Abstract
Protein-protein interactions represent a new class of exciting but challenging drug targets, because their large, flat binding sites lack well defined pockets for small molecules to bind. We report here a methodology for chemical synthesis and screening of large combinatorial libraries of bicyclic peptides displayed on rigid small-molecule scaffolds. With planar trimesic acid as the scaffold, the resulting bicyclic peptides are effective for binding to protein surfaces such as the interfaces of protein-protein interactions. Screening of a bicyclic peptide library against tumor necrosis factor-alpha (TNFα) identified a potent antagonist that inhibits the TNFα-TNFα receptor interaction and protects cells from TNFα-induced cell death. Bicyclic peptides of this type may provide a general solution for inhibition of protein-protein interactions.
Keywords: Combinatorial library, bicyclic peptide, tumor necrosis factor-alpha, protein-protein interaction, macrocycle
INTRODUCTION
Protein-protein interactions (PPIs) are of central importance in essentially all biochemical pathways, including those involved in disease processes. PPIs therefore represent a large class of new, exciting drug targets.1 However, PPIs are considered the prototypical “undruggable” or “challenging” targets for the conventional small-molecule approach, because PPIs usually involve large, flat interfaces, with which small molecules usually do not make enough points of contact to impart high affinity or specificity. For some of these PPIs, small-molecule inhibitors have been successfully developed by targeting the so-called “hot spots” at the interaction interface.1-4 A more general approach is to develop specific antibodies against the PPI interface.5-7 Non-immunoglobulin protein scaffolds have also been engineered into specific binders to target proteins through library screening and/or in vitro evolution.8-11 Antibodies and protein binders possess large binding surfaces of their own and are capable of making multiple contacts with a target surface (e.g., those involved in PPIs). Unfortunately, protein-based drugs are impermeable to the mammalian cell membrane; as such they are generally limited to targeting extracellular proteins and are not orally available. Recently, others12-18 and we19,20 have begun a third approach by generating macrocyclic compounds (e.g., cyclic peptides and peptidomimetics) as PPI inhibitors. These macrocycles typically have molecular weights between 500 and 2000 and occupy a largely untapped therapeutic space, often referred to as the “middle space”.21 Because of their relatively large sizes and ability to make multiple points of contact with a flat surface, macrocycles effectively compete with proteins for binding to flat surfaces and yet retain many of the pharmacokinetic properties of small molecules such as membrane permeability.22-24 Compared to protein drugs, macrocycles have greater metabolic stability, less likelihood of eliciting immune response, and lower cost of production. To further rigidify the structures and improve the binding affinity/specificity and metabolic stability, bicyclic peptides and peptoids have also been generated.25-32 Since rational design of macrocyclic inhibitors against PPIs is difficult, a popular approach has involved synthesizing and screening large libraries of bicyclic peptides and peptoids. To date, bicyclic peptide libraries have only been synthesized ribosomally by phage or mRNA display and are largely limited to proteinogenic amino acids (and certain unnatural α-L-amino acids) as building blocks.27-29
Tumor necrosis factor-alpha (TNFα) is a pleiotropic inflammatory cytokine of a variety of functions, many of which are not yet fully understood.33 TNFα is responsible for cachexia, wasting in patients with chronic diseases such as cancer and tuberculosis34 and is implicated in the development of septic shock and multi-organ failure in severely infected patients.35 It is also responsible for numerous chronic inflammatory disorders such as rheumatoid arthritis, ankylosing spondylitis, inflammatory bowel disease, psoriasis, hidradenitis suppurativa, and refractory asthma.36 These disorders are currently treated with protein inhibitors, including monoclonal antibodies infliximab (Remicade), adalimumab (Humira) or certolizumab pegol (Cimzia), and a circulating receptor fusion protein etanercept (Enbrel). These proteins bind specifically to TNFα and prevent its interaction with TNFα receptors (TNFRs). These biologic drugs are administered by in-hospital intravenous injections. Considerable efforts have been made over the past two decades to develop small-molecule inhibitors against TNFα, which have the potential to be administered orally. However, these efforts have so far led to only a few weak inhibitors.37-42 In this work, we developed a general methodology for chemical synthesis and screening of large combinatorial libraries of bicyclic peptides displayed on rigid small-molecule scaffolds. Chemical synthesis permitted the incorporation of unnatural amino acids (e.g., D-amino acids) and potentially non-peptidic building blocks into the bicyclic molecules, increasing their structural diversity and metabolic stability. Screening of a bicyclic peptide library against TNFα identified a compound that inhibits the TNFα-TNFR interaction and protects cells from TNFα-induced cell death.
RESULTS AND DISCUSSION
Design and Synthesis of Bicyclic Peptide Library
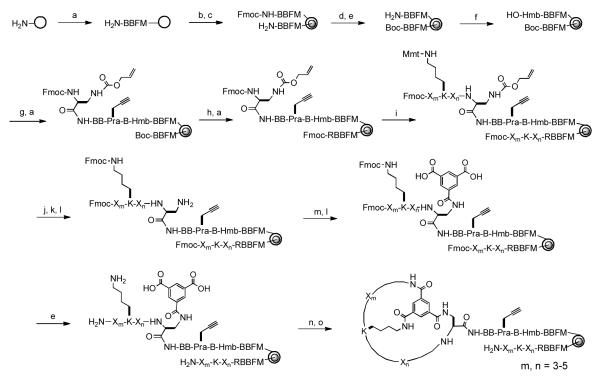
Antibodies recognize specific antigens by utilizing six small loops, called the “complementarity determining regions”. By grafting two or more flexible loops onto rigid protein scaffolds, other investigators have engineered protein binders of antibody-like affinity and specificity.8-11 We envisioned that displaying peptidic loops on rigid small-molecule scaffolds should also generate molecules that rival antibodies for binding affinity and specificity. To develop inhibitors against PPIs, we chose a planar structure as the scaffold, which should bias the resulting bicyclic peptides towards an overall “planar” (as opposed to globular) shape. An overall planar geometry would maximize the surface area of the molecules and therefore their ability to interact with flat protein surfaces. To test the validity of this approach, we designed a bicyclic peptide library by “wrapping” a peptide sequence of 6-10 random residues around a trimesoyl group (Fig. 1). Peptide cyclization was mediated by the formation of three amide bonds between trimesic acid and the N-terminal amine, the side chain of a C-terminal L-2,3-diaminopropionic acid (Dap), and the side chain of a fixed lysine within the random region. The resulting bicyclic peptides contained 3-5 random residues in each ring. The random sequence was constructed with a 25-amino acid set that included 10 proteinogenic amino acids [Ala, Arg, Asp, Gln, Gly, His, Ile, Ser, Trp, and Tyr], 5 nonproteinogenic α-L-amino acids [L-4-fluorophenylalanine (Fpa), L-norleucine (Nle), L-ornithine (Orn), and L-phenylglycine (Phg)], and 10 α-D-amino acids [D-2-naphthylalanine (D-Nal), D-Ala, D-Asn, D-Glu, D-Leu, DLys, D-Phe, D-Pro, D-Thr, and D-Val]. These building blocks were selected based on their structural diversity, metabolic stability, and commercial availability. Although not included here, non-peptidic building blocks are also compatible with our library synthesis and decoding method.19 This library has a theoretical diversity of 1.0 × 1014. In practice, the library size is limited by the amount of resin that can be conveniently employed and typically on the order of 107 (vide infra). Despite the fact that only a small fraction of all possible structures can be synthesized, we felt that it is critical to sample a large structural space. Once an active compound is identified, its affinity and specificity for the target protein may be improved by synthesizing and screening a second-generation library consisting of analogs of the initial hit. Inclusion of the unnatural amino acids increases the structural diversity and metabolic stability of the library compounds but necessitates chemical synthesis of the library.
Figure 1.
Synthesis of bicyclic peptide library. Reagents and Conditions: (a) Standard Fmoc/HATU chemistry; (b) soak in water; (c) 0.4 equiv Fmoc-OSu in Et2O/CH2Cl2; (d) di-t-butyl dicarbonate; (e) piperidine; (f) 4-hydroxybenzoic acid/HBTU/HOBT; (g) Fmoc-β-Ala-OH/DIC; (h) 50% TFA in DCM; (i) split-and-pool synthesis by Fmoc/HATU chemistry; (j) 2% TFA in DCM (6x); (k) Fmoc-OSu/ DIPEA in DCM; (l) Pd(PPh3)4; (m) diallyl protected trimesic acid/HATU; (n) PyBOP/HOBT/DIPEA; (o) modified reagent K.
The main challenge associated with screening chemically synthesized bicyclic peptide libraries is the structural determination of hit compounds. To overcome this difficulty, we synthesized the bicyclic peptide library in the one bead-two compound (OBTC) format on 2.0 g of TentaGel microbeads (90 μm; ~100 pmol peptide/bead; 2.86 × 106 beads/g). Each library bead was topologically segregated into two different layers, with the outer layer displaying a unique bicyclic peptide and the inner layer containing the corresponding linear peptide as an encoding tag (Figure 1). This was achieved by quickly suspending wet beads in 1:1 (v/v) DCM/Et2O containing 0.5 equivalent of N-(9-fluorenylmethoxycarbonyloxy)succinimide (Fmoc-OSu).43,44 Because the organic solvent is immiscible with water, only peptides on the bead surface were exposed to and reacted with Fmoc-OSu. The beads were washed with DMF and the remaining free N-terminal amines inside the beads were protected with a Boc group. After removal of the Fmoc group, a p-hydroxymethylbenzoic acid (Hmb) linker was added (for selective release of the bicyclic peptide), followed by the addition of β-Ala, L-propargylglycine (Pra), two β-Ala, and Fmoc-L-Dap(Alloc)-OH. The Pra residue serves as a handle for selective labeling of the bicyclic peptide via click chemistry (vide infra). The Dap residue permits attachment of the bicyclic peptide to the solid support as well as providing a side chain for peptide cyclization. The N-terminal Boc group was then removed from the inner peptides by treatment with trifluoroacetic acid (TFA) and an arginine residue was added to provide a fixed positive charge, which facilitates later peptide sequencing by mass spectrometry. The random region linear peptide synthesis, the Mmt group was removed using 2% TFA in DCM and replaced with an Fmoc group (the Mmt group was partially removed during deprotection of the Alloc group). The Alloc group on the C-terminal Dap was removed by treatment with Pd(PPh3)4 and the exposed sidechain amine was acylated with diallyl trimesic acid. Finally, the allyl (on the trimesoyl moiety) and Fmoc protecting groups (on the N-terminus and the lysine side chain) were removed and the surface peptides were cyclized by treatment with benzotriazol-1-yl-oxy-tris(pyrrolidino)phosphonium hexafluorophosphate (PyBOP). The peptides inside the beads were unaffected by the cyclization procedure due to lack of the Dap residue and remained in the linear form to serve as encoding tags. Note that macromolecular targets (e.g., proteins) cannot diffuse into the bead interior and thus the linear encoding peptides do not interfere with library screening. The symmetry of the trimesoyl unit ensured that a single bicyclic product was formed on each bead.
Library Screening against TNFα
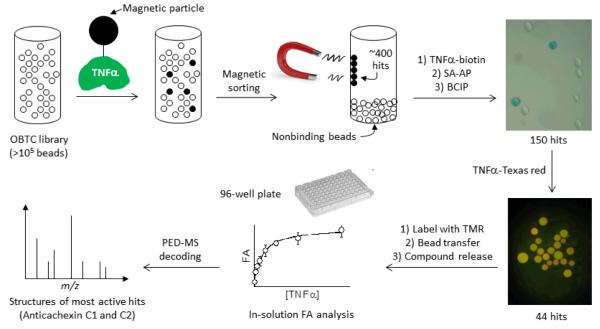
The bicyclic peptide library was subjected to four rounds of screening against recombinant TNFα that contained an N-terminal ybbR tag48 (MVLDSLEFIASKL) and had been specifically labeled at the ybbR tag with a biotin or fluorescent dye molecule. During the first round, 100 mg of the bicyclic peptide library (~3 × 105 beads) was incubated with biotinylated TNFα (0.8 μM) and streptavidin-coated magnetic particles (Figure 2). The resulting magnetic beads (~400 beads) were isolated from the library by magnetic sorting,49,50 during which the positive beads were attracted to the wall while the negative beads settled to the bottom of the container. The ~400 beads were washed, incubated again with the biotinylated TNFα (1.5 μM), and subjected to a second round of screening using an was synthesized by the split-and-pool method45-47 and an Nε-4-methoxytrityl (Mmt)-protected lysine was added in the middle of the random sequence to provide a side-chain amine for peptide cyclization. Following completion of the on-bead enzyme-linked assay and a streptavidin-alkaline phosphatase (SA-AP) conjugate. Binding of TNFα to a bead recruited SA-AP to the bead surface and upon the addition of 5-bromo-4-chloro-3-indolyl phosphate (BCIP), produced a turquoise colored precipitate on that bead. This procedure resulted in 150 intensely colored beads, which were manually isolated with a micropipet and incubated with Texas-red labeled TNFα (0.3 μM). The 44 most fluorescent beads were selected under a fluorescence microscope.
Figure 2.
Scheme showing the steps involved in peptide library screening against TNFα.
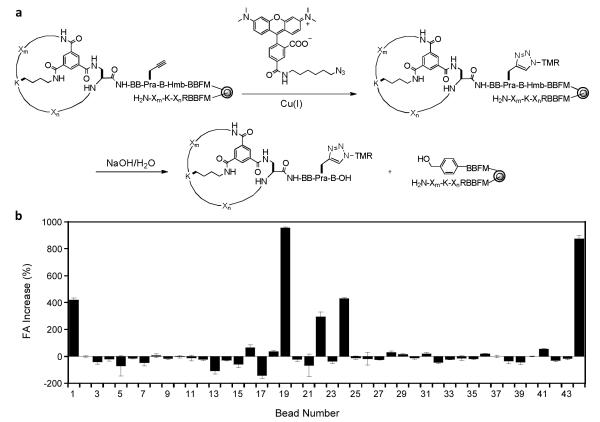
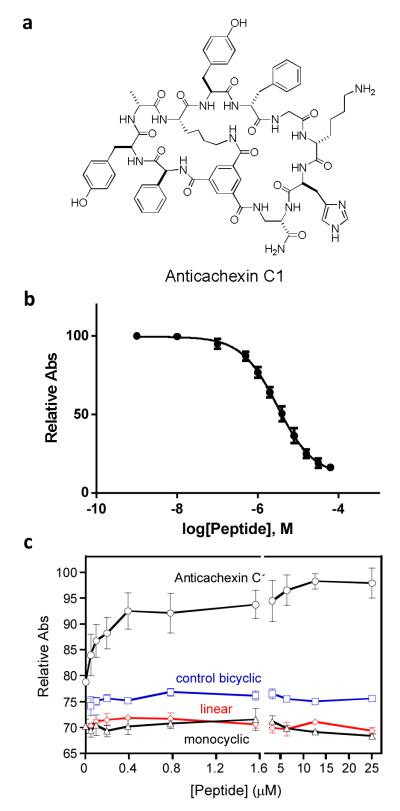
Finally, the 44 beads were treated with tetramethylrhodamine (TMR) azide in the presence of Cu(I), resulting in selective labeling of the bicyclic peptides at the Pra residue (Figures 2 and 3a). The beads were then placed into individual microcentrifuge tubes (1 bead/tube) and the TMR-labeled bicyclic peptide was released from each bead by treatment with 0.1 M NaOH, which selectively hydrolyzed the Hmb ester linkage associated with the bicyclic peptide. After neutralization, the released bicyclic peptide from each bead was tested for binding to TNFα in solution by fluorescence anisotropy (FA).51,52 Each of the 44 bicyclic peptides (~100 nM) was incubated with 5 μM TNFα and the 12 bicyclic peptides that showed ≥15% anisotropy increase (relative to the control without TNFα) (Figure 3b) were further analyzed at varying concentrations of TNFα (0-18 μM) to determine their dissociation constants (KD). Six peptides (bead No. 1, 16, 22, 24, 36, and 41) had KD values ranging from 0.8 to 7.8 μM (Fig. S1), while the other peptides showed no significant TNFα binding (bead No. 18, 19, 28, 29, 31, and 44). Next, for the 6 binding peptides, their corresponding beads containing the linear encoding peptides were retrieved from the micro-centrifuge tubes and subjected to partial Edman degradation-mass spectrometry (PED-MS) analysis.53 Two of the beads (hits No. 1 and 36) produced mass spectra of sufficient quality, from which unambiguous, complete sequences of bicyclo(Phg-Tyr-d-Ala-Lys-Tyr-d-Phe-Gly-d-Lys-His-Dap) and bicyclo(Ala-d-Phe-Trp-d-Thr-Gln-Lys-Nle-d-Leu-Ala-His-Dap) were obtained (Figure 4a and S2). These compounds are named as Anticachexin C1 and C2 thereafter, respectively.
Figure 3.
Solution-phase screening of initial hits (4th round). (a) Selective labeling of bicyclic peptides with TMR and their release from individual beads by base hydrolysis. (b) Evaluation of the 44 released bicyclic peptides for binding to TNFα in solution by fluorescent anisotropy using a fixed concentration of TNFα (5 μM) and TMR-labeled bicyclic peptide (100 nM).
Figure 4.
(a) Structure of Anticachexin C1. (b) Inhibition of TNFα-TNFR1 interaction by Anticachexin C1. The absorbance values on the y axis, which reflect the amount of TNFR1 bound to immobilized TNFα in the presence of increasing concentrations of Anticachexin C1, are relative to that in the absence of peptide inhibitor (100%). (c) Protection of WEHI-13VAR cells against TNFα (0.04 ng/mL)-induced cell death by Anticachexin C1 (0-25 μM). The absorbance values on the y axis, which reflect the number of live cells, are relative to that of DMSO control (no TNFα, no peptide).
The fact that only a relatively small number of the hits derived from on-bead screening (6 out of 44 beads) show strong binding to TNFα in solution suggests that most of the initial hits were weak binders or false positives, a problem commonly associated with on-bead screening. Most likely, the high ligand density on the library beads (~100 mM) resulted in multi-dentate interactions (i.e., simultaneous interaction of a single TNFα molecule with two or more resin-bound bicyclic peptides) and high avidity.54 False negatives are also possible as a result of several factors (e.g., poor aqueous solubility of a bicyclic peptide, inefficient release of a bicyclic peptide from resin by 0.1 M NaOH due to its strong noncovalent binding to the hydrophobic TentaGel resin, and/or strong binding of a bicyclic peptide to bovine serum albumin which was present in all FA assays). Elimination of these false negative compounds at this stage is actually desirable, as they are likely very hydrophobic and may bind non-specifically to many proteins. This highlights the importance of our library design, which permits selective release of the bicyclic peptide and therefore solution-phase binding analysis and avoids the need to individually resynthesize all 44 initial hits.
Binding Affinity and Specificity of Hit Compounds for TNFα
Anticachexin C1, C2, and the linear and monocyclic variants of Anticachexin C1 were resynthesized with a fluorescein isothiocyanate (FITC) label (Fig. S3), purified by HPLC, and assayed against TNFα by FA analysis. Anticachexin C1 and C2 bound to TNFα with KD values of 0.45 and 1.6 μM, respectively (Fig. S3). The linear C1 variant exhibited weak binding to TNFα (KD >10 μM), whereas the monocyclic peptide showed no significant binding affinity. Bicyclo(Arg-Arg-Arg-Arg-Nal-Phe-Dap-Ser-d-Val-Pro-pTyr-His-Dap), a control peptide unrelated to TNFα, also showed no detectable binding to TNFα. These results demonstrate that both the peptide sequence and the overall bicyclic structure are critical for binding to the target protein. To determine whether Anticachexin C1 and C2 are specific ligands of TNFα, we tested them for binding to five arbitrarily selected proteins, including bovine serum albumin (BSA), a glutathione-S-transferase-PLCγ SH2 domain fusion (GST-SH2), protein phosphatase PTP1B, HIV capsid protein, and a GST-BRCT fusion protein. Anticachexin C1 showed weak binding to BSA (KD~34 μM), but not to any of the other proteins, while Anticachexin C2 was less selective and showed substantial binding to BSA and GST-SH2 proteins (KD values of 7.4 and 1.5 μM, respectively; Fig. S4). Finally, unlabeled Anticachexin C1 and C2 inhibited the binding of FITC-labeled Anticachexin C2 to TNFα in a concentration-dependent manner (IC50 values of ~1 and ~4 μM, respectively) (Fig. S5), suggesting that both compounds bind to the same (or overlapping) site on TNFα. Because of its higher affinity and specificity for TNFα, Anticachexin C1 was selected for further biological tests.
Inhibition of TNFα-TNFR interaction by Anticachexin C1
TNFα signaling begins with the binding of the TNFα trimer to the extracellular domain of TNFR1, triggering the release of an inhibitory protein, silencer of death domains, from the intracellular domain of TNFR1.33 To test whether Anticachexin C1 inhibits the interaction between TNFα and TNFR1, biotinylated TNFα was immobilized onto a Neutravidin-coated 96-well microtiter plate. The plate was incubated with 0.5 nM horse radish peroxidase (HRP)-conjugated TNFR1 in the presence of varying concentrations of Anticachexin C1. After washing, the amount of HRP-TNFR1 bound to each well was quantitated by ELISA.55 Anticachexin C1 inhibited the TNFα-TNFR1 interaction in a concentration-dependent manner, with an IC50 value of 3.1 ± 0.3 μM (Fig. 4b).
Protection against TNFα-Induced Cell Death
The ability of Anticachexin C1 to protect cells against TNFα-induced cell death was tested with cultured WEHI-13VAR fibroblasts, which are highly sensitive to TNFα in the presence of actinomycin-D.56 The cells were treated with a fixed concentration of TNFα (0.04 ng/ml) and varying concentrations of Anticachexin C1 (0-25 μM) and the fraction of live cells was quantitated by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Anticachexin C1 protected the cells from TNFα-induced cell death in a concentration-dependent manner, whereas the corresponding monocyclic and linear peptides did not (Fig. 4c). As expected, the control bicyclic peptide that does not bind TNFα had no protective effect. The MTT assay was also conducted at a fixed concentration of Anticachexin C1 (50 μM) but varying concentrations of TNFα (0-250 ng/mL). TNFα exhibited an LD50 value of 0.46 ng/mL in the absence of TNFα inhibitor; in the presence of 50 μM Anticachexin C1, the LD50 value was shifted to 1.8 ng/mL (Fig. S6).
CONCLUSION
In this study, we report the first chemical synthesis and screening of a large combinatorial library of bicyclic peptides against a macromolecular target of biomedical significance. Compared to previous methods for bicyclic peptide library synthesis,27-29 which involve ribosomal peptide synthesis followed by chemical cyclization, our method has the advantage that it allows the incorporation of any unnatural amino acid or non-peptidic building blocks, greatly increasing the structural diversity and metabolic stability of the cyclic peptides. Chemical synthesis also allows for the use of orthogonal protecting groups, which in turn permits more “forcing” reaction conditions to drive the desired cyclization reaction to completion and prevents any undesired cyclization reaction from occurring. We demonstrate that bicyclic peptides displayed on a rigid planar scaffold are effective for binding to protein surfaces such as PPI interfaces. With a KD value of 0.45 μM, Anticachexin C1 is the most potent non-protein TNFα inhibitor reported to date. The bicyclic peptide library may be readily screened against other protein and nucleic acid targets.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grants from the National Institutes of Health (GM062820 and CA312855).
Footnotes
Supporting Information Experimental details and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes The authors declare no competing financial interests.
REFERENCES
- (1).Wells J, McClendon C. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- (2).Yin H, Hamilton AD. Angew. Chem. Int. Ed. 2005;44:4130–4163. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- (3).Garner AL, Janda KD. Curr. Topics Med. Chem. 2011;11:258–280. doi: 10.2174/156802611794072614. [DOI] [PubMed] [Google Scholar]
- (4).Morelli X, Bourgeas R, Roche P. Curr. Opin. Chem. Biol. 2011;15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- (5).Ockey DA, Gadek TR. Expert Opin. Therapeutic Patents. 2002;12:393–400. [Google Scholar]
- (6).Loregian A, Palu G. J. Cell. Physiol. 2005;204:750–762. doi: 10.1002/jcp.20356. [DOI] [PubMed] [Google Scholar]
- (7).Tanaka T, Rabbitts TH. Cell Cycle. 2008;7:1569–1574. doi: 10.4161/cc.7.11.6061. [DOI] [PubMed] [Google Scholar]
- (8).Koide A, Bailey CW, Huang X, Koide S. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- (9).Beste G, Schmidt FS, Stibora T, Skerra A. Proc. Natl. Acad. Sci. USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Rutledge SE, Volkman HM, Schepartz A. J. Am. Chem. Soc. 2003;125:14336–14347. doi: 10.1021/ja034508o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Steiner D, Forrer P, Plückthun A. J. Mol. Biol. 2008;382:1211–1227. doi: 10.1016/j.jmb.2008.07.085. [DOI] [PubMed] [Google Scholar]
- (12).Leduc AM, Trent JO, Wittliff JL, Bramlett KS, Briggs SL, Chirgadza NY, Wang Y, Burris TP, Spatola AF. Proc. Natl. Acad. Sci. USA. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Fasan R, Dias RLA, Moehle K, Zerbe O, Vrijbloed JW, Obrecht D, Robinson JA. Angew. Chem. Int. Ed. 2004;43:2109–2112. doi: 10.1002/anie.200353242. [DOI] [PubMed] [Google Scholar]
- (14).Millward SW, Fiacco S, Austin RJ, Roberts RW. ACS Chem. Biol. 2007;2:625–634. doi: 10.1021/cb7001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Tavassoli A, Lu Q, Gam J, Pan H, Benkovic SJ, Cohen SN. ACS Chem. Biol. 2008;3:757–764. doi: 10.1021/cb800193n. [DOI] [PubMed] [Google Scholar]
- (16).Ardi VC, Alexander LD, Johnson VA, McAlpine SR. ACS Chem. Biol. 2011;6:1357–1366. doi: 10.1021/cb200203m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Yamagishi Y, Shoji I, Miyagowa S, Kawakami T, Katoh T, Goto Y. Chem. Biol. 2011;18:1562–1570. doi: 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- (18).Dewan V, Liu T, Chem K, Qian Z, Xiao Y, Kleiman L, Mahasenan KV, Li C, Matsuo H, Pei D, Musier-Forsyth K. ACS Chem. Biol. 2012;7:761–769. doi: 10.1021/cb200450w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu X, Upadhyaya P, Villalona-Calero MA, Briesewitz R, Pei D. Med. Chem. Commun. 2013;4:378–382. doi: 10.1039/C2MD20329D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhou H, Liu L, Huang J, Bernard D, Karatas H, Navarro A, Lei M, Wang S. J. Med. Chem. 2013;56:1113–1123. doi: 10.1021/jm3015298. [DOI] [PubMed] [Google Scholar]
- (21).Terrett N. MedChemComm. 2013;4:474–475. [Google Scholar]
- (22) (a).Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J. Am. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]; (b) White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SSF, Kalgutkar AS, Bauman JN, Zhang YZ, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. Nat. Chem. Biol. 2011;7:810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chatterjee J, Gilon C, Hoffman A, Kessler H. Acc. Chem. Res. 2008;41:1331–1342. doi: 10.1021/ar8000603. [DOI] [PubMed] [Google Scholar]
- (24).Qian Z, Liu T, Liu Y-Y, Briesewitz R, Barrios AM, Jhiang SM, Pei D. ACS Chem. Biol. 2013;8:423–431. doi: 10.1021/cb3005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Sun Y, Lu G, Tam JP. Org. Lett. 2001;3:1681–1684. doi: 10.1021/ol015889i. [DOI] [PubMed] [Google Scholar]
- (26).Virta P, Lonnberg HJ. J. Org. Chem. 2003;68:8534. doi: 10.1021/jo0344945. [DOI] [PubMed] [Google Scholar]
- (27).Heinis C, Rutherford T, Freund S, Winter G. Nat. Chem. Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- (28).Chen S, Morales-Sanfrutos J, Angelini A, Cutting B, Heinis C. ChemBioChem. 2012;13:1032–1038. doi: 10.1002/cbic.201200049. [DOI] [PubMed] [Google Scholar]
- (29).Sako Y, Morimoto J, Murakami H, Suga H. J. Am. Chem. Soc. 2008;130:7232–7234. doi: 10.1021/ja800953c. [DOI] [PubMed] [Google Scholar]
- (30).Timmerman P, Barderas R, Desmet J, Altschuh D, Shochat S, Hollestelle MJ, Höppener JWM, Monasterio A, Casal JL, Meloen RH. J. Biol. Chem. 2009;284:34126–34134. doi: 10.1074/jbc.M109.041459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lee JH, Kim H-S, Lim H-S. Org. Lett. 2011;13:5012–5015. doi: 10.1021/ol201773f. [DOI] [PubMed] [Google Scholar]
- (32).Vollrath SBL, Brase S, Kirshenbaum K. Chem. Sci. 2012;3:2716–2731. [Google Scholar]
- (33).Chen G, Goeddel DV. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- (34).Kawakami M, Cerami A. J. Exp. Med. 1981;154:631–639. doi: 10.1084/jem.154.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Beutler B, Mahoney J, Le Trang N, Pekala P, Cerami A. J. Exp. Med. 1985;161:984–995. doi: 10.1084/jem.161.5.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Esposito E, Cuzzocrea S. Curr. Med. Chem. 2009;16:3152–3167. doi: 10.2174/092986709788803024. [DOI] [PubMed] [Google Scholar]
- (37).Alzani R, Corti A, Grazioli L, Cozzi E, Ghezzi P, Marcucci F. J. Biol. Chem. 1993;268:12526–12529. [PubMed] [Google Scholar]
- (38).He MM, Smith AS, Oslob JD, Flangan WM, Braisted AC, Whitty A, Cancilla MT, Wang J, Lugovskoy AA, Yoburn JC, Fung AD, Farrington G, Eldredge JK, Day ES, Cruz LA, Cachero TG, Miller SK, Friedman JE, Choong IC, Cunningham BC. Science. 2005;310:1022–1025. doi: 10.1126/science.1116304. [DOI] [PubMed] [Google Scholar]
- (39).Chan DS, lee H, Yang F, Che C, Wong CCL, Abagyan Ruben., Leung C, Ma D. Angew. Chem. Int. Ed. 2010;49:2860–2864. doi: 10.1002/anie.200907360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Choi H, Lee Y, Park H, Oh D-S. Bioorg. Med. Chem. Lett. 2010;20:6195–6198. doi: 10.1016/j.bmcl.2010.08.116. [DOI] [PubMed] [Google Scholar]
- (41).Buller F, Zhang Y, Scheuermann J, Schafer J, Buhlmann P, Neri D. Chem. Biol. 2009;16:1075–1086. doi: 10.1016/j.chembiol.2009.09.011. [DOI] [PubMed] [Google Scholar]
- (42).Leung CH, Chan DS, Kwan MH, Cheng Z, Wong CY, Zhu GY, Fong WF, Ma DL. ChemMedChem. 2011;6:765–768. doi: 10.1002/cmdc.201100016. [DOI] [PubMed] [Google Scholar]
- (43).Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. J. Am. Chem. Soc. 2006;128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- (44).Liu R, Marik J, Lam KS. J. Am. Chem. Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- (45).Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- (46).Houghten RA, Pinilla C, Blondelle SE, Appel JR, Dooley CT, Cuervo JH. Nature. 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- (47).Furka A, Sebestyen F, Asgedom M, Dibo G. Int. J. Pep. Prot. Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- (48).Yin J, Straight PD, McLoughlin SM, Zhou Z, Lin AJ, Golan DE, Kelleher NL, Kolter R, Walsh CT. Proc. Natl. Acad. Sci. USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kodadek T, Bachhawat-Sikder K. Mol. BioSyst. 2006;2:25–35. doi: 10.1039/b514349g. [DOI] [PubMed] [Google Scholar]
- (50).Hu BH, Jones MR, Messersmith PB. Anal. Chem. 2007;79:7275–7285. doi: 10.1021/ac070418g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Hintersteiner M, Kimmerlin T, Kalthoff F, Stoeckli M, Garavel G, Seifert J-M, Meisner N-C, Uhl V, Buehler C, Weidemann T, Auer M. Chem. Biol. 2009;16:724–735. doi: 10.1016/j.chembiol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- (52).Liu T, Qian Z, Xiao Q, Pei D. ACS Comb. Sci. 2011;13:537–546. doi: 10.1021/co200101w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Thakkar A, Wavreille A-S, Pei D. Anal. Chem. 2006;78:5935–5939. doi: 10.1021/ac0607414. [DOI] [PubMed] [Google Scholar]
- (54).Chen X, Tan PH, Zhang Y, Pei D. J. Comb. Chem. 2009;11:604–611. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Martin TL, Mufson EJ, Mesulam MM. J. Histochem. Cytochem. 1984;32:793. doi: 10.1177/32.7.6736628. [DOI] [PubMed] [Google Scholar]
- (56).Khabar KS, Siddiqui S, Armstrong JA. Immunol. Lett. 1995;46:107–110. doi: 10.1016/0165-2478(95)00026-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.