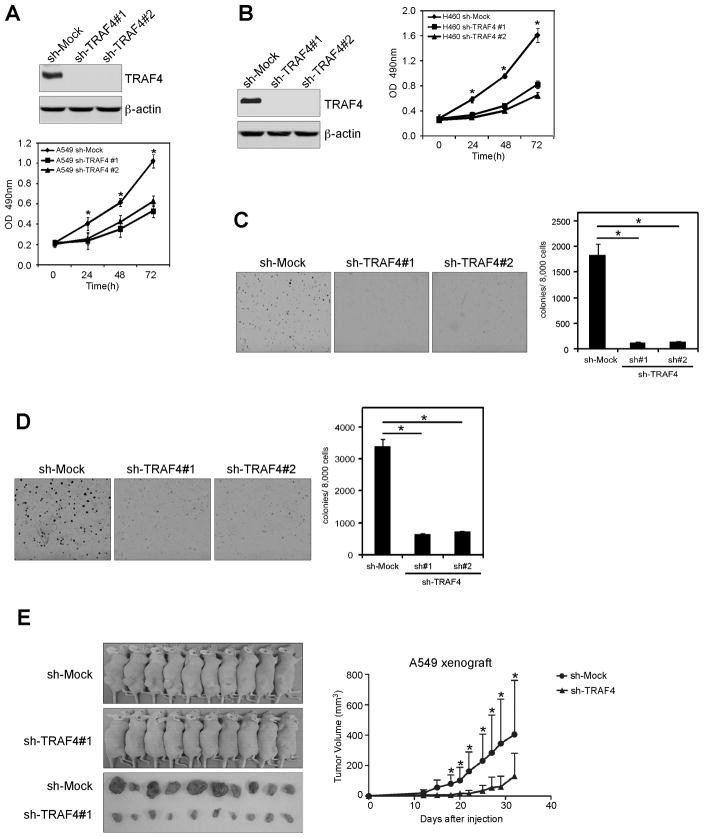
Based on our observations, we hypothesized that TRAF4 may affect the tumorigenic properties in lung cancer. Thus, we generated stable knockdown TRAF4 A549 (Fig. 3A, upper) and H460 (Fig. 3B, left) cells and measured anchorage-dependent growth. Results showed that knockdown TRAF4 inhibited proliferation of A549 (Fig. 3A, lower) or H460 cells (Fig. 3B, right). Knocking down TRAF4 expression also suppressed anchorage-independent growth of A549 (Fig. 3C) and H460 (Fig. 3D) lung cancer cells. We extended our study to examine an additional 5 lung cancer cell lines, including H520, HCC827, H1650, H1299 and H1975 cells. Results indicated that knockdown TRAF4 inhibited proliferation and anchorage-independent growth of all lung cancer cell lines examined (Fig. S1). Furthermore, we found that xenograft growth of A549 cells in athymic nude mice was also attenuated after TRAF4 knockdown (Fig. 3E). These results suggest that blocking TRAF4 expression significantly reduces the tumorigenic properties of lung cancer cells.
Figure 3.
Knocking down TRAF4 expression in human lung cancer cells reduces their tumorigenic properties. A and B, knockdown TRAF4 attenuates A549 anchorage-dependent and -independent growth. Stable knockdown of TRAF4 in A549 cells was performed. A, MTS and B, soft agar assays were performed. C and D, knockdown TRAF4 attenuates anchorage-dependent and anchorage-independent growth of H460 cells. Stable knockdown of TRAF4 in H460 cells was performed as for (A). C, MTS and D, soft agar assays were performed. E, knockdown of TRAF4 reduces tumorigenic properties of A549 human lung cancer cells. Representative photographs of mice from each group injected with A549-shMock or A549-shTRAF4 cells. Tumors dissected from each group are shown (lower). Tumor growth curve of mice injected with A549-shMock or A549-shTRAF4 cells is shown (right). Data are represented as means ± S.D of each group. The asterisk (*) indicates a significant difference (p < 0.05) in tumor volume (mm3).
TRAF4 is required for Akt activation and recruitment to the membrane
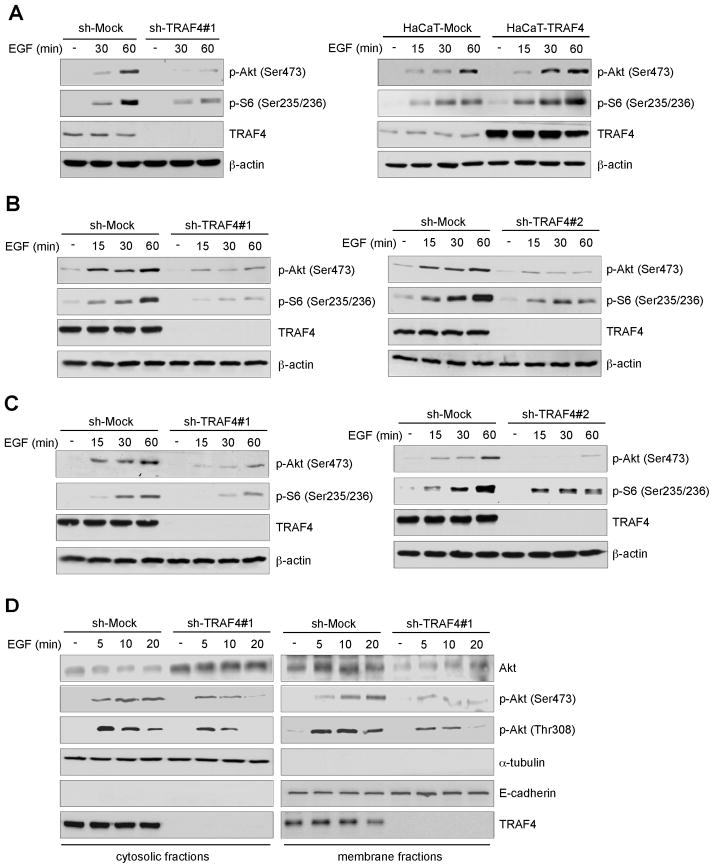
Unlike other TRAF family members, TRAF4 has not yet been clearly associated with a specific receptor or signaling pathway. To examine the function of TRAF4 in EGF signaling, we assessed the activation of the EGF pathway in knockdown TRAF4 HaCaT cells treated with EGF. Results showed that knocking down TRAF4 expression impaired Akt phosphorylation and its downstream kinase S6 (Fig. 4A, left) and overexpression of TRAF4 promoted phosphorylation of Akt and S6 (Fig. 4A, right) with EGF treatment. We also investigated the effect of TRAF4 on EGF signaling in lung cancer cells. Indeed, TRAF4 knockdown strikingly inhibited EGF-induced Akt signaling in A549 (Fig. 4B) and H460 (Fig. 4C) lung cancer cells. Recruitment of Akt to the cell membrane is critical for Akt activation. We next investigated whether knockdown of TRAF4 affected Akt membrane translocation. Results showed that TRAF4 is not only found in the cytoplasm, but is also expressed on the cell membrane. In shMock cells, EGF treatment induced Akt membrane translocation and activation; and phosphorylation of Akt on both Thr308 and Ser473 was dramatically up-regulated after EGF stimulation. However, knockdown of TRAF4 inhibited EGF-induced Akt membrane recruitment, which corresponded with a decrease in phosphorylation of Akt on both Thr308 and Ser473 (Fig. 4D). Immunofluorescence staining results showed that Akt membrane translocation induced by EGF stimulation was inhibited in TRAF4 knockdown cells (Fig. S2).
Figure 4.
TRAF4 regulates EGF-induced Akt activation. A, TRAF4 regulates Akt activation in HaCaT cells. A, TRAF4-knockdown stable cells (left) and TRAF4-overexpressing stable cells (right) were starved for 36 h and then treated with EGF (0, 50 ng/ml) for various times and Western blotting was performed. B and C, TRAF4 regulates Akt activation in A549 and H460 lung cancer cells. B, A549-shMock and A549-shTRAF4 cells or C, H460-shMock and H460-shTRAF4 cells were starved overnight and then treated with EGF (0, 50 ng/ml) for various times and harvested for Western blotting. D, TRAF4 regulates recruitment of Akt to the membrane. A549-shMock and A549-shTRAF4 cells were starved overnight and treated with EGF (0, 50 ng/ml) for various times. Membrane and cytosolic fractions were isolated and analyzed by Western blot.
Because E3 ligase TRAF6 and Skp2 are important for growth factors IGF-1 and EGF-induced Akt ubiquitination and activation (13, 14), respectively, we further determined whether EGF-induced Akt activation and membrane recruitment depend on TRAF6 in lung cancer. As was expected, EGF-induced Akt activation in A549 (Fig. S3A) and H460 (Fig. S3B) lung cancer cells as well as membrane recruitment (Fig. S3C) were not significantly different between shMock and shTRAF6 cells, which is consistent with a previous study reporting that TRAF6 is unnecessary for EGF-induced Akt activation (14). Next, we wanted to know whether Skp2 or TRAF4 is most important for EGF-induced Akt activation in lung cancer. Thus, we compared the level of TRAF4, Skp2 and Akt phosphorylation in 8 lung cancer cell lines and one normal lung cell line. Unexpectedly, unlike TRAF4 that is highly expressed in lung cancer cells, compared to normal lung cells, Skp2 exhibited a relatively similar, low expression level in all of the lung cancer cell lines examined. Additionally, the TRAF4 high expression was accompanied by Akt hyperphosphorylation (Fig. S4A). Moreover, knockdown of Skp2 in lung cancer cells had no effect on EGF-induced Akt activation (Fig. S4B), which suggested that TRAF4, but not Skp2, is required for Akt activation in lung cancer.
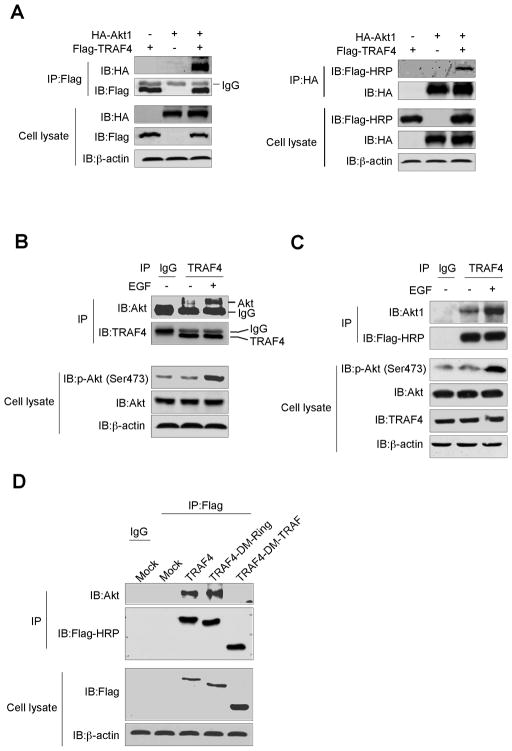
TRAF4 interacts with Akt1
To investigate the mechanism explaining how TRAF4 regulates Akt activity, we first studied the interaction between TRAF4 and Akt1. Flag-TRAF4 and HA-Akt1 were co-transfected into 293T cells and immunoprecipitated with anti-Flag or anti-HA to detect HA-Akt1 or Flag-TRAF4 in the IP complex (Fig. 5A). In addition, the endogenous Akt1 and TRAF4 interaction was observed in A549 (Fig. 5B) and HaCaT cells (Fig. 5C) and was at least partially dependent on EGF treatment. These results demonstrated that TRAF4 is a novel protein-binding partner with Akt1. To identify the domain responsible for mediating the interaction between TRAF4 and Akt, we constructed Flag-TRAF4-Ring domain deletion (Flag-TRAF4-DM-Ring) and Flag-TRAF4-TRAFc domain deletion (Flag-TRAF4-DM-TRAF) mutants (Fig. S5A) and generated HaCaT cells stably overexpressing TRAF4-WT or TRAF4 mutants (Fig. S5B). Flag-TRAF4 was immunoprecipitated with anti-Flag and the endogenous level of Akt was detected in the IP complex. The result showed that the TRAFc domain mediated the interaction between TRAF4 and Akt1 (Fig. 5D). We also showed that the TRAF4 Ring domain or TRAFc domain deletion mutant lost the ability to promote cell transformation (Fig. S5C, D). Collectively, our data demonstrated that TRAF4 binds with Akt1 through the TRAFc domain and its physiological function relies on the Ring and TRAFc domains.
Figure 5.
TRAF4 binds to Akt1. A, TRAF4 interacts with Akt1. Flag-TRAF4 and HA-Akt1 were co-transfected into 239T cells. At 36 h after transfection, cell lysates were harvested and Flag-TRAF4 or HA-Akt1 was immunoprecipitated with anti-Flag or anti-HA. Western blotting was performed. B, TRAF4 interacts with endogenous Akt1. A549 cells were starved and treated with EGF (0, 50 ng/ml) for 10 min. Cell extracts were immunoprecipitated with anti-TRAF4 or control IgG. The immunoprecipitated complex was detected by Western blotting with anti-Akt. C, TRAF4 interacts with Akt1 in HaCaT cells stably overexpressing TRAF4. Cells were starved and treated with EGF (0, 50 ng/ml) for 10 min. Cell extracts were immunoprecipitated with anti-Flag or control IgG. The immunoprecipitated complex was detected by Western blotting with an Akt1 or Flag-HRP antibody. D, the TRAF domain mediates the interaction between TRAF4 and Akt1. Stable HaCaT cells overexpressing a TRAF4-Ring domain-deletion or a TRAF4-TRAF domain deletion were generated. Lysates from stable cells were immunoprecipitated with anti-Flag or control IgG. The immunoprecipitated complex was detected by Western blotting with an Akt or Flag-HRP antibody.
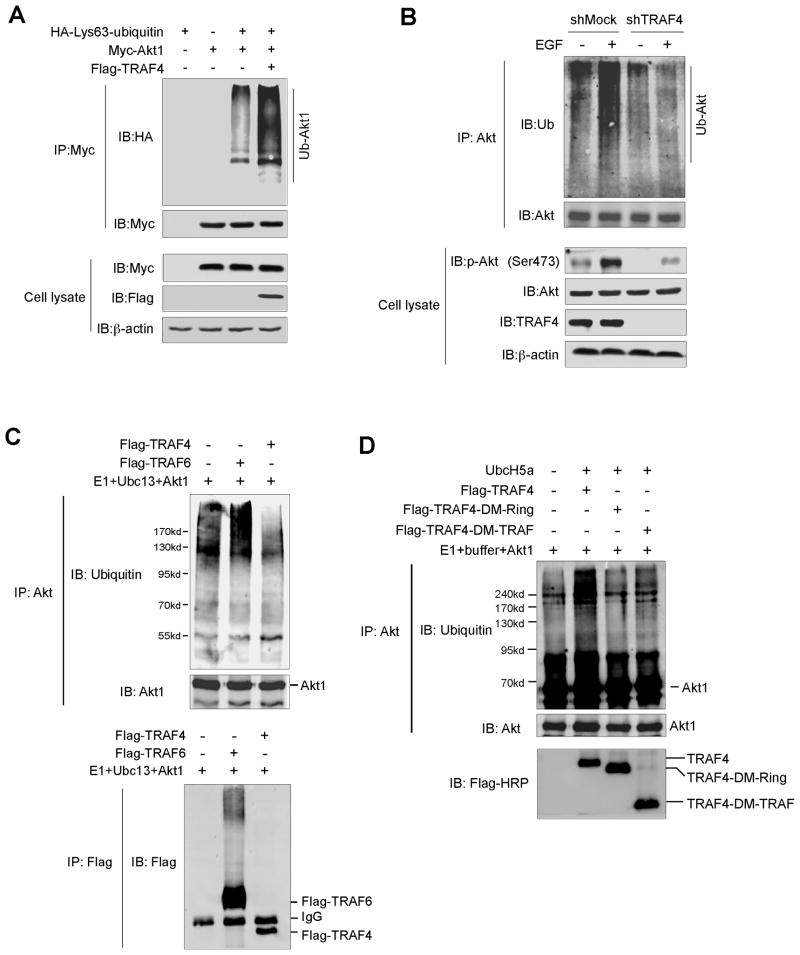
TRAF4 is required for Akt1 Ub-Lys63 ubiquitination
Previous results showed that TRAF4 is required for recruitment of Akt to the cell membrane for its activation. Because TRAF4 interacts with Akt (Fig. 5), we hypothesized that TRAF4 might function as an E3 ligase to mediate Akt activation through ubiquitination. Indeed, TRAF4 overexpression induced Akt1 Ub-Lys63 mediated ubiquitination in 293T cells (Fig. 6A). Notably, we found that knockdown of TRAF4 abrogated EGF-induced Akt ubiquitination in A549 cells (Fig. 6B). Ubc13 is a well-known E2 ubiquitin-conjugating enzyme and is required for E3 ligase activity, including TRAF2 and TRAF6 mediated Ub-Lys63 ubiquitination (24, 25). To investigate whether TRAF4 has a potential E3 ligase activity, an in vitro ubiquitination assay was performed. The result showed that Ubc13 together with TRAF6 induced Akt1 ubiquitination and TRAF6 auto-ubiquitination. However, Ubc13 is not available for TRAF4-induced Akt1 ubiquitination (Fig. 6C). Instead, we observed that another E2 ubiquitin-conjugating enzyme, UbcH5a (26, 27), is suitable for TRAF4-induced Akt1 ubiquitination in vitro (Fig. 6D). Importantly, we also noticed that the TRAF4-Ring domain deletion and TRAF4-TRAFc domain deletion failed to induce Akt1 unbiquitination. This is consistent with the observation that the Ring domain in TRAF4 is a functional domain for E3 ligase activity and the TRAFc domain mediates the interaction between TRAF4 and Akt1. We next examined whether knockdown of the putative TRAF4 target Akt1 affects HaCaT cell transformation and lung cancer cell growth in soft agar. As expected, knockdown of Akt1 (Fig. S6A) inhibited HaCaT cell proliferation (Fig. S6B) and EGF-induced neoplastic transformation (Fig. S6C). We also observed a similar impairment in cell proliferation and colony formation ability in various types of human lung cancer cells, including H1650, H1299, A549 and H460 cells (Fig. S7A–D). These results indicate that TRAF4 might possess an E3 ligase activity and is required for Akt1 ubiquitination.
Figure 6.
TRAF4 is required for Lys63-mediated Akt1 polyubiquitination. A, TRAF4 induces Lys63-linked Akt1 polyubiquitination. 293T cells were co-transfected with constructs as indicated. At 48 h after transfection, cells were harvested and then immunoprecipitated with anti-Myc. Ubiquitinated Akt1 was visualized by Western blot using anti-HA. B, TRAF4 affects endogenous Akt ubiquitination upon EGF treatment. A549-shMock and A549-shTRAF4 cells were starved overnight and treated with EGF (0, 50 ng/ml). Cell lysates were harvested and Akt was immunoprecipitated and immunoblotted with anti-ubiquitin to capture polyubiquitinated Akt. C, TRAF6 and Ubc13 mediate Akt1 ubiquitination. The ubiquitination reaction buffer, E1, Ubc13 and TRAF4 or TRAF6 were incubated with Akt1 and polyubiquitinated Akt1, TRAF6 or TRAF4 was detected by Western blotting. D, TRAF4 and UbcH5a mediate Akt1 ubiquitination. The ubiquitination reaction buffer, E1, E2 (UbcH5a) and TRAF4 (WT) or TRAF4 mutants (TRAF4-DM-Ring and TRAF4-DM-TRAF) were incubated with Akt1 and polyubiquitinated Akt1 was detected by Western blotting.
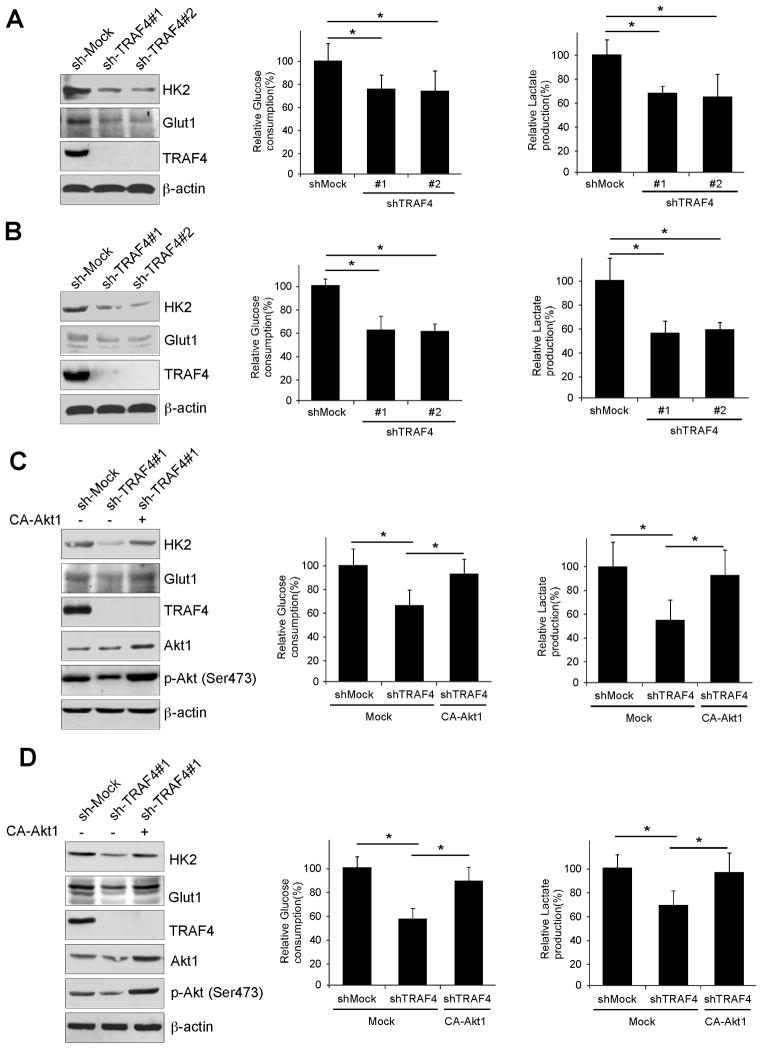
TRAF4 regulates lung cancer cellular glycolysis that is dependent on Akt activity
The Akt signaling pathway is a hub in the regulation of cancer metabolism (28). We speculated that TRAF4 might be involved in the regulation of lung cancer cellular glucose metabolism. Indeed, we observed that knockdown of TRAF4 inhibited the expression of Glut1 and HK2 (Fig. 7A, B, left), and also attenuated glucose uptake (Fig. 7A, B, middle) and lactate production (Fig. 7A, B, right) in both A549 and H460 lung cancer cells. The inhibitory effect on glucose metabolism induced by knocking down TRAF4 expression was not a result of differences in cell numbers (Fig. S8A, B). In order to further determine whether the regulation of glycolysis by TRAF4 is dependent on Akt activity, we transfected constitutively activated Akt1 (CA-Akt1) into knockdown TRAF4 stable A549 and H460 cells. CA-Akt1 transfection up-regulated Glut1 and HK2 expression (Fig. 7C, D, left). Moreover, CA-Akt1 rescued the deficient glucose uptake (Fig. 7C, D, middle) and lactate production (Fig. 7C, D, right) in TRAF4 knockdown lung cancer cells. These results suggest that TRAF4 regulates glycolysis in human lung cancer cells, and these biochemical processes are partly mediated by TRAF4 modulation of Akt activation.
Figure 7.
Akt activation is required for TRAF4-regulated glycolysis in human lung cancer. A and B, TRAF4 regulates glycolysis in A549 and H460 lung cancer cells. Western blotting was performed to detect Glut1 and HK2 in sh-Mock and sh-TRAF4 cells (left). The levels of glucose consumption (middle) and lactate production (right) were examined in these cells. C and D, overexpression of constitutively active Akt (CA-Akt1) rescues glycolysis in knockdown TRAF4-A549 (C) and -H460 lung cancer cells (D). The CA-Akt1 plasmid was transfected into TRAF4 stable knockdown lung cancer cells. Western blot analysis was performed to detect Glut1 and HK2 expression (left). Glucose consumption (middle) and lactate production (right) were determined as in (A, B).
Discussion
TRAF4 was initially identified in human breast carcinoma in 1995 (19). Previous studies revealed that TRAF4 is engaged in several signaling pathways. For instance, TRAF4 binds with MEKK4 to regulate JNKs activation (29) and also interacts with NOD2 to inhibit innate immune responses (30, 31). TRAF4 plays a role in restricting IL-17 signaling and Th17-mediated disease (32). TRAF4 mediates GITR-induced NF-κB activation (33) and TRAF4 can localize on the membrane to regulate cellular adherent junctions (34). Although TRAF4 is highly expressed in human carcinomas (22), its biological functions in tumorigenesis remain unclear.
Our results showed that TRAF4 is overexpressed in human lung cancer cell lines and tissues. To study the function of TRAF4, we generated a stable TRAF4 knockdown HaCaT cell line and TRAF4 knockdown attenuated EGF-induced transformation (Fig. 2A). TRAF4 overexpression promoted EGF-induced cell transformation and even without EGF, TRAF4 overexpression induced anchorage-independent growth of HaCaT cells (Fig. 2C). Additionally, TRAF4 enhanced H-RasG12V-induced 3T3 cell transformation (Fig. 2D). Furthermore, we demonstrated that TRAF4 is required for the malignant phenotype of human lung cancer cells, including proliferation, colony growth in soft agar, and the ability to form tumors in athymic nude mice (Fig. 3, S1). These results suggested that TRAF4 plays an important role in human lung carcinogenesis. However, a prior study identified TRAF4 as a p53-regulated pro-apoptotic gene in a p53 temperature-sensitive cell line Vm10 (23). TRAF4 overexpression in p53 wildtype tumor cells, U2OS and HCT116, induced apoptosis and inhibited colony formation (23). This apparent discrepancy suggests that the same gene can display various biological functions that are dependent upon the form of stimulation and cell type.
EGFR-mediated signaling transduction is almost always deregulated in human lung cancer, especially the hyperactivation of its downstream kinases, including the Ras-Raf-MEK-ERKs and PI3K-Akt pathways, which notably accelerate malignant progression and induce drug resistance (35–37). We found that TRAF4 knockdown impaired EGF-induced Akt activation (Fig. 4A–C), but had no effect on EGFR or ERK1/2 activity (data not shown). Our results revealed that EGF-induced Akt membrane recruitment was decreased in TRAF4 knockdown lung cancer cells (Fig. 4E, S2). These results led us to conclude that TRAF4 is involved in regulating the Akt signaling pathway.
Recent studies indicated that Lys63-linked ubiquitination of Akt is critical in the process of Akt membrane recruitment and activation in response to growth factors (11, 13, 14). In addition, TRAF2 reportedly regulates RSK2 activation through Lys63-linked ubiquitin in EGF signaling (38). Consistent with structural characteristics of other TRAF protein family members, TRAF4 has an N-terminal Ring domain followed by a series of zinc fingers. The Ring domain of TRAF proteins confers E3 ubiquitin ligase capability (39). However, only two TRAF family members, TRAF6 (40) and TRAF2 (41), were demonstrated to possess E3 ligase activity. We found that TRAF4 binds with Akt1 through the C-terminal TRAFc domain, and importantly, this binding was also enhanced by EGF treatment (Fig. 5). Moreover, this is the first report to show that TRAF4 is required for Akt ubiquitination (Fig. 6). We examined the role of Skp2 in EGF-triggered Akt ubiquitination and activation in human lung cancer. Unexpectedly, Skp2 expression was not different in human lung cancer cells compared with normal lung cells. Notably, knockdown of Skp2 had no effect on EGF-induced Akt activation (Fig. S4). Our results imply that TRAF4, and not Skp2, is the major mediator for Akt ubiquitination and activation in human lung cancer.
To sustain unlimited growth, most tumor cells consume glucose through the glycolytic pathway as their major energy source to rapidly generate ATP even in the presence of oxygen (28). This phenomenon, called the Warburg effect, is an emerging hallmark of cancer (42). The underlying mechanisms explaining the deregulation of glucose metabolism in cancer cells have not yet been completely elucidated, but Akt is known to regulate vital metabolic enzymes or transporters, such as hexokinase 2 (HK2) and glucose transporter 1 (Glut1) (43–45). We first demonstrated that TRAF4 mediates glycolysis in human lung cancer by regulating the expression of Glut1 and HK2 through Akt signaling (Fig. 7), confirming the importance of TRAF4-mediated Akt activation in lung tumorigenesis and lung cancer metabolic reprogramming.
Overall, we demonstrated that TRAF4 regulates Akt ubiquitination and activation, and plays a critical role in human lung carcinogenesis. TRAF4 is overexpressed in human lung cancer and is essential for lung cancer cells to sustain tumorigenic properties, such as colony formation, glycolysis and xenograft tumor growth. These results suggest that TRAF4 a good molecular target for human lung cancer prevention and treatment.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by The Hormel Foundation and National Institute’s of Health grants CA120388, CA081064 and ES016548. National Basic Research Program of China (2011CB504305), National Natural Science Foundation of China (30930101 and 81161120410), The National High Technology Research and Development Program of China (2012AA02A501).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Katlic MR, Facktor MA, Berry SA, McKinley KE, Bothe A, Jr, Steele GD., Jr ProvenCare lung cancer: a multi-institutional improvement collaborative. CA Cancer J Clin. 2011;61:382–96. doi: 10.3322/caac.20119. [DOI] [PubMed] [Google Scholar]
- 3.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 4.Janku F, Stewart DJ, Kurzrock R. Targeted therapy in non-small-cell lung cancer--is it becoming a reality? Nat Rev Clin Oncol. 2010;7:401–14. doi: 10.1038/nrclinonc.2010.64. [DOI] [PubMed] [Google Scholar]
- 5.Brazil DP, Park J, Hemmings BA. PKB binding proteins. Getting in on the Akt. Cell. 2002;111:293–303. doi: 10.1016/s0092-8674(02)01083-8. [DOI] [PubMed] [Google Scholar]
- 6.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627–44. doi: 10.1038/nrd2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brognard J, Clark AS, Ni Y, Dennis PA. Akt/protein kinase B is constitutively active in non-small cell lung cancer cells and promotes cellular survival and resistance to chemotherapy and radiation. Cancer Res. 2001;61:3986–97. [PubMed] [Google Scholar]
- 9.Memmott RM, Dennis PA. The role of the Akt/mTOR pathway in tobacco carcinogen-induced lung tumorigenesis. Clin Cancer Res. 2010;16:4–10. doi: 10.1158/1078-0432.CCR-09-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–83. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 11.Yang WL, Wu CY, Wu J, Lin HK. Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 2010;9:487–97. doi: 10.4161/cc.9.3.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin K. The Akt DUBbed InAktive. Sci Signal. 2013;6:pe1. doi: 10.1126/scisignal.2003864. [DOI] [PubMed] [Google Scholar]
- 13.Yang WL, Wang J, Chan CH, Lee SW, Campos AD, Lamothe B, et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–8. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan CH, Li CF, Yang WL, Gao Y, Lee SW, Feng Z, et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kedinger V, Rio MC. TRAF4, the unique family member. Adv Exp Med Biol. 2007;597:60–71. doi: 10.1007/978-0-387-70630-6_5. [DOI] [PubMed] [Google Scholar]
- 16.Blaise S, Kneib M, Rousseau A, Gambino F, Chenard MP, Messadeq N, et al. In vivo evidence that TRAF4 is required for central nervous system myelin homeostasis. PLoS One. 2012;7:e30917. doi: 10.1371/journal.pone.0030917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Regnier CH, Masson R, Kedinger V, Textoris J, Stoll I, Chenard MP, et al. Impaired neural tube closure, axial skeleton malformations, and tracheal ring disruption in TRAF4-deficient mice. Proc Natl Acad Sci U S A. 2002;99:5585–90. doi: 10.1073/pnas.052124799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiels H, Li X, Schumacker PT, Maltepe E, Padrid PA, Sperling A, et al. TRAF4 deficiency leads to tracheal malformation with resulting alterations in air flow to the lungs. Am J Pathol. 2000;157:679–88. doi: 10.1016/S0002-9440(10)64578-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Regnier CH, Tomasetto C, Moog-Lutz C, Chenard MP, Wendling C, Basset P, et al. Presence of a new conserved domain in CART1, a novel member of the tumor necrosis factor receptor-associated protein family, which is expressed in breast carcinoma. The Journal of biological chemistry. 1995;270:25715–21. doi: 10.1074/jbc.270.43.25715. [DOI] [PubMed] [Google Scholar]
- 20.Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R. Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res. 1996;56:3886–90. [PubMed] [Google Scholar]
- 21.Kornegoor R, Moelans CB, Verschuur-Maes AH, Hogenes MC, de Bruin PC, Oudejans JJ, et al. Oncogene amplification in male breast cancer: analysis by multiplex ligation-dependent probe amplification. Breast Cancer Res Treat. 2012;135:49–58. doi: 10.1007/s10549-012-2051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Camilleri-Broet S, Cremer I, Marmey B, Comperat E, Viguie F, Audouin J, et al. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene. 2007;26:142–7. doi: 10.1038/sj.onc.1209762. [DOI] [PubMed] [Google Scholar]
- 23.Sax JK, El-Deiry WS. Identification and characterization of the cytoplasmic protein TRAF4 as a p53-regulated proapoptotic gene. The Journal of biological chemistry. 2003;278:36435–44. doi: 10.1074/jbc.M303191200. [DOI] [PubMed] [Google Scholar]
- 24.Fukushima T, Matsuzawa S, Kress CL, Bruey JM, Krajewska M, Lefebvre S, et al. Ubiquitin-conjugating enzyme Ubc13 is a critical component of TNF receptor-associated factor (TRAF)-mediated inflammatory responses. Proc Natl Acad Sci U S A. 2007;104:6371–6. doi: 10.1073/pnas.0700548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG. Site-specific Lys-63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of I kappa B kinase activation. The Journal of biological chemistry. 2007;282:4102–12. doi: 10.1074/jbc.M609503200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensen JP, Bates PW, Yang M, Vierstra RD, Weissman AM. Identification of a family of closely related human ubiquitin conjugating enzymes. The Journal of biological chemistry. 1995;270:30408–14. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- 27.Iwai K, Yamanaka K, Kamura T, Minato N, Conaway RC, Conaway JW, et al. Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc Natl Acad Sci U S A. 1999;96:12436–41. doi: 10.1073/pnas.96.22.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–37. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 29.Abell AN, Johnson GL. MEKK4 is an effector of the embryonic TRAF4 for JNK activation. The Journal of biological chemistry. 2005;280:35793–6. doi: 10.1074/jbc.C500260200. [DOI] [PubMed] [Google Scholar]
- 30.Marinis JM, Hutti JE, Homer CR, Cobb BA, Cantley LC, McDonald C, et al. IkappaB kinase alpha phosphorylation of TRAF4 downregulates innate immune signaling. Molecular and cellular biology. 2012;32:2479–89. doi: 10.1128/MCB.00106-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marinis JM, Homer CR, McDonald C, Abbott DW. A novel motif in the Crohn’s disease susceptibility protein, NOD2, allows TRAF4 to down-regulate innate immune responses. The Journal of biological chemistry. 2011;286:1938–50. doi: 10.1074/jbc.M110.189308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zepp JA, Liu C, Qian W, Wu L, Gulen MF, Kang Z, et al. Cutting edge: TNF receptor-associated factor 4 restricts IL-17-mediated pathology and signaling processes. J Immunol. 2012;189:33–7. doi: 10.4049/jimmunol.1200470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esparza EM, Arch RH. TRAF4 functions as an intermediate of GITR-induced NF-kappaB activation. Cellular and molecular life sciences: CMLS. 2004;61:3087–92. doi: 10.1007/s00018-004-4417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathew SJ, Rembold M, Leptin M. Role for Traf4 in polarizing adherens junctions as a prerequisite for efficient cell shape changes. Molecular and cellular biology. 2011;31:4978–93. doi: 10.1128/MCB.05542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–81. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 36.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–74. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbst RS, Fukuoka M, Baselga J. Gefitinib--a novel targeted approach to treating cancer. Nat Rev Cancer. 2004;4:956–65. doi: 10.1038/nrc1506. [DOI] [PubMed] [Google Scholar]
- 38.Peng C, Zhu F, Wen W, Yao K, Li S, Zykova T, et al. Tumor necrosis factor receptor-associated factor family protein 2 is a key mediator of the epidermal growth factor-induced ribosomal S6 kinase 2/cAMP-responsive element-binding protein/Fos protein signaling pathway. The Journal of biological chemistry. 2012;287:25881–92. doi: 10.1074/jbc.M112.359521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee NK, Lee SY. Modulation of life and death by the tumor necrosis factor receptor-associated factors (TRAFs) J Biochem Mol Biol. 2002;35:61–6. doi: 10.5483/bmbrep.2002.35.1.061. [DOI] [PubMed] [Google Scholar]
- 40.Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–66. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465:1084–8. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Yecies JL, Manning BD. Transcriptional control of cellular metabolism by mTOR signaling. Cancer Res. 2011;71:2815–20. doi: 10.1158/0008-5472.CAN-10-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallace DC. Mitochondria and cancer: Warburg addressed. Cold Spring Harb Symp Quant Biol. 2005;70:363–74. doi: 10.1101/sqb.2005.70.035. [DOI] [PubMed] [Google Scholar]
- 45.Sommermann TG, O’Neill K, Plas DR, Cahir-McFarland E. IKKbeta and NF-kappaB transcription govern lymphoma cell survival through AKT-induced plasma membrane trafficking of GLUT1. Cancer Res. 2011;71:7291–300. doi: 10.1158/0008-5472.CAN-11-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.