Abstract
BlockLogo is a web-server application for visualization of protein and nucleotide fragments, continuous protein sequence motifs, and discontinuous sequence motifs using calculation of block entropy from multiple sequence alignments. The user input consists of a multiple sequence alignment, selection of motif positions, type of sequence, and output format definition. The output has BlockLogo along with the sequence logo, and a table of motif frequencies. We deployed BlockLogo as an online application and have demonstrated its utility through examples that show visualization of T-cell epitopes and B-cell epitopes (both continuous and discontinuous). Our additional example shows a visualization and analysis of structural motifs that determine specificity of peptide binding to HLA-DR molecules. The BlockLogo server also employs selected experimentally validated prediction algorithms to enable on-the-fly prediction of MHC binding affinity to 15 common HLA class I and class II alleles as well as visual analysis of discontinuous epitopes from multiple sequence alignments. It enables the visualization and analysis of structural and functional motifs that are usually described as regular expressions. It provides a compact view of discontinuous motifs composed of distant positions within biological sequences. BlockLogo is available at: http://research4.dfci.harvard.edu/cvc/blocklogo/ and http://methilab.bu.edu/blocklogo/
Keywords: T-cell epitope, B-cell epitope, protein-protein interaction, block entropy, sequence variability and conservation
1. Introduction
Sequence logos are useful tools for visual display of conservation and variability in a multiple sequence alignment (MSA) of DNA, RNA, or protein sequences (T D Schneider and Stephens, 1990). Individual nucleotides or residues in each position in an MSA are displayed by stacking the characters, where the height of each character corresponds to its frequency relative to the frequencies all characters in that position, and the height of the stack is determined by the total information content (Shannon, 1948). Sequence logos aid the interpretation of sequence data by visualization of conserved motifs representing various functional or structural properties. Examples of motifs that have been visually analyzed using sequence logos are: transcription factors (Wade et al., 2004), enzyme DNA sequences (Goll and Bestor, 2005), proteolytic cleavage sites (Mahrus et al., 2008), T-cell epitopes (Bryson et al. 2009; Olsen et al. 2011), and the analysis of targets of neutralizing antibodies in HIV (Sun et al., 2008), among others. Sequence logos display stacked motifs with the most frequent residues shown at the bottom and the least frequent motif displayed on the top of the stack. Sequence logos visualize biological sequence motifs where the height of the logo element represents its log-transformed frequency displayed in bits of information. Logos often do not display low-frequency motifs because their heights are below useful resolution.
The most popular sequence logo web server is WebLogo (Crooks et al., 2004). It enables users to generate standard sequence logos for DNA, RNA, and protein sequences. In addition to the WebLogo web server, several specialized logo generators have been developed to visualize specific motifs or functional sequence units that are unapparent from the standard sequence logos. Examples of extensions to the basic sequence logo are: RNA structure logo (J Gorodkin et al., 1997) which combines the standard sequence logo with information about base pairing and mutual information of base pairs; enoLOGOS (Workman et al., 2005) which displays energy measurements, probability matrices and alignment matrices in addition to the standard sequence logo; two-sample logo (Vacic et al., 2006) which displays comparative sequence logos for two sets of MSA; CorreLogo (Bindewald et al., 2006) calculates mutual information of nucleotides in different positions to determine correlation and potential base pairing; Phylo-mLogo (Shih et al., 2007) creates sequence logos for comparison of phylogenetically distinct clades within an MSA of DNA sequences; Blogo (Li et al., 2008) displays a sequence logo with statistically significant bias of individual positions; RNAlogo (T.-H. Chang et al., 2008) extends the RNA structure logo with a graphical representation of secondary structure; PoreLogo (Oliva et al., 2009) uses sequence logos and 3D protein structures to visualize motifs of channels in transmembrane proteins; iceLogo (Colaert et al., 2009) provides a probability-based visualization by allowing users to define reference sequences of the sample’s origin; Seq2Logo (Thomsen and Nielsen, 2012) offers the capacity to visualize amino acid sequence profiles in terms of amino acid enrichment and depletion; RIlogo (Menzel et al., 2012) for visualization of RNA-RNA interactions; and CodonLogo (Sharma et al., 2012) which enables visualization of conserved codon patterns. The BlockLogo web server (Figure 1) enables visualization of continuous and discontinuous immune epitopes and various sequence motifs. To our knowledge, it is the first logo web server that specifically enables visualization and analysis of immunologically relevant motifs.
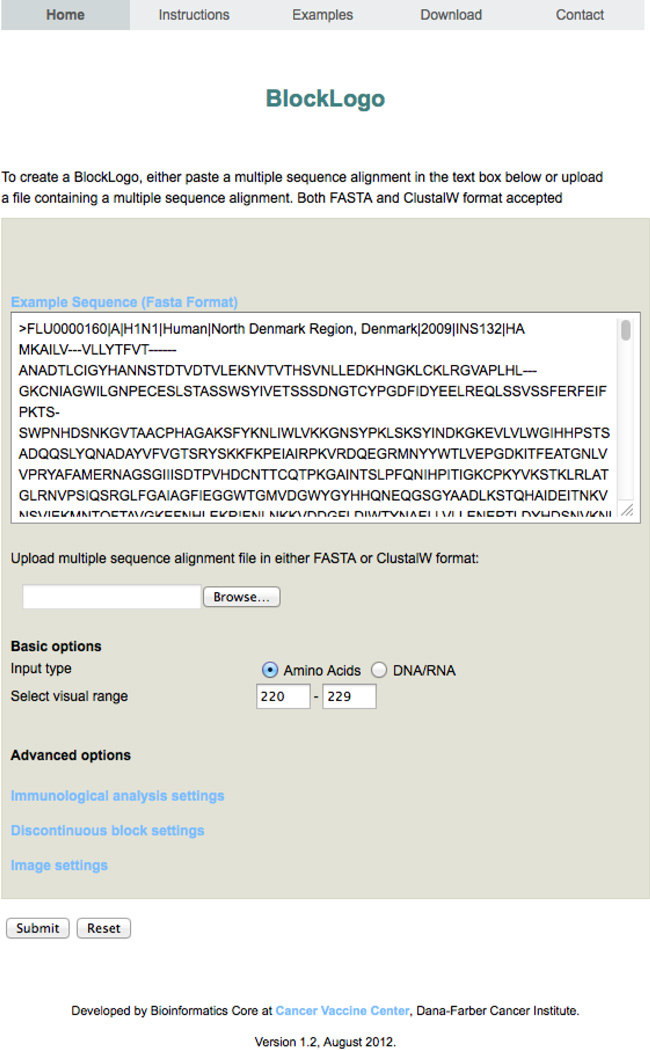
Figure 1.
The front page of BlockLogo with an example of input for visualization of region 220–229 of MSA of influenza A HA. Numbering is relative to the MSA alignment position and the input in this example is in the FASTA format.
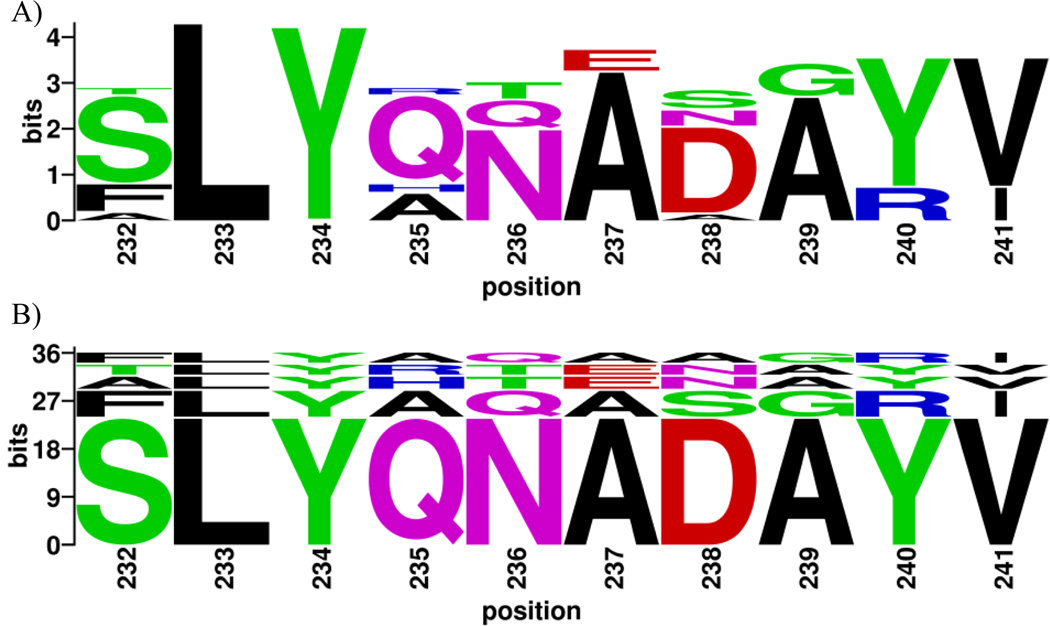
WebLogo is suitable for visualization of immunological motifs such as immune epitopes. A main limitation of the standard sequence logo for this type of application is that sequence logos carry no information about the relationship between the residues in the logo, but treat each residue as an individual independent position. Often, such logos have limited interpretability. For example, the sequence logo of influenza A HA peptide 232–241 (Figure 2A) shows variability that can be encoded by as many as 3,072 different peptides (4×1×1×4×3×2×4×2×2×2, corresponding to the number of different residues in each position). The BlockLogo presented in Figure 2B and Table 1 shows, at a glance, that the vast majority of actual sequence diversity is produced by only five peptides that can be read directly from BlockLogo. The actual number of different peptides that have produced sequence logo displayed in Figure 2A is seven, as shown in Table 1. The peptides visible in this BlockLogo have frequencies >6%, while each of the two peptides not readable from BlockLogo have frequency of <1%. Sequence logos can be useful for visualizing individual anchor position variability of MHC binding peptides, however since many motifs, such as T-cell epitopes, are recognized as linear peptides rather than individual residues, they should be visualized as continuous sequence blocks or fragments. A typical MHC class I T-cell epitopes may be between 8 and 11 amino acids long. MHC class II epitopes can be longer than 30 amino acids but they bind MHC through a nine amino acids long binding core (Reinherz et al., 1999). The input to the BlockLogo web server tool is an MSA of nucleotides, of short peptides of equal length, or of a user-defined subset of positions (here termed a “block”) within an MSA of longer protein sequences. The user-defined positions from within an MSA (i.e. positions derived from the continuous or discontinuous motifs) define the blocks. The information content (Shannon entropy) and relative frequency of each block are calculated, and the sequences printed in the BlockLogo, stacked according to frequency, from the most to the least frequent, from the bottom to the top of the stack. An extension of BlockLogo enables prediction of the binding affinity of identified peptides for a selection of common HLA molecules using the netMHC prediction algorithms (Lundegaard et al. 2011; Nielsen et al. 2007) that have been experimentally validated for accuracy.
Figure 2.
(A) Sequence logo plot of the residues in the 10-residue block starting at position 232 of the Influenza virus HA protein generated using WebLogo. (B) BlockLogo of the peptides in the 232–241 block. The residue position in the MSA is shown on the X-axis, and the information content is shown on the Y-axis. See Table 1 for peptide frequencies and HLA binding affinity predictions.
Table 1.
HLA binding predictions of each peptide present in the block of 10-mer peptides starting at position 232 in an MSA of influenza HA proteins. Prediction were performed for HLA A*02:01 allele, but can be done for a number of alleles (see materials and methods). The table is also included in the BlockLogo web server output if HLA binding predictions are selected upon submission.
| # | Peptide | Frequency (%) | Accumulated frequency (%) |
Predicted binding affinity (nM) |
|---|---|---|---|---|
| 1 | SLYQNADAYV | 64.65 | 64.65 | 11.45 |
| 2 | FLYAQASGRI | 14.14 | 78.79 | 24.37 |
| 3 | ALYHTENAYV | 7.07 | 85.86 | 9.74 |
| 4 | TLYRTENAYV | 6.06 | 91.92 | 20.51 |
| 5 | FLYAQAAGRI | 6.06 | 97.98 | 22.23 |
| 6 | FLHAQASGRI | 1.01 | 98.99 | 130.48 |
| 7 | SLYQNADSYV | 1.01 | 100.00 | 19.39 |
2. Materials and Methods
2.1. Variability and conservation metrics
Calculation of information content of individual positions in an MSA of homologous protein sequences is based on Shannon entropy (Shannon, 1948). Similarly, Shannon entropy can be calculated for each motif within a defined block. Each block contains W unique motifs of length l in a dataset of N sequences. The formula used for calculation of block entropy is (Olsen et al., 2011):
| (1) |
Where H(Bx) is the total entropy of a block of motifs starting at position x, w is a unique motif in the space of W unique motifs in block Bx. Pw(x) is the frequency of motif w at position x. In standard sequence logos, the theoretical maximum entropy of single position in a protein sequence is log220 ≈ 4.32 bits (corresponding to equal representation of all 20 amino acids), so each amino acid in a position can be represented by its fractional information content of that position (4.32 - H(x)). The theoretical maximum entropy of a block is 34.58 bits for 8-mer motifs (log2208), ~38.90 bits for 9-mers (log2209), and ~43.22 bits for 10-mers (log22010). The maximum bit value on the Y-axis is calculated according to the input alphabet (RNA, DNA, or amino acids) and the selected block length. The height of displayed BlockLogo is scaled to match the height of the sequence logo.
The fractional information content of each unique motif, w, in each block, Bx, is calculated using the formula (Olsen et al., 2011):
| (2) |
where H(w) is the entropy of peptide w; Pw(x) is the frequency of peptide w; and H(Bx) is the total information content of the block B starting at position x, in the MSA. The blocks (peptides or nucleotide fragments, or discontinuous motifs) are displayed in order from the most frequent to the least frequent block starting from the base of the X-axis.
2.2. Prediction of T-cell epitopes
The HLA binding affinity of peptides is predicted using NetMHC 3.0 (Lundegaard et al., 2011) and NetMHCII 2.2 (Nielsen et al., 2007). These algorithms were chosen based on their high accuracy determined in our previous studies of the accuracy of online HLA binding prediction servers (Lin et al. 2008a; Lin et al. 2008b; Zhang et al. 2011). When the HLA binding prediction option is selected, netMHC is used to predict HLA class I binders if the selected block is of length 8–11, and for HLA class II binders if the selected block is of length 13–25.
2.3. Visualization of continuous peptides
Conservation of continuous immunological motifs, such as T-cell epitopes, can be easily visualized and characterized using BlockLogo. To display this information, the user must submit an MSA of homologous protein sequences, and select a continuous range for visualization. The hemagglutinin (HA) sequences used to generate the examples presented in this article were collected from FluKB (http://research4.dfci.harvard.edu/cvc/flukb/). All the sequences were aligned using MAFFT (Katoh and Standley, 2013). Example data and their outputs are available at http://research4.dfci.harvard.edu/cvc/blocklogo/HTML/examples.php and at the mirror site http://methilab.bu.edu/blocklogo/HTML/examples.php.
2.4. Visualization of discontinuous peptides
In some cases the investigated motifs within an MSA are not linear peptides. For example, the residues forming a B-cell epitope are typically discontinuous positions within a protein sequence. To display a discontinuous motif, the user needs to define the set of positions from the MSA selected for visualization. By indicating the epitope positions in the uploaded MSA, discontinuous epitopes are extracted from the sequences, converted into virtual strings, and then processed by the BlockLogo and WebLogo enabling cross-comparison. The discontinuous BlockLogo and sequence logo have the MSA positions indicated below the stacked logos. For examples of discontinuous motifs, the neutralizing HA antibody F10, HLA DRB1 binding pocket 1 β chain were visualized. The information of neutralizing antibody F10 and validated strains were collected from the literature (Sui et al., 2009). B-cell epitopes were defined using two measurements: the accessible surface area (ASA) loss (Chothia, 1974) and the minimum distance between antibody and antigen atoms (McConkey et al., 2003). Residues with more than 20% ASA loss between the HA monomer and HA/antibody complex, and residues with atoms located within 4 Å of the F10 antibody atoms were considered to be part of the B-cell epitope. The F10 neutralizing epitope was defined from the F10-HA structure (PDB ID: 3FKU) (Figures 3 and 4). The HA protein sequence in FluKB with highest similarity to the HA sequence in the F10-HA was chosen using BLAST search (Altschul et al., 1990). The MAFFT tool (Katoh and Standley, 2013) was used to generate the MSA of all HA proteins in FluKB (29,113 complete HA protein sequences). The epitope positions defined by Sui et al. (2009) were mapped to the MSA. Then, a motif was extracted with residues on these positions for each sequence The information of HLA DRB1 binding pocket were collected from the literature (Chelvanayagam, 1997) and the HLA-DRB1 sequences were extracted from the IMGT/HLA database (Robinson et al., 2013). The example data and their output are available at BlockLogo under the examples tab.
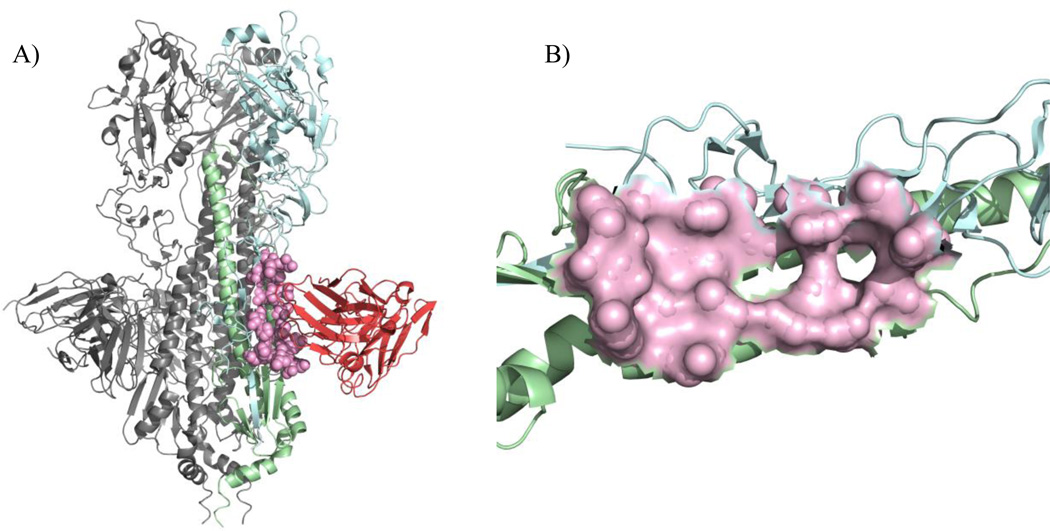
Figure 3.
(A) The structure of influenza A HA protein with the neutralizing antibody F10 (PDB ID: 3FKU) and its conformational epitope shown in pink, corresponding to residues 12, 32, 34, 36, 292, 293, 294 and 319 in chain A, and 18, 19, 20, 21, 38, 41, 42, 45, 49, 52, 53 and 56 in chain B. (B) The discontinuous epitope on HA protein recognized by F10.
Figure 4.
(A) Sequence logo of neutralizing epitopes for the broadly neutralizing antibody F10 within 29,113 influenza A virus HA proteins. (B) BlockLogo of the discontinuous residues representing the F10 neutralizing epitope. The numbering in these two figures corresponds to the residue positions in the MSA of the HA proteins. Table 2 shows the motif frequencies along with the corresponding neutralization assay results.
In both WebLogos and BlockLogos, the colors of the amino acids correspond to their chemical properties; polar amino acids (G, S, T, Y, C, Q, and N) are shown in green, basic amino acids (K, R, and H) are shown in blue, acidic amino acids (D and E) are shown in red, and hydrophobic amino acids (A, V, L, I, P, W, F, and M) are shown in black.
2.5. Software implementation
BlockLogo is written in Perl and uses Encapsulated PostScript format. The logos are created from open source templates available through the WebLogo web site (Crooks et al., 2004). The program uses the open source package ImageMagick (www.imagemagick.org) to convert the images to the supported formats.
3. User interface
The user is prompted to copy/paste an MSA, or upload a file containing an MSA, in standard FASTA or ClustalW formats. Users can select a block from the MSA by specifying the start and end positions of the subset, or a series of individual positions corresponding to positions of a discontinuous motif. The motifs that have a gap in any of the positions within the specified range will be excluded by default. In the analysis of discontinuous motifs, the sequences with gaps in specified positions will be included in the analysis if user selects this option.
Motifs of low frequency may not be visible when displayed by BlockLogo – this is a property of all logo visualizations. If the image height in pixels multiplied by the percent occurrence of a motif is less than 3, the low resolution of these low frequency motifs makes it difficult to see them within the logo. We therefore enabled the user to define image options (image format and size in pixels), which will alter the appearance and resolution of the logo and the resulting size of the image file. The logo on the results site is scaled to fit the size of the browser window. Clicking the logo on the result page will display the logo in the user defined format and size to enable generation of publication-quality figures. A standard sequence logo generated using locally installed WebLogo is printed below the BlockLogo to enable the comparison of two images.
On-the-fly prediction of HLA class I and II binding affinities in the defined block can be performed by selecting “predict epitopes in block” option together with user input of target HLA alleles. The results are displayed as a listing of HLA alleles including a table with detailed information on the predicted epitopes. The home page of BlockLogo server is shown in Figure 1.
4. Example applications
4.1. Conservation of influenza A T-cell epitopes
To illustrate the utility of BlockLogo, we analyzed a block of peptides in 29,113 influenza virus HA protein sequences, containing approximately 36.1 bits of information. All peptides in the block of 10-mers, starting at position 232 were predicted to bind to HLA A*02:01 with similar affinities. The relative frequencies of individual peptides within the viral population cannot be determined from the standard sequence logo produced with WebLogo (Figure 2A), but are clear from the BlockLogo (Figure 2B). Table 1 lists seven different peptides visualized in Figure 2 that represent the complete list of motifs found in the MSA, along with their individual frequencies, cumulative frequencies, and their predicted binding affinities to HLA A*02:01. The most frequent peptide in this block is present in the 64.65% of viral population – this information is not obvious from the standard sequence logo analysis. The combination of MHC binding prediction and BlockLogo visualization reveals this particular region to be highly antigenic, and thus potentially valuable in polyvalent vaccine designs. This peptide is not a known T-cell epitope and it is of potential interest since it is a predicted binder of high affinity to HLA A*02:01 and is highly conserved among HA.
4.2. Conservation of influenza A cross-neutralizing B-cell epitopes
The BlockLogo can also be used to display motifs as virtual peptides composed of a selection of discontinuous sites within a protein. This function can be applied to visualize conservation of B-cell epitopes, which can, for example, be used for representation and characterization of cross-neutralizing viral B-cell epitopes described in (Xu et al., 2010). The discontinuous BlockLogo (Figure 3B) displays the diversity of residues forming conformational epitope that is recognized by the broadly neutralizing antibody F10 (Sui, et al., 2009) shown in Figure 3A and Figure 3B. This BlockLogo shows the conservation/variability of F10 B-cell epitopes identified within the alignment of 29,113 sequences of full-length influenza HA proteins. The length of the block determines the information content of this motif, which is approximately 68 bits of information. The BlockLogo (Figures 4B) is a better indicator of the diversity of F10 neutralizing epitopes than the traditional sequence logo (Figure 4A), since it includes information about the frequency of actual sequences of naturally occurring epitopes. The description of ten most frequent epitopes (discontinuous peptides), including the influenza subtype of origin and the status of experimental binding validation is given in Table 2. The complete list of motifs in this example comprises 112 peptides. The details of this example with the full list of motifs can be accessed at the BlockLogo web site.
Table 2.
Ten most frequent influenza A HA discontinuous peptides on neutralizing epitope region recognized by neutralizing antibody F10 in FluKB (29,113 complete HA protein sequences). The table shows the amino acids of the epitope, HA subtype, frequency within the data set, and validation status - escape variants are those strains not neutralized by the F10.
| # | Discontinuous peptide | Subtype | Frequency (%) | Accumulated frequency (%) |
Validation |
|---|---|---|---|---|---|
| 1 | HHVLSLPTVDGWLTQITVNI | H1 | 24.61 | 24.61 | N/A |
| 2 | HNTLDKPTVDGWLTQINLNI | H3 | 15.64 | 40.25 | Escape |
| 3 | HHQISMPTVDGWKTQITVNI | H5 | 13.49 | 53.74 | Neutralized |
| 4 | HHVLSLPTVDGWQTQITVNI | H1 | 7.61 | 61.35 | N/A |
| 5 | QHKLTLPVVAGWRTQITVNV | H9 | 3.93 | 65.28 | Neutralized |
| 6 | HNTLDKPTIDGWLTQINLNI | H3 | 3.51 | 68.79 | N/A |
| 7 | THALSKPNIAGWLTQITLNS | B | 2.95 | 71.74 | N/A |
| 8 | HHVLSLPTIDGWQTQITVNI | H1 | 2.61 | 74.35 | Neutralized |
| 9 | HHVLNKTTIDGWRTQITVNI | H6 | 1.99 | 76.34 | N/A |
| 10 | HTQLTKPTIDGWLTQINLNI | H4 | 1.59 | 77.93 | N/A |
4.3. Variability of HLA-DRB1 binding pocket P1
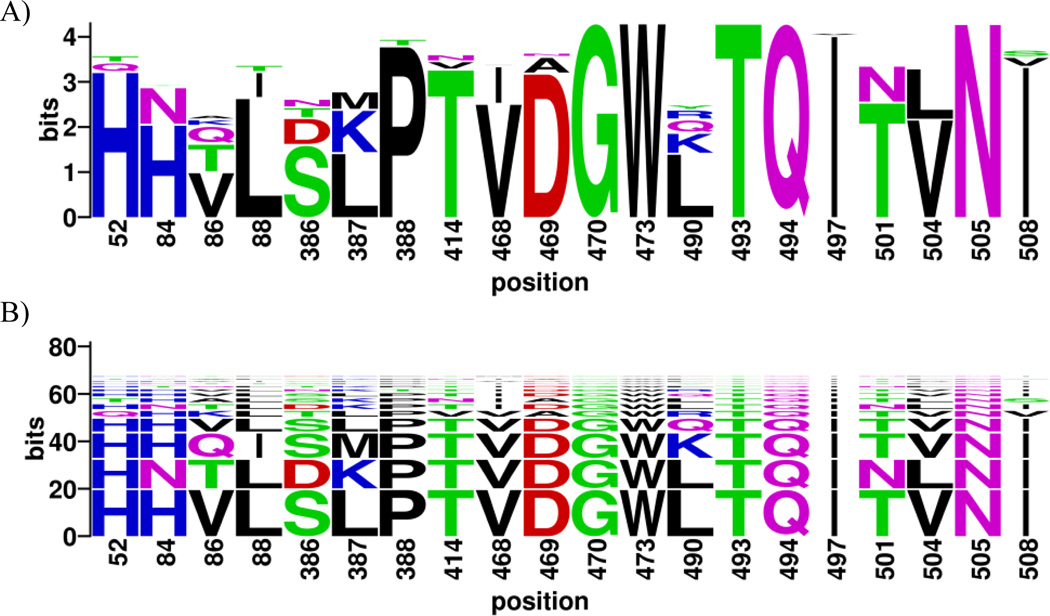
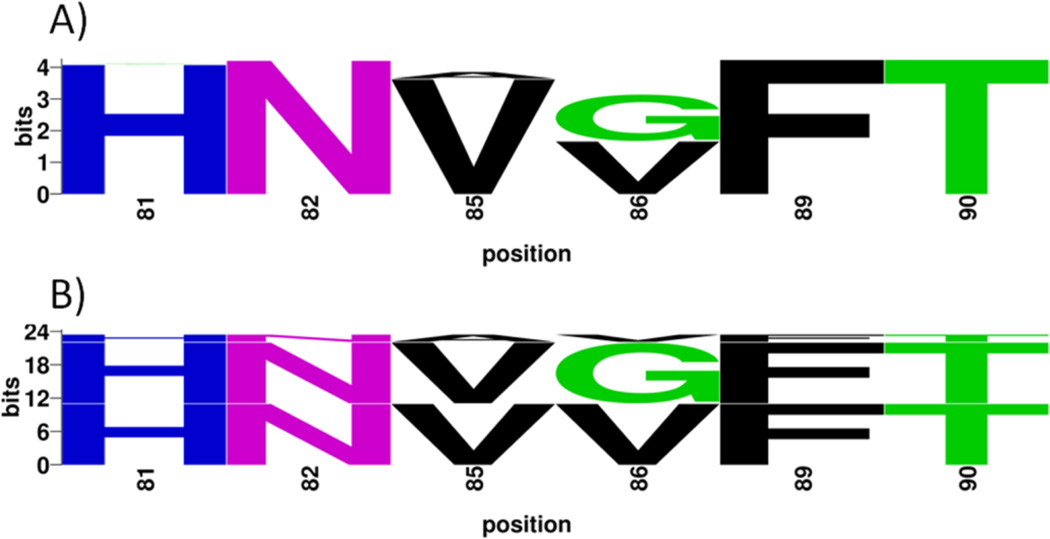
The usage of BlockLogo can easily be extended beyond T and B-cell epitopes to predict and visualize other peptide-protein interactions, and structural and functional motifs. For example, BlockLogo can be used to visualize variation in known structural motifs, such as HLA class II binding pocket 1 (P1) of HLA-DR, defined by variable β1 chain and invariant HLA-DR α chain. Pocket P1 accommodates the primary anchor of class-II HLA-DR binding peptides. Positions that define binding pockets for large number of HLA-DR molecules were described earlier (Chelvanayagam, 1997). These sequence motifs can be used to determine preferences for the primary anchor residue of binding peptides and shared specificities. The variability of the HLA DRB1 P1 pocket sequences among all known HLA-DRB1 alleles (Robinson et al, 2013) is visualized in Figure 5. Table 3 lists these motifs, their frequencies in the population, and serogroups in which they are found. Six variable positions (positions 81, 82, 85, 86, 89 and 90 in the alignment) constitute the pocket P1. Of 959 HLA-DRB1 protein sequences containing 23.9 bits of information, three motifs (HNVVFT, HNVGFT, and HNAVFT) account for 97% of the HLA-DRB1 sequences, seven motifs are represented each by a set of 2–5 sequences, and seven motifs are represented by a single sequence (Table 3). The vast majority of alleles from a particular serogroup contain a major motif (approximately 90% of the alleles) and a small number (approximately 10%) have a minor motif. Motif HNVVFT is a major signature of DRB1*03, 13, 14, and 15 serogroups; HNVGFT is a major signature for DRB1*01, 04, 07, 08, 09, 10, 11, and 16 serogroups; and HNAVFT is a major signature for DRB1*12 serogroup. In addition, motif HNVVFT is a minor signature of DRB1*04 and 11 serogroups; HNVGFT is a minor signature for DRB1*03, 14 and 15 serogroups; and HNAVFT is a minor signature for the DRB1*1 serogroup. Other motifs are observed in HLA alleles that are extremely rare in the general population (less than 1%). These results show that the fine specificity of primary anchor binding is determined by three major structural motifs and these motifs are unequally distributed between the serogroups.
Figure 5.
Visualization of diversity of binding pocket 1 βchain in DRB1 alleles using sequence logo (A) and BlockLogo (B). See Table 3 for the frequencies and alleles for each motif.
Table 3.
Frequency and allele distribution of discontinuous motifs in binding pocket 1 β chain of the DRB1 protein from the MSA of 947 DRB1 sequences. The “NA” stands for rare alleles where major and minor serogroups could not be defined, with their observed serotypes given in the brackets. These rare alleles represent 2.85% of all HLA-DRB1 sequences that are likely to have different binding specificities of peptide repertoires than those that belong to the major serogroups.
| # | Discontinuous peptide |
Frequency (%) | Accumulated frequency (%) |
Number of sequences |
DRB1 serogroup signatures |
|---|---|---|---|---|---|
| 1 | HNVVFT | 45.62 | 45.62 | 432 | 03, 13, 14, 15 (major) 04, 11 (minor) |
| 2 | HNVGFT | 45.51 | 91.13 | 431 | 01, 04, 07, 08, 09, 10, 11, 16 (major) 03, 14, 15 (minor) |
| 3 | HNAVFT | 6.02 | 97.15 | 57 | 12 (major) 01 (minor) |
| 4 | YNVVFT | 0.53 | 97.68 | 5 | NA (04, 14, 15) |
| 5 | YNVGFT | 0.42 | 98.10 | 4 | NA (04, 11, 15) |
| 6 | HNVDFT | 0.32 | 98.42 | 3 | NA (07, 11, 13) |
| 7 | HSVVFT | 0.21 | 98.63 | 2 | NA (03, 13) |
| 8 | HNVSFT | 0.21 | 98.84 | 2 | NA (08, 13) |
| 9 | HNVAFT | 0.21 | 99.05 | 2 | NA (03) |
| 10 | HNVMFT | 0.21 | 99.26 | 2 | NA (13, 14) |
| 11 | RNVVFT | 0.11 | 99.37 | 1 | NA (15) |
| 12 | HNFGFT | 0.11 | 99.47 | 1 | NA (13) |
| 13 | HNLGFT | 0.11 | 99.58 | 1 | NA (11) |
| 14 | QNVGFT | 0.11 | 99.68 | 1 | NA (11) |
| 15 | HNIGFT | 0.11 | 99.79 | 1 | NA (11) |
| 16 | HNIVFT | 0.11 | 99.89 | 1 | NA (03) |
| 17 | DNVGFT | 0.11 | 100.00 | 1 | NA (01) |
5. Conclusion and discussion
BlockLogo is a novel sequence logo tool optimized for visualization of user-defined continuous and discontinuous motifs, fragments, and peptides. Paired with the prediction of HLA binding, BlockLogo is a useful tool for rapid assessment of the immunological potential of selected regions within an MSA, such as those containing human pathogen sequences or tumor antigens alignments. The BlockLogo tool provides an easily interpretable visual representation of the immunological status and frequency for each predicted epitope. The observed frequencies of epitopes and their corresponding receptors (T or B-cell receptors) are vital for vaccine design, since the selection and combination of targets determine the pathogen coverage and the host population coverage of the vaccine. Since continuous epitopes are recognized as peptides rather than individual amino acids, traditional sequence logos do not show the specific peptides and their corresponding frequencies that are found in the analyzed sequences. BlockLogo thus provides a more precise and more informative representation of these motifs. Experimental approaches for identification and validation of sequence motifs useful as vaccine targets involving multiple HLA alleles and pathogen proteomes are laborious and costly. BlockLogo complements wet lab experimental methods by enabling pre-screening of key antigenic regions that are likely to contain vaccine targets. Previously (Olsen et al., 2011), we demonstrated the usefulness of BlockLogo representation for visualizing and capturing linear motifs of T-cell epitope candidates from large multiple sequence alignments. We have extended that work to enable the analysis and visualization of variability of discontinuous motifs, such as B-cell epitopes, protein-protein interaction sites, and receptor-ligand sites. To our knowledge, BlockLogo is the first logo generator that allows users to create logos based on a custom character set, consisting of either continuous or discontinuous motifs. BlockLogo is available at: http://research4.dfci.harvard.edu/cvc/blocklogo and http://methilab.bu.edu/blocklogo/.
Highlights.
We developed a tool for visualization of linear and non-linear immunological motifs
Utility is demonstrated for neutralizing influenza B cell epitopes
Utility is demonstrated for allergenic and hypoallergenic Bet v 1 allergens
Utility is demonstrated for variability of HLA-DRB1 binding pocket P1
The BlockLogo tool is available at http://research4.dfci.harvard.edu/cvc/blocklogo/
Acknowledgments
Funding
LRO was funded by the Novo Nordisk Foundation; UJK was funded by Oticon Foundation; C. Simon was funded by the Novo Scholarship Programme; GLZ, JS, VB, and ELR acknowledge funding from NIH grant U01 AI 90043.
Abbreviations
- HA
Hemagglutinin
- HLA
Human leukocyte antigen
- MHC
Major histocompatibility complex
- MSA
Multiple Sequence Alignment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bindewald E, Schneider Thomas D, Shapiro Ba. CorreLogo: an online server for 3D sequence logos of RNA and DNA alignments. Nucleic acids research. 2006;34:W405–W411. doi: 10.1093/nar/gkl269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson S, Julien J-P, Hynes RC, Pai EF. Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications. Journal of virology. 2009;83:11862–11875. doi: 10.1128/JVI.01604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T-H, Horng J-T, Huang H-D. RNALogo: a new approach to display structural RNA alignment. Nucleic acids research. 2008;36:W91–W96. doi: 10.1093/nar/gkn258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelvanayagam G. A roadmap for HLA-DR peptide binding specificities. Human immunology. 1997;58:61–69. doi: 10.1016/s0198-8859(97)00185-7. [DOI] [PubMed] [Google Scholar]
- Chothia C. Hydrophobic bonding and accessible surface area in proteins. Nature. 1974;248:338–339. doi: 10.1038/248338a0. [DOI] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K. Improved visualization of protein consensus sequences by iceLogo. Nature methods. 2009;6:786–787. doi: 10.1038/nmeth1109-786. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annual review of biochemistry. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gorodkin J, Heyer LJ, Brunak S, Stormo GD. Displaying the information contents of structural RNA alignments: the structure logos. Computer applications in the biosciences: CABIOS. 1997;13:583–586. doi: 10.1093/bioinformatics/13.6.583. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular biology and evolution. 2013 doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yang B, Liang S, Wang Y, Whiteley C, Cao Y, Wang X. BLogo: a tool for visualization of bias in biological sequences. Bioinformatics (Oxford, England) 2008;24:2254–2255. doi: 10.1093/bioinformatics/btn407. [DOI] [PubMed] [Google Scholar]
- Lin HH, Ray S, Tongchusak S, Reinherz, Ellis L, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC immunology. 2008a;9:8. doi: 10.1186/1471-2172-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HH, Zhang GL, Tongchusak S, Reinherz, Ellis L, Brusic V. Evaluation of MHC-II peptide binding prediction servers: applications for vaccine research. BMC bioinformatics. 2008b;9(Suppl 12):S22. doi: 10.1186/1471-2105-9-S12-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. Journal of immunological methods. 2011;374:26–34. doi: 10.1016/j.jim.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey BJ, Sobolev V, Edelman M. Discrimination of native protein structures using atom-atom contact scoring. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3215–3220. doi: 10.1073/pnas.0535768100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel P, Seemann SE, Gorodkin RILogo: visualizing RNA-RNA interactions. Bioinformatics (Oxford, England) 2012 Jan;28:2523–2526. doi: 10.1093/bioinformatics/bts461. [DOI] [PubMed] [Google Scholar]
- Nielsen M, Lundegaard C, Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC bioinformatics. 2007;8:238. doi: 10.1186/1471-2105-8-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R, Thornton JM, Pellegrini-Calace M. PoreLogo: a new tool to analyse, visualize and compare channels in transmembrane proteins. Bioinformatics (Oxford, England) 2009;25:3183–3184. doi: 10.1093/bioinformatics/btp545. [DOI] [PubMed] [Google Scholar]
- Olsen LR, Zhang GL, Keskin DB, Reinherz Ellis L, Brusic V. Conservation analysis of dengue virus T-cell epitope-based vaccine candidates using peptide block entropy. Frontiers in immunology. 2011;2:1–15. doi: 10.3389/fimmu.2011.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinherz EL, Tan K, Tang L, Kern P, Liu J, Xiong Y, Hussey RE, Smolyar A, Hare B, Zhang R, Joachimiak A, Chang HC, Wagner G, Wang J. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science (New York, N.Y.) 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- Robinson J, Halliwell Ja, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA database. Nucleic acids research. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic acids research. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. A mathematical theory of communication. Bell System Technical Journal. 1948;27:379–423. 623–656. [Google Scholar]
- Sharma V, Murphy DP, Provan G, Baranov PV. CodonLogo: a sequence logo-based viewer for codon patterns. Bioinformatics (Oxford, England) 2012;28:1935–1936. doi: 10.1093/bioinformatics/bts295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AC-C, Lee DT, Peng C-L, Wu Y-W. Phylo-mLogo: an interactive and hierarchical multiple-logo visualization tool for alignment of many sequences. BMC bioinformatics. 2007;8:63. doi: 10.1186/1471-2105-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z-YJ, Oh KJ, Kim M, Yu J, Brusic V, Song L, Qiao Z, Wang Jia-huai, Wagner Gerhard, Reinherz Ellis L. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28:52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Thomsen MCF, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and twosided representation of amino acid enrichment and depletion. Nucleic acids research. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics (Oxford, England) 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- Wade JT, Hall DB, Struhl K. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004;432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo, Gary D, Benos PV. enoLOGOS: a versatile web tool for energy normalized sequence logos. Nucleic acids research. 2005;33:W389–W392. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE, Wilson IA. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science (New York, N.Y.) 2010;328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GL, Ansari HR, Bradley P, Cawley GC, Hertz T, Hu X, Jojic N, Kim Y, Kohlbacher O, Lund O, Lundegaard C, Magaret Ca, Nielsen M, Papadopoulos H, Raghava GPS, Tal V-S, Xue LC, Yanover C, Zhu S, Rock MT, Crowe JE, Panayiotou C, Polycarpou MM, Duch W, Brusic V. Machine learning competition in immunology - Prediction of HLA class I binding peptides. Journal of immunological methods. 2011;374:1–4. doi: 10.1016/j.jim.2011.09.010. [DOI] [PubMed] [Google Scholar]