Abstract
Several genome-wide association studies (GWAS) for bipolar disorder (BD) have found a strong association of the Ankyrin3 (ANK3) gene. This association spans numerous linked single nucleotide polymorphisms (SNPs) in a ~250 kb genomic region overlapping ANK3. The associated region encompasses predicted regulatory elements as well as two of six validated alternative first exons, which encode distinct protein domains at the N-terminus of the protein also known as ankyrin-G (AnkG). Using RNA Ligase-Mediated Rapid Amplification of cDNA Ends (RLM-RACE) to identify novel transcripts in conjunction with a highly sensitive, exon-specific multiplexed mRNA expression assay, we detected differential regulation of distinct ANK3 transcription start sites (TSSs) and coupling of specific 5’ ends with 3’ mRNA splicing events in post-mortem human brain and human stem cell-derived neural progenitors and neurons. Furthermore, allelic variation at the BD–associated SNP rs1938526 correlated with a significant difference in cerebellar expression of a brain-specific ANK3 transcript. These findings suggest a brain-specific cis-regulatory transcriptional effect of ANK3 may be relevant to BD pathophysiology.
Keywords: ANK3, Ankyrin-G (AnkG), Axon initial segment, bipolar disorder, stem cells, human neurons
INTRODUCTION
Bipolar disorder is a common, chronic disorder affecting 1–3% of the adult population. Decades of family and twin studies demonstrate high heritability and sibling recurrence risks1; until recently linkage and candidate gene studies have not identified consistently associated loci. Advances in our knowledge of human genetic variation, such as the data provided by the Human Genome project, HapMap project and subsequent genome-wide association studies (GWAS), have led to a wealth of new genetic associations for complex genetic diseases2. Our group and others have recently performed GWAS of bipolar disorder (BD). These studies identified genome-wide significant association (P = 9.1×10−9) between DNA variants in the Ankyrin 3 gene (ANK3) and susceptibility to BD3,4 and were subsequently replicated in several studies, including the discovery sample in the recent large Psychiatric GWAS Consortium (PGC) analysis5,6. In the early Ferreira et al study associated SNP, rs10994336 (near ANK3), had a frequency of 7.1% in Caucasian patients with BD and 5.3% in controls, conferring a disease odds ratio of 1.353. In the recent PGC analysis, the most associated SNP was rs10994397 was in LD with the previous SNP although approximately 100kb 5’. These data provide a strong rationale for further genetic and biological investigation of ANK3.
The human ANK3 gene spans ~700 kb of genomic DNA on chromosome 10q21.2, encoding multiple transcripts up to 17,020 nucleotides in length (44 exons, RefSeq ID NM020987)7. ANK3 expression shows tissue and cell-specific variation, and encodes multiple protein isoforms (AnkG isoforms, e.g. at least six in skeletal muscle8). These isoforms include distinct combinations of highly conserved protein domains, including a membrane-binding domain consisting of 23 ankyrin repeats, a spectrin binding domain, a death domain, and several domains of unknown function. As suggested by the existence of these domains, AnkG is a multifunctional protein, which forms a critical component of the axon initial segment (AIS) and of nodes of Ranvier, with described functions in the development and physiological regulation of neural activity9–12. The association of ANK3 with BD is consistent with the idea that the expression of ANK3 may change neuronal development or activity in ways that increase an individual’s susceptibility to BD.
The primary association signal (5’ BD risk allele) overlaps two previously described first exons of ANK3 (exon1b and exon1e)7,13 and their associated promoters, but does not overlap the 3’ portions of ANK3. A second association signal has been reported near the 3’ end of ANK3 (3’ BD risk allele). In previous research, the mouse homolog of exon1b was shown to be brain-specific (b = brain), whereas exon1e is ubiquitously expressed (e = everywhere)13. The GWAS for BD detected association for SNPs located within a ~250 kb region of linkage disequilibrium (r2 < 0.5) including ANK3 exon1e, exon1b and the surrounding promoter and intronic territory. The most strongly associated imputed SNP is rs10994336 (P = 9.1 X 10−9), while the most associated directly genotyped SNP in our samples is rs1938526 (P = 1.3 × 10−8)4. In the recent PGC BD study6 rs1938526 is only 21kb 5’ of the most strongly associated SNP (rs10994397; P = 7.1×10−9) and has a genomic-control corrected P = 2.0×10−8. The restricted territory of the association suggests that functional effects of the genetic variant are also linked to this 5’ region of ANK3. A second independent association signal was detected with a peak at rs98041905,6, but will not be addressed further in this manuscript. In the absence of associated coding changes despite considerable investigation (data not shown), we hypothesized that BD risk variants could exert their functional effects by altering the regulation of ANK3 expression, either through altered promoter activity or splice site choice.
In this study, we sought to address transcriptional regulation of distinct ANK3 isoforms in the human brain, as well as assessing the correlation of rs1938526 with expression levels of specific mRNA transcripts. Due to the large diversity of transcripts, we first identified and characterized distinct transcriptional start sites (TSS) by RNA Ligase-Mediated Rapid Amplification of cDNA Ends (RLM-RACE). Using a highly sensitive, multiplexed, exon-specific custom mRNA expression assay, we quantified differential regulation of ANK3 transcripts and splice variants in postmortem human brain samples from three brain regions, in the developing human brain, and in human neural progenitors and neurons derived from induced pluripotent stem cells in vitro. We also report a correlation between cerebellar expression levels of a particular brain-specific ANK3 isoform and genetic variation at ANK3 associated with BD.
METHODS
SNP analysis of DNA from post-mortem brains
Genome-wide SNP data (Affymetrix 5.0) was previously collected for all samples in the Stanley Medical Research Institute (SMRI) Brain Bank collection. RNA was used from tissue from frontal cortex (BA9), anterior cingulate (BA24) and cerebellum (lateral cerebellar hemisphere). All microarray datasets are publicly available from the Stanley Online Genomics database (http://www.stanleygenomics.org). The genotype of individuals was ascertained for a subset of SNPs indicated by the BD GWAS dataset in Ferreira et al.4, in order to select samples for inclusion. The primary selection criterion for analysis was carrier status at the most strongly disease-associated directly genotyped SNP at the ANK3 locus is rs1938526 (C/T), which is in linkage disequilibrium (LD) with the most strongly associated imputed SNP, rs10994336 (r2 = 0.65). The limited degree of LD between the top genotyped SNP and the top imputed SNP preclude a confident imputation and association between the top imputed SNP and the expression data. Eleven postmortem individuals were heterozygous at rs1938526 (C/T), and 29 matching homozygous major allele (T/T) individuals were selected for comparison.
ANK3 RLM-RACE
RLM-RACE utilizes enzymatic steps to effectively replace the 5’ caps of full-length mRNAs with a synthetic RNA linker, followed by random-primed cDNA synthesis. RLM-RACE was performed according to manufacturer’s instructions (Ambion Inc.), using two specific 3’ primers targeting ANK3 exon5 to target long isoforms and ANK3 exon31 to target short isoforms. These 3’ primer positions were selected for optimal annealing temperatures and an expected distance of under 1000 nucleotides from the synthetic 5’ RACE linker, based on previously annotated ANK3 transcripts. Subsequent PCR with transcript-specific and 5’ linker-specific primers selectively amplified 5’ ends of ANK3 mRNAs (see Supplementary Information for sequences). RLM-RACE was conducted with total RNA samples from human adult frontal cortex, hippocampus, thalamus, cerebellum, whole brain, heart and kidney, obtained from Ambion Inc, and the MGH Brain Bank. The PCR products were then cloned into the pGEM-T vector and multiple clones were bidirectionally sequenced (T7 and SP6) to determine the 5’ ends of the transcripts (minimum of 3 clones sequenced for each of 21 RLM-RACE PCR products).
Production of synthetic ANK3 target RNA
A synthetic plasmid was designed with all fifteen ANK3 target sequences in linear order, with 50 nucleotide nonhomologous spacers (ERCC sequences). The resulting 2250 nucleotide sequence was created and cloned into a pEX-1 vector by Blue Heron, Inc. The plasmid was amplified and linearized using a 3’ PciI restriction endonuclease site. In vitro transcription was performed in duplicate reactions with the MegaScript T7 kit (Ambion Inc.), and RNA products were analyzed by standard gel electrophoresis, Bioanalyzer 2100, and Nanostring nCounter assays with a custom codeset at several serially diluted concentrations (0.5 fM, 5 fM, 50 fM and 500 fM).
Collection and RNA extraction from fetal tissues
Discarded fetal tissues were collected in accordance with the Partners Healthcare IRB and the Brigham and Women’s Perinatology Pathology Service. Fetal samples were only available from a limited range of developmental stages, and were derived from normal material without known genetic abnormalities. The tissue was collected within 24 hours of surgical procedures flash frozen on dry ice, then stored at −80°C until RNA extraction. Some samples were frozen with RNA-Later (Qiagen, Inc.). Whole brain RNA extraction was performed with standard protocols (RNEasy or miRNEasy, Qiagen Inc.), quantified by Nanodrop or Bioanalyzer, and stored at −80°C.
Cell culture of proliferative human neural progenitors and differentiated neurons
EnStem-A™ human neural progenitor cells (NPCs) derived from WA09 (H9) human embryonic stem cells were obtained from a commercial source (EMD-Millipore), expanded in neural expansion medium [70% DMEM (Invitrogen), 30% Ham's F-12 (Mediatech) supplemented with 1% B-27 (Invitrogen), 20 ng/ml each EGF (Sigma) and bFGF (R&D Systems)] on 20 µg/ml poly-ornithine (Sigma)/5 µg/ml laminin (Sigma) coated tissue culture-treated 6-well plates (BD-Falcon). Terminal neural differentiation was achieved by plating expanded cells at a seeding density of 60,000 cells per cm2 on poly-ornithine/laminin plates as above in neural expansion medium lacking both EGF and bFGF mitogens to induce mitotic withdrawal and differentiation with medium replacement every 3–5 days for 18 days as ascertained by the appearance of distinct neural morphology. Cell samples were collected by manual scraping, followed by centrifugation pelleting to remove aspirate, followed by quick-freezing on dry ice. Total RNA and miRNA was purified using miRNeasy columns (Qiagen) as per vendor instructions, quantified by Nanodrop (Thermo Scientific) and stored at −80°C for further analyses.
Quantitative expression assay
A custom nCounter probeset (Nanostring Inc.) composed of 15 ANK3 probes along with an additional set of 210 probes (Supplementary Table 1) to control genes and other genes unrelated to this study was hybridized with 100 ng samples of total RNA (or synthetic ANK3 target RNA, described above) and processed according to the manufacturer’s instructions.
ANK3 expression and splice isoform analysis
RNA samples (1 to 2 µg) were obtained from the three brain regions (frontal cortex, cingulate cortex, and cerebellum) available at the time from the Stanley Medical Research Institute Array Collection. Samples were processed from 40 post-mortem individuals (11 ANK3 minor allele carrier samples (C/T), 29 homozygous major allele samples (T/T); see Supplementary Information for full details). Triplicate 100 ng RNA samples were processed for the three brain regions from each individual, and imaged for digital quantification of expression using the Nanostring nCounter system.
Data Processing and analysis
The Nanostring nCounter system provides a digital count for each target ID, as well as positive and negative spike-in target controls. Raw expression data was compiled into a data matrix with the InforSense software suite (ID Business Solutions, Ltd.), baseline subtracted to the median value of the eight negative spike-in controls and normalized to the six positive spike-in controls. Subsequently, the ANK3 probe values were normalized using the relative hybridization factors derived from the synthetic target experiments and global expression values were normalized using Variance Stabilization Normalization (described in more detail in Supplementary Information).
RESULTS
Detection of transcriptional start sites by RLM-RACE
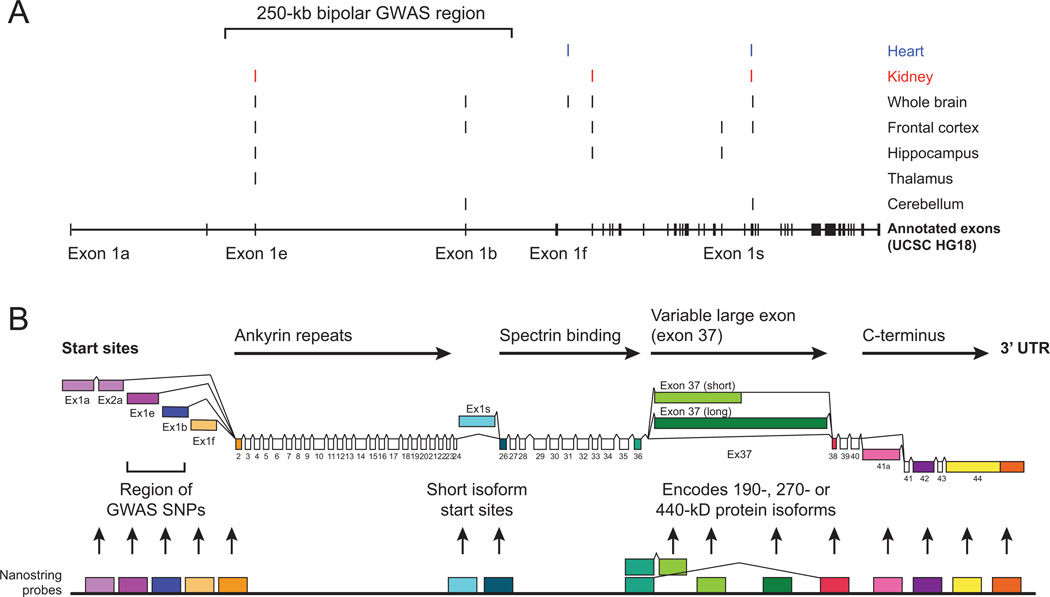
ANK3 encodes multiple alternatively spliced transcripts, including at least 53 known exons (mRNA lengths ranging between ~5 kb and ~17 kb)7,8. We used RLM-RACE, a modified version of 5’-RACE, to detect potential novel 5’ termini and additional splice transcripts using two specific 3’ primers, which anneal to ANK3 exon5 to target long transcripts and ANK3 exon31 to target short transcripts. This procedure was used to determine alternative transcription start sites (TSSs) for ANK3 mRNAs from adult human frontal cortex, hippocampus, thalamus, and cerebellar RNA, as well as from whole brain, heart and kidney RNA samples. Distinct PCR product bands were excised, cloned and a minimum of three cloned inserts from each of the 21 PCR products were sequenced in both orientations (Figure 1).
Figure 1. ANK3 transcription start sites and alternative splicing.
A) ANK3 transcription start sites (TSS) were identified by 5’ RLM-RACE with mRNA isolated from different human tissue samples, including heart, kidney and five different brain samples. Start sites are plotted relative to human ANK3 mRNA clones from the NCBI database, displayed in the lower section of the plot. Transcription start sites and putative promoters for exon1b and exon1e are within a ~250 kb region containing multiple SNPs which were associated with bipolar disorder in recent GWAS, marked in black at the top of the chart. B) Reference diagram of ANK3 transcript isoforms, and corresponding protein-coding domains. The diagram incorporates the alternative first exons from the transcription initiation sites described above, as well as alternative splice isoforms annotated in NCBI for human and mouse ANK3 loci. Differential splicing of the variable large exon (exon37) produces mRNA encoding 190, 270 or 440 kD AnkG isoforms. Fifteen Nanostring nCounter probes were designed to target distinct exons or splice junctions (arbitrary color code for ease of reference).
Six TSSs were detected for ANK3 in brain samples, including exon1e, exon1b, exon1f, exon2, exon1s, and exon26. Four of these start sites were also found in heart or kidney samples; exon 1b and exon1s were detected only in brain tissues, but not in heart or kidney. The RLM-RACE data, together with previously annotated alternative splice variation at exon37 and exon41, indicated multiple complex patterns of ANK3 transcript expression in the brain, which required effective methods to quantify expression of the distinct transcripts in parallel.
Quantitative analysis of ANK3 transcript expression
Pilot experiments indicated the utility of the Nanostring nCounter14 system for quantitative analysis of ANK3 isoform expression, which provides extremely accurate and replicable digital counts of target mRNA transcripts. This technology is based on direct hybridization and quantification of mRNA molecules with fluorescently “bar-coded” probes. Based on our RLM-RACE and publicly available data, we designed a custom Nanostring probeset including 15 ANK3 exon-specific probes as well as 210 probes for other mRNA targets, including controls for normalization. This number of probes provided a sufficient basis for normalization of expression data between samples and across brain regions (Supplementary Figure 1, Supplementary Table 1). The 15 ANK3 exon-specific probes fall into three main classes; probes for distinct first exons which correspond to unique TSSs (ANK3 exon1a, exon1e, exon1b, exon1f, and exon1s), probes for alternative internal splice variants (ANK3 exon37 short, long or skipped splice (i.e. exon36–38) variants, and exon 41a), and shared exons (ANK3 exon2, exon26, exon42, exon44 short 3’ untranslated region (UTR) and exon44 long 3’ UTR.
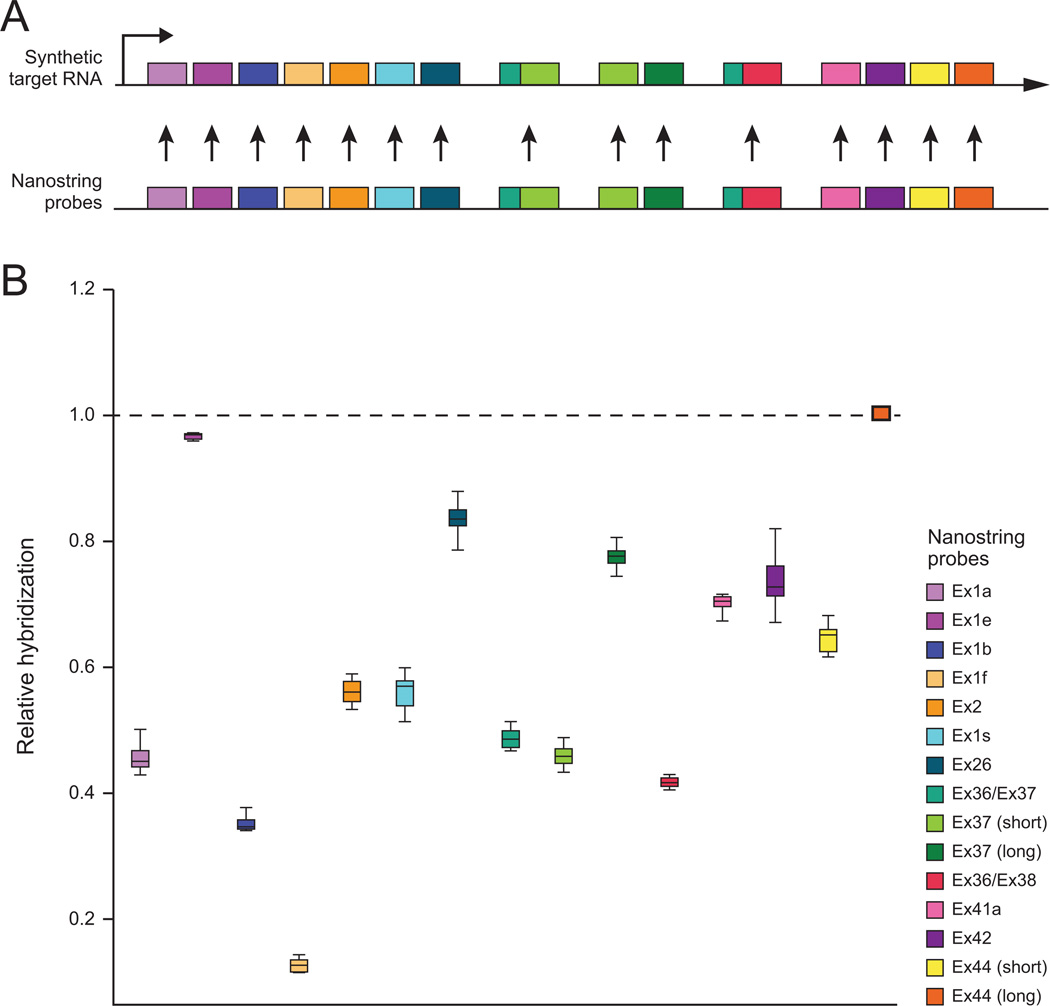
The Nanostring nCounter system provides accurate probe detection counts that can be compared across samples; however, each probe has different hybridization kinetics and thus data from different probes cannot immediately be compared. In order to compare expression levels of different ANK3 transcripts, we devised a method to calculate a relative hybridization factor (RHF) in order to compare the detection count data across each of the 15 ANK3 probes in our Nanostring probeset (see Supplementary Information for description). In the case of ANK3, for each sample, we derived a hybridization ratio for each probe relative to the probe with the highest number of counts, i.e. the highest relative hybridization (the probe for ANK3 exon44 long 3’ UTR sequence, which we designated as having RHF = 1). This probe therefore should show the lowest level of variation between technical replicates; it is serendipitous that it is also the terminal target sequence on our in vitro transcript. The ratio was consistent across a range of concentrations from ~0.5 femtoMolar (fM) to ~500 fM input RNA, with minimal deviation for all 15 probes (i.e. the standard deviation of each RHF was <3% from the average for the 6 samples ranging from ~50 fM to ~500 fM input RNA) (Figure 2). We derived the average ratio for each probe across eight IVT reactions at ~5 fM to ~500 fM, which we subsequently used as the probe-specific RHFs. The primary data for each brain RNA sample was then modified for the ANK3 probes by adjusting each value by the resulting probe-specific RHFs, thus providing a method for quantitative comparison of distinct ANK3 probes (and targeted transcripts) across samples.
Figure 2. Relative hybridization efficiency of ANK3 probes.
A) Plasmid DNA containing all 15 ANK3 target sequences in linear order was transcribed in vitro (Synthetic Target RNA) and hybridized with a custom nCounter probeset in triplicate for ~0.5, ~5, ~50 and ~500 fM concentrations. B) Average relative hybridization value for each of the 15 ANK3 probes. The average values were derived from data for 2 synthetic transcript hybridization samples at 5 femtoMolar (fM), 3 samples at 50 fM, and 3 samples at 500 fM. The variance in relative counts between these samples was less than 5%, indicating robust linear scaling of probe counts across this range of input concentrations. Synthetic target RNA hybridization to ANK3 probes yielded consistent relative hybridization coefficients for each probe calculated relative to the terminal probe for ANK3 exon44 (yellow, circled in black). These values were used for the calculation of probe-specific relative hybridization factors (RHFs).
Analysis of ANK3 expression and regulation in three brain regions
Using the methods described above, we characterized developmental and tissue-specific differences in the expression and alternative splicing of ANK3 to facilitate subsequent analysis of expression differences associated with the ANK3 BD risk haplotype. For this purpose, we compared post-mortem adult brain samples selected on the basis of a genotyped marker for the BD-associated variant; risk-associated individuals are heterozygous (C/T) and non-risk individuals are homozygous (T/T) at rs1938526.
Expression assays were performed in triplicate with total RNA from frontal cortex, cingulate cortex and cerebellum samples derived from 40 post-mortem adult brains (from the Stanley Medical Research Institute, SMRI)15. The post-mortem samples were derived from autopsies with various causes of death, and include individuals diagnosed with bipolar disorder, schizophrenia, and matched controls. Eleven individuals were selected (from the SMRI array collection) based on genotyping data indicating heterozygosity at rs1938526 (C/T), and 29 further homozygous major allele (T/T) samples were chosen for comparison after careful matching on age, sex and diagnosis (Supplementary Table 2). There were no samples homozygous for the minor allele.
To compare ANK3 expression data across multiple samples and tissues, we serially processed the raw data with four manipulations. The first two stages were normalization to positive control spike-in probes targeting exogenous spiked-in RNA, and baseline subtraction using probe counts for negative control spike-in probes. The third process involved multiplying the ANK3 probe counts by the RHF for each ANK3 probe as described above. The final process involved a normalization step across samples using a variance stabilization normalization (VSN) algorithm implemented in R, commonly used for expression array data16 and recently used for Nanostring nCounter data17. For these comparisons of expression across tissues, we normalized the data across all three tissues (subsequent analyses normalized each tissue separately, or normalized adult brain samples with fetal and in vitro samples).
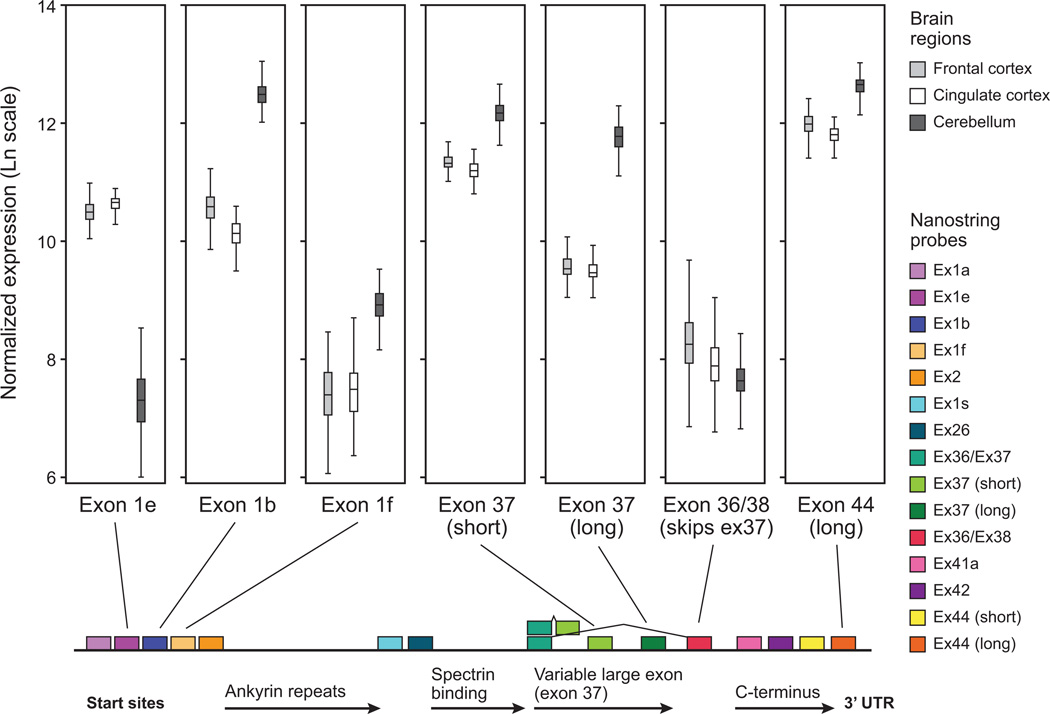
Two primary results emerged from the initial analysis comparing ANK3 transcript expression in different brain regions (Figure 3). Overall expression in cerebellum was about 2-fold higher than in frontal or cingulate cortex (as measured by probes for shared exons, such as exon44 / 3’ UTR). This correlates with changes in promoter function, resulting in a difference between cortical and cerebellar expression levels of isoforms with different transcriptional start sites. For example, isoforms containing exon1e or exon1b were expressed at roughly equal levels in frontal cortex and cingulate cortex, and the two promoters together account for the majority of ANK3 expression. However, in cerebellum, ANK3 exon1b was expressed at approximately 4-fold higher levels than in frontal or cingulate cortex, whereas expression of exon1e was dramatically lower in cerebellum than cortex (roughly 4-fold). Expression of exon1f and exon1s transcripts (shorter transcripts from 3’ TSSs) was also up-regulated in the cerebellum and was expressed at higher levels in cerebellum than exon1e, although they remained at approximately 10-fold lower levels than exon1b.
Figure 3. Differential expression of ANK3 isoforms across brain regions.
Distinct ANK3 isoforms are differentially expressed in frontal cortex, cingulate cortex, and cerebellum. Isoforms including exon1e or exon1b are expressed at similarly high levels in frontal and cingulate cortex, whereas cerebellum downregulates exon1e, and upregulates exon1b isoforms. Exon1f is expressed at low levels in frontal and cingulate cortex, and is upregulated in cerebellum although less abundantly than exon1b. Inclusion of the shorter splice variant of exon37 is preferred in frontal and cingulate cortex (encoding the 270 kD AnkG protein), whereas inclusion of the longer exon37 is upregulated in cerebellum (encoding the 440 kD AnkG protein). Splice variants that do not include exon37 (encoding the 190 kD AnkG protein) are present at low levels in all brain regions.
Second, it appeared that samples with high exon1b probe counts also had high probe counts for the long splice form of exon37, which suggests (but does not prove) that inclusion of these exons is molecularly coupled. This effect was most pronounced in cerebellum. By comparing the probe count for the long ANK3 mRNA isoforms (i.e. that include the long (~8 kb) form of exon37) to general ANK3 probes such as exon44 (3’ UTR), more than half of the transcripts in cerebellum appear to include the long splice variant (encoding a ~440 kD protein). In contrast, frontal cortex and cingulate cortex include the long splice variant in less than a quarter of ANK3 transcripts. Furthermore, all of the three brain regions express similarly low levels of transcripts that skip exon37 entirely, i.e. splicing directly from exon36 to exon38 (encoding a ~190 kD protein). This transcript variant was detected in higher proportions in other tissue samples (data not shown).
Specific effect of BD-associated haplotype on ANK3 isoform expression in cerebellum
We next determined if the expression differences were correlated with ANK3 genotype status at the BD-associated SNP, rs1938526. Comparison of ANK3 expression profiles between post-mortem adult brain samples which are heterozygous (C/T) and homozygous (T/T) at rs1938526 indicated differences in the expression of specific transcripts. Statistical comparison indicated a lower average expression in C/T heterozygotes as measured by 7 of 15 distinct ANK3 probes in cerebellum samples, with significant p-values (P < 0.05) for ANK3 exon1b, exon1f, exon26, exon37_short, exon42, exon44_short and exon44_long. This effect was not detected in frontal cortex or cingulate cortex samples. Six of these probes correspond to exon sequences that could be selectively included in exon1b-initiated medium-length and long splice isoforms (i.e. targets for exon1b, exon37, and shared probes). This finding suggests that in individuals heterozygous for rs1938526, the brain-specific ANK3 exon1b promoter from the BD-associated haplotype produces lower transcript levels, at least in a subset of cerebellar cells.
Interestingly, although no haplotype-dependent difference in expression was detected for ANK3 transcripts derived from the exon1e TSS, there was an additional significant difference in expression of exon1f in the same cerebellar samples. Since exon1f transcripts are expressed at lower levels overall, our current data cannot resolve whether exon1f transcripts include longer exon37 splice variants and contribute to the signal detected with exon37 probes. However, since overall expression of the exon1f exon was 8 to 10-fold lower than exon1b and several shared exons, the exon1f TSS presumably does not contribute as strongly as exon1b TSS to the significant expression effect seen for exon37 and other probes that target shared exons.
Developmental regulation of ANK3 expression
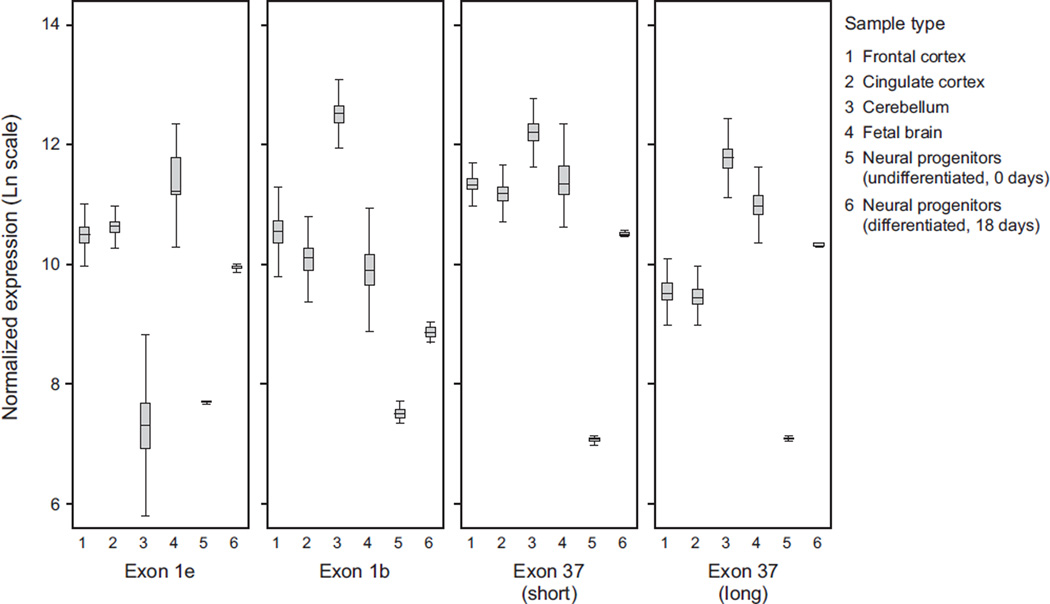
We next investigated whether expression of ANK3 isoforms displays a distinct pattern of developmental regulation during fetal development, and during the transition from the proliferative state of human stem-cell derived neural progenitor cells (NPCs) to lineage-committed, differentiated, post-mitotic neurons. We observed a relatively consistent pattern of ANK3 isoform expression in fetal brain samples from gestational week 13, 15, 17, 19, and 21. Fetal samples were only available from this limited range of developmental stages, and were not derived from known genetically abnormal samples. The overall consistency of the fetal expression data across these gestational time points provided a good indication of its reliability, and allowed for derivation of average ANK3 expression values across this gestational period. The overall level of ANK3 expression from samples within this developmental period was similar to adult expression; however, fetal brain samples showed a pattern of ANK3 isoform expression which was distinct from the adult frontal cortex, cingulate cortex and cerebellar samples. Fetal brain ANK3 isoform expression was dominated by the exon1e start site, with ~4-fold lower level of expression from the exon1b promoter. Furthermore, most fetal transcripts included the long splice form of exon37 (Figure 5).
Figure 5. Developmental regulation of ANK3 isoforms.
Average ANK3 isoform expression is displayed for 6 sample types, for data from the exon1e, exon1b, exon37 (short) and exon37 (long) probes. The samples are 1) adult frontal cortex (n=40), 2) adult cingulate cortex (n=40), 3) adult cerebellum (n=40), 4) fetal brain samples ranging from 13 to 21 weeks gestational age (n=5), 5) EnStem proliferative neural progenitor cells (NPCs) (n = 1) and 6) differentiated NPCs (differentiated for 18 days) (n = 1).
In order to further investigate the mechanism(s) of transcriptional regulation of distinct ANK3 transcripts, we compared ANK3 expression in human NPCs in the proliferative state and after 18 days of differentiation. We observed ~25% lower overall ANK3 expression in proliferative NPCs than in fetal or adult samples, with a substantially different expression profile (Figure 5). Proliferative NPCs expressed ~50% ANK3 transcripts with exon1e and exon1b, and ~50% short isoforms which appeared to initiate at ANK3 exon26 (or an uncharacterized alternative start site, but not exon1s). Furthermore, ANK3 transcripts expressed in proliferative NPCs did not include exon37, as indicated by elevated counts from the probe for the exon36–exon38 splice junction, and low counts from all three probes targeting exon37.
After 18 days of in vitro differentiation, the ANK3 transcript expression profile converted to a pattern that is similar to the fetal samples described above, although with ~50% lower overall ANK3 expression than in the fetal or adult samples (Figure 5). Compared to the proliferative NPCs, the differentiated neurons exhibited higher expression of ANK3 exon1e and exon1b transcripts, and higher expression of the long splice form of exon37. This indicates a regulatory change of ANK3 transcription and splicing during the transition from proliferative NPCs to differentiated neurons.
Discussion
Developmental and tissue-specific regulation of ANK3 isoforms
ANK3 encodes multiple differentially regulated isoforms that play distinct roles across multiple tissues. Alternative ANK3 mRNAs encode distinct protein variants, and could alter 5’ or 3’ UTR mechanisms involved in the localization, translational regulation or stability of the mRNA transcripts. Inclusion of alternative exons could change cellular processes through downstream effects on AnkG. Based on the location of the associated ANK3 regions and diversity of known ANK3 mRNA sequences in the NCBI database, we initially characterized additional ANK3 transcripts and possible 5’ exons by RLM-RACE. We detected six distinct TSS for ANK3 in brain, of which two appear to be brain-specific (exon1b TSS, and exon1s TSS for a short isoform which does not include the ankyrin repeats). Human brain tissue-specific regulation of distinct ANK3 transcripts and the extent to which alternative first exons are subsequently coupled to alternatively spliced 3’ exons had not (to our knowledge) been previously investigated. This study used a novel method for quantitative comparison of ANK3 transcript isoforms to investigate differential expression across brain regions (frontal cortex, cingulate cortex and cerebellum), and to investigate developmental regulation of transcript expression.
Frontal cortex and cingulate cortex both expressed roughly equal levels of exon1e and exon1b, and appeared to include the short splice variant of exon37 in most transcripts, encoding the 270 kD AnkG protein isoform. In contrast, the cerebellum showed a ~4 fold increase in expression of exon1b transcripts, accounting for most transcripts, and a similar increase in the long splice variant of exon37 putatively encoding the 440 kD AnkG protein isoform13, accounting for at least 50% of transcripts. Exon1e and exon1f were expressed at far lower levels than exon1b in cerebellum. Therefore, it can be concluded that the majority of ANK3 transcripts in cerebellum included exon1b and encoded roughly equivalent amounts of the 270 kD and 440 kD AnkG protein isoforms.
Since ANK3 exon1b showed restricted brain-specific expression, as previously reported for the mouse13, and exon1b and the long splice variant of exon37 showed similar patterns and levels of expression, our data suggest that a substantial portion of the neuron-specific functions of ANK3 are mediated by isoforms encoded by transcripts that include exon1b and the long splice variant of exon37. However, it remains unclear whether transcription initiation at the ANK3 exon1b promoter determines downstream splicing regulation of exon37. Furthermore, our investigation used RNA from whole tissue samples did not separate expression from the different neuronal cell types. Future expression data from different neural subtypes will provide greater resolution of ANK3 expression patterns.
We detected different patterns of ANK3 expression in fetal human brain samples as well as in human NPCs and neurons differentiated in vitro. The data suggest that human NPCs predominantly express a short isoform of ANK3 that does not include exon 37. In contrast, the 18-day in vitro differentiated human neurons show expression similar to fetal brain samples, with predominant expression of exon1e isoforms that include the long form of exon37 (encoding ~440 kD AnkG proteins). Taken together, these findings indicate that future investigation of differentiated human neurons from patient-specific induced pluripotent stem cells could provide an informative model for investigation of ANK3 regulation and function during neurodevelopment.
Cis-regulatory effect of BD-associated variant on ANK3 exon1b transcripts
The correlation between allelic status at SNP rs1938526 and cerebellar expression levels of the brain-specific ANK3 exon1b isoform (as well as the less highly expressed exon1f isoform), provides evidence that the BD GWAS risk haplotype effects gene expression. It is premature to speculate on the mechanism by which the GWAS association results may contribute to disease pathophysiology, but correlation with a localized effect on specific transcript types in a specific brain region provides a potential avenue for further targeted investigation. It remains to be determined whether the cis-regulatory effect on ANK3 exon1b (and exon1f) transcripts is specific to the cerebellum or found in other brain regions potentially involved in BD. Alternatively, the signal may simply be most clear in cerebellum due to the dominance of exon1b-initiated ANK3 transcripts in this region, as well as the limited degree of cellular diversity (and therefore lower hypothetical dilution of RNA from affected vs. unaffected cell types). Similar effects may exist in specific neural subtypes in other brain regions, including frontal and cingulate cortices examined in this study, but may not be readily detected in RNA samples extracted from the diverse mixtures of cell types in these tissues.
The rs10994336 risk genotype has been associated with risk-taking and startle response differences18, deleterious effects on sustained attention19,20, and with structural variation in the anterior limb of the internal capsule as well as impaired set-shifting and increased risk-taking21. Recent behavioral investigation in mouse models with Ank3 exon1b haploinsufficiency and with regionally targeted Ank3 knockdown indicate similar effects on increasing risk-taking behavior and changes in other behaviors reminiscent of bipolar symptoms (Leussis and Petryshen, personal communication). Although recent publications have detected several different cis-eQTL associations at the ANK3 locus, our data are the first to indicate a brain-specific cis-eQTL expression effect of the 5’ BD risk-associated haplotype. In one cis-eQTL expression analysis, allelic expression imbalance (AEI) of ANK3 was investigated in lymphoblastoid lines which express extremely low levels of ANK3; AEI was detected for a set of SNPs which were not in linkage disequilibrium with rs10994336 or rs1938526 (r2 = 0.08)22. In another publication, using post-mortem human superior temporal gyrus samples, homozygous major allele status (C/C) at the 3’ BD risk-associated SNP rs9804190 was associated with lower overall expression of ANK3. However, rs9804190 (Ch10: 61509737) is 460kb away from rs1938526 (Ch10: 61970289), and they are not in linkage disequilibrium (r2 = 0.024)23. In addition, a low frequency (0.007) non-synonymous coding variant was detected in exon41 near the 3’ end of ANK3, but it is also not in LD with rs10994336 or rs193852624.
There is a growing understanding of an important role for the cerebellum in emotional and behavioral regulation in humans25, and emerging evidence of altered cerebellar volumes in bipolar disorder26, and evidence for altered neurotrophic factor signaling in the cerebellar cortex27. Given evidence summarized in the work of Stoodley and Schmahmann28, for the topographical organization of the cerebellum into specialized regions including those for affective processing, more detailed investigation of cerebellar subregions should help determine the ANK3 transcript expression is specific to those areas that have been implicated in control of affective behavior from both imaging studies and studies of patients with cerebellar damage.
Specific function of the alternative ANK3 transcripts
The cellular effect of ANK3 variation ultimately depends on the quantity and activity of distinct AnkG proteins, their localization, post-translational modification, and protein-protein interactions. While exon1e is broadly expressed in multiple tissue types, exon1b is restricted to brain tissues13. This restriction is likely due to a conserved Neuron-Restrictive Silencer Element (NRSE)29 found ~5 kb 5’ of exon1b, which is known to recruit chromatin-modifying complexes involved in the epigenetic regulation of gene expression. ANK3 exon1b knockout mice show aberrant Purkinje cell synaptic development and display ataxic locomotion13,30 suggesting that exon1b is important in cerebellar functioning. Recent publications indicate substantial plasticity of the AnkG-dependent axon initial segment (AIS) in response to altered neural activity, thus suggesting that the AnkG proteins (and perhaps ANK3 promoters) could be dynamically responsive to cellular signaling processes31–33.
In conclusion, our initial characterization of ANK3 brain expression patterns across brain regions and developmental stages provides a foundation for further expression analyses. Our comparison of these patterns between heterozygous carriers of the BD-associated ANK3 allele and individuals homozygous for the major allele suggests a brain-specific functional correlate of the common genetic variant associated with BD. Mouse brain microarray experiments suggest that lithium treatment alters the level of ANK3 expression34,35, with possible relevance to lithium’s therapeutic effects. The BD drug lamotrigine modulates voltage-gated sodium channels that are localized to the AIS by AnkG36, consistent with a role for ANK3 in biochemical pathways targeted for pharmacological treatment of BD. A better understanding of the endogenous expression and functional variation in human ANK3 provides one avenue for investigating pathways and mechanisms of bipolar disorder and therapeutic development.
Supplementary Material
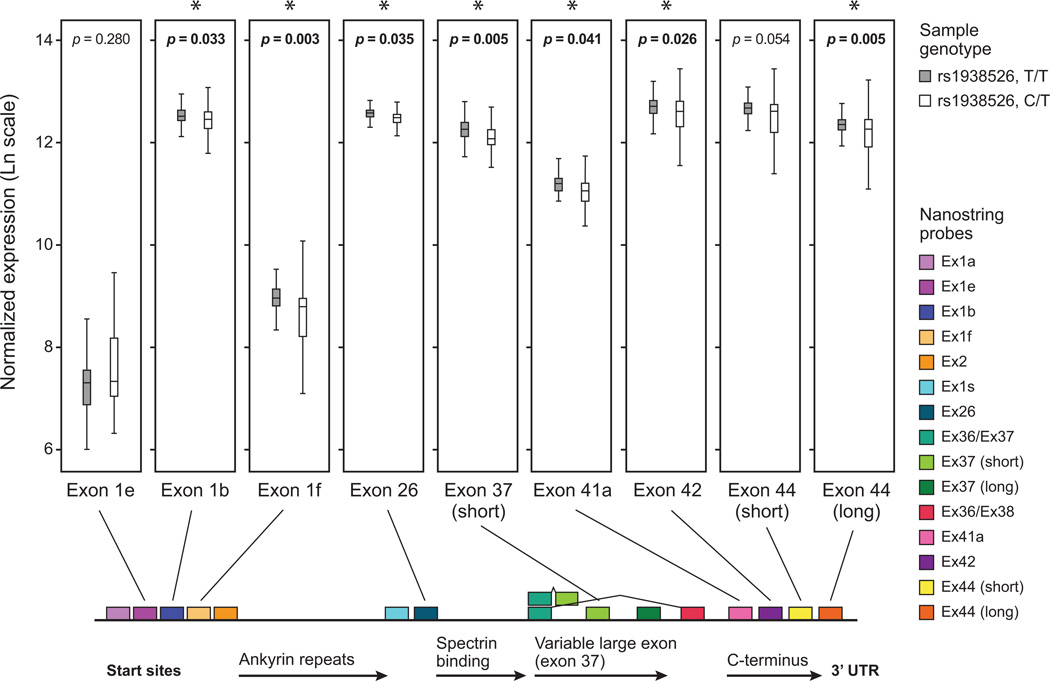
Figure 4. Effect of BD-associated ANK3 regulatory haplotype on expression of ANK3 isoforms in cerebellum.
Seven distinct probes for ANK3 indicate significantly lower average expression in cerebellum samples from individuals who are heterozygous for the BD-associated SNP rs1938526, as compared to individuals who are homozygous for the major allele. A consistent trend toward lower expression in heterozygotes is indicated by all seven probes (exon1b (p = 0.033), exon1f (p = 0.003), exon26 (p = 0.035), exon 37 (short) (p = 0.005), exon42 (p = 0.026), and exon44 (long 3’ UTR) (p = 0.005). In contrast, ANK3 exon1e does not show differential expression.
Acknowledgements
We gratefully acknowledge the Stanley Medical Research Institute (Maree Webster) for providing RNA and tissue samples from their post-mortem human brain collection and the Massachusetts General Hospital Brain Bank (Charles Vanderburg) for generously providing post-mortem human brain samples. Funding was provided by the Stanley Medical Research Institute and from the National Institute Of Mental Health (R33MH087896). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute Of Mental Health or the National Institutes of Health. We thank Drs. Tracey Petryshen and Melanie Leussis for ongoing discussions and helpful review of this manuscript.
Footnotes
Conflict of Interest
None reported.
References
- 1.Craddock N, Sklar P. Genetics of bipolar disorder: successful start to a long journey. Trends Genet. 2009;25:99–105. doi: 10.1016/j.tig.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nothen MM, et al. A genome- wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J, Pearl J, Strohmaier J, Breuer R, et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry. 2009;14:487–491. doi: 10.1038/mp.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklar P, Ripke S, Scott LJ, Andreassen OA, et al. Psychiatric GWAS Consortium Bipolar Disorder Working Group..Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–2359. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- 8.Hopitzan AA, Baines AJ, Ludosky MA, Recouvreur M, Kordeli E. Ankyrin-G in skeletal muscle: tissue-specific alternative splicing contributes to the complexity of the sarcolemmal cytoskeleton. Exp Cell Res. 2005;309:86–98. doi: 10.1016/j.yexcr.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Bennett V, Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol Med. 2008;14:28–36. doi: 10.1016/j.molmed.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Pan Z, Kao T, Horvath Z, Lemos J, Sul JY, Cranstoun SD, Bennett V, Scherer SS, Cooper EC. A common ankyrin-G-based mechanism retains KCNQ and NaV channels at electrically active domains of the axon. J Neurosci. 2006;26:2599–2613. doi: 10.1523/JNEUROSCI.4314-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohler PJ, Bennett V. Defects in ankyrin-based cellular pathways in metazoan physiology. Front Biosci. 2005;10:2832–2840. doi: 10.2741/1739. [DOI] [PubMed] [Google Scholar]
- 12.Bennett V, Chen L. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr Opin Cell Biol. 2001;13:61–67. doi: 10.1016/s0955-0674(00)00175-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhou D, Lambert S, Malen PL, Carpenter S, Boland LM, Bennett V. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J Cell Biol. 1998;143:1295–1304. doi: 10.1083/jcb.143.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 15.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH. The Stanley Foundation Brain collection and neuropathology consortium. Schizophr Res. 2000;44:151–155. doi: 10.1016/S0920-9964(99)00192-9. [DOI] [PubMed] [Google Scholar]
- 16.Huber W, Von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(suppl. 1):S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 17.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roussos P, Giakoumaki SG, Georgakopoulos A, Robakis NK, Bitsios P. The CACNA1C and ANK3 risk alleles impact on affective personality traits and startle reactivity but not on cognition or gating in healthy males. Bipolar Disord. 2011;13:250–259. doi: 10.1111/j.1399-5618.2011.00924.x. [DOI] [PubMed] [Google Scholar]
- 19.Ruberto G, Vassos E, Lewis CM, Tatarelli R, Girardi P, Collier D, Frangou S. The cognitive impact of the ANK3 risk variant for bipolar disorder: initial evidence of selectivity to signal detection during sustained attention. PLoS One. 2011;6:e16671. doi: 10.1371/journal.pone.0016671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hatzimanolis A, Smyrnis N, Avramopoulos D, Stefanis CN, Evdokimidis I, Stefanis NC. Bipolar disorder ANK3 risk variant effect on sustained attention is replicated in a large healthy population. Psychiatr Genet. 2012 Apr 11; doi: 10.1097/YPG.0b013e328353ae79. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linke J, Witt SH, King AV, Nieratschker V, Poupon C, Gass A, Hennerici MG, Rietschel M, Wessa M. Genome-wide supported risk variant for bipolar disorder alters anatomical connectivity in the human brain. Neuroimage. 2012;59:3288–3296. doi: 10.1016/j.neuroimage.2011.10.083. [DOI] [PubMed] [Google Scholar]
- 22.Quinn EM, Hill M, Anney R, Gill M, Corvin AP, Morris DW. Evidence for cis-acting regulation of ANK3 and CACNA1C gene expression. Bipolar Disord. 2010;12:440–445. doi: 10.1111/j.1399-5618.2010.00817.x. [DOI] [PubMed] [Google Scholar]
- 23.Roussos P, Katsel P, Davis KL, Bitsios P, Giakoumaki SG, Jogia J, Rozsnyai K, Collier D, Frangou S, et al. Molecular and Genetic Evidence for Abnormalities in the Nodes of Ranvier in Schizophrenia. Arch Gen Psychiatry. 2012;69:7–15. doi: 10.1001/archgenpsychiatry.2011.110. [DOI] [PubMed] [Google Scholar]
- 24.Dedman A, McQuillin A, Kandaswamy R, Sharp S, Anjorin A, Gurling H. Sequencing of the ANKYRIN 3 gene (ANK3) encoding ankyrin G in bipolar disorder reveals a non-conservative amino acid change in a short isoform of ankyrin G. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:328–335. doi: 10.1002/ajmg.b.32030. [DOI] [PubMed] [Google Scholar]
- 25.Villanueva R. The cerebellum and neuropsychiatric disorders. Psychiatry Res. 2012 Mar 20; doi: 10.1016/j.psychres.2012.02.023. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 26.Baldaçara L, Nery-Fernandes F, Rocha M, Quarantini LC, Rocha GG, Guimarães JL, Araújo C, Oliveira I, Miranda-Scippa A, Jackowski A. Is cerebellar volume related to bipolar disorder? J Affect Disord. 2011;135:305–309. doi: 10.1016/j.jad.2011.06.059. [DOI] [PubMed] [Google Scholar]
- 27.Soontornniyomkij B, Everall IP, Chana G, Tsuang MT, Achim CL, Soontornniyomkij V. Tyrosine kinase B protein expression is reduced in the cerebellumof patients with bipolar disorder. Journal of Affective Disorders. 2011;133:646–654. doi: 10.1016/j.jad.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoodley CJ, Schmahmann JD. Evidence for topographic organization in the cerebellum of motor control versus cognitive and affective processing. Cortex. 2010;46:831–844. doi: 10.1016/j.cortex.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schoenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1995;267:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 30.Ango F, di Cristo G, Higashiyama H, Bennett V, Wu P, Huang ZJ. Ankyrin-based subcellular gradient of neurofascin, an immunoglobulin family protein, directs GABAergic innervation at purkinje axon initial segment. Cell. 2004;119:257–272. doi: 10.1016/j.cell.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010;465:1070–1074. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuba H, Oichi Y, Ohmori H. Presynaptic activity regulates Na(+) channel distribution at the axon initial segment. Nature. 2010;465:1075–1078. doi: 10.1038/nature09087. [DOI] [PubMed] [Google Scholar]
- 33.Grubb MS, Shu Y, Kuba H, Rasband MN, Wimmer VC, Bender KJ. Short- and long-term plasticity at the axon initial segment. J Neurosci. 2011;31:16049–16055. doi: 10.1523/JNEUROSCI.4064-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McQuillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mRNA to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenet Genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- 35.Nanavati D, Austin DR, Catapano LA, Luckenbaugh DA, Dosemeci A, Manji HK, Chen G, Markey SP. The effects of chronic treatment with mood stabilizers on the rat hippocampal post-synaptic density proteome. J Neurochem. 2011;119:617–629. doi: 10.1111/j.1471-4159.2011.07424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lang DG, Wang CM, Cooper BR. Lamotrigine, phenytoin and carbamazepine interactions on the sodium current present in N4TG1 mouse neuroblastoma cells. J Pharmacol Exp Ther. 1993;266:829–835. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.