Abstract
The E3 ubiquitin ligase CHIP (C-terminus of Hsc70 Interacting Protein, a 70 kDa homodimer) binds to the molecular chaperone Hsc70 (a 70 kDa monomer) and this complex is important in both the ubiquitination of Hsc70 and the turnover of Hsc70-bound clients. Here we used NMR spectroscopy, bio-layer interferometry, and fluorescence polarization to characterize the Hsc70-CHIP interaction. We found that CHIP binds tightly to two molecules of Hsc70 forming a 210 kDa complex, with a Kd of approximately 60 nM, and that the IEEVD motif at the C-terminus of Hsc70 (residues 642–646) is both necessary and sufficient for binding. Moreover, the same motif is required for CHIP-mediated ubiquitination of Hsc70 in vitro, highlighting its functional importance. Relaxation-based NMR experiments on the Hsc70-CHIP complex determined that the two partners move independently in solution, similar to “beads on a string”. These results suggest that a dynamic C-terminal region of Hsc70 provides for flexibility between CHIP and the chaperone, allowing the ligase to “search” a large space and engage in productive interactions with a wide range of clients. In support of this suggestion, we find that deleting residues 623–641 of the C-terminal region, while retaining the IEEVD motif, caused a significant decrease in the efficiency of Hsc70 ubiquitination by CHIP.
Heat shock cognate 70 (Hsc70) is the constitutively expressed member of the Hsp70 family of molecular chaperones of Mr 70 kDa. As a central component of the protein quality control system, Hsc70 works together with a list of co-chaperones to facilitate the refolding and degradation of misfolded proteins.1–4 One of the key co-chaperones of Hsc70 is the carboxyl-terminus of Hsc70-interacting protein (CHIP), a 70 kDa homodimer which is an E3 ubiquitin ligase that facilitates the ubiquitination and degradation of Hsc70-bound “client” proteins.5 This concerted activity between Hsc70 and CHIP has been shown to reduce the levels of disease-associated and misfolded client proteins, such as the polyglutamine expanded androgen receptor, the microtubule-binding protein tau and the cystic fibrosis transmembrane conductance regulator.6–9 In addition to transferring ubiquitin to Hsc70-bound clients, CHIP also modifies the chaperone itself by the addition of both canonical and non-canonical polyubiquitin chains.10,11 Ubiquitination of Hsc70 increases its chaperone activity,12 promotes endosomal trafficking13 and enhances turnover of the chaperone.14 Together, these findings have led to much interest in understanding this important protein-protein interaction.
Previous studies have shown that the C-terminal eight amino acids of Hsc70, GPTIEEVD-COOH, are necessary for the interaction with CHIP.15,16 Interestingly, a similar motif, TSRMEEVD-COOH is found at the C-terminus of heat shock protein 90 (Hsp90)17 and CHIP is thought to work with both Hsc70 and Hsp90 to promote degradation of misfolded proteins.18 The CHIP interface that binds to both chaperones is known as a tetratricopeptide repeat (TPR) domain. TPR domains consist of 3 to 16 degenerate 34 amino acid motifs that form anti-parallel α-helical hairpins.19,20 This structurally conserved domain is commonly found to facilitate protein-protein interactions in a variety of cellular contexts.21,22 Informative co-crystal structures with the TPR domain of CHIP 23 or another co-chaperone, Hsp70-Hsp90 organizing protein (Hop)24, and a C-terminal peptide of Hsc70 suggest that the GPTIEEVD sequence binds in an extended confirmation along a shallow groove parallel to the α-helical axis of the TPR domain. The main electrostatic interactions believed to hold the peptide in place form a “carboxylate clamp” between Lys and Asn side-chains of the TPR domain and the last residue Asp side-chain and backbone carboxylate of the peptide.23,24
Despite several insights into the Hsc70-CHIP complex, many important questions remain. We were particularly interested in whether regions of Hsc70 outside the EEVD motif might be involved in binding CHIP. Previous hydrogen-deuterium exchange experiments revealed that the GPTIEEVD peptide and full length Hsc70 cause similar changes in amide-hydrogen protection within CHIP,25 suggesting that the peptide motif by itself is sufficient to mediate the complete interaction. While the binding of full-length Hsc70 to CHIP has yet to be studied in rigorous detail, regions outside of the TPR domain in Hop are believed to be involved in binding to full-length Hsp70,17,26 raising the possibility that this is also true for Hsc70 and CHIP. We additionally wanted to explore the structure of the Hsc70-CHIP complex in solution to understand how CHIP might be able to promote the ubiquitination of both Hsc70 and client proteins bound to the chaperone. This functional promiscuity would presumably require CHIP to sample a relatively wide area around Hsc70 and we were interested in how this might be accomplished. These inquiries have ramifications for our understanding of both the Hsc70- and Hsp90-CHIP complex, as both are likely to have similar binding modes and patterns of ubiquitination.27
Towards those goals, we characterized the interaction of Hsc70 binding to CHIP by biolayer interferometry (BLI), fluorescence polarization (FP) and NMR. These findings confirmed that the EEVD motif is required for binding to CHIP, and that full-length Hsc70 binds only ~6-fold tighter to CHIP than the isolated peptide suggesting that very little additional interactions beyond the IEEVD take place. Our solution-state NMR studies show that full-length Hsc70 and CHIP move independently of each other in the complex, similar to “beads on a string”. Additional interactions between CHIP and Hsp70 beyond EEVD, which may account for the extra 1.08 kCal/Mole in binding free energy, must therefore be transient, and not localized. These results refine our understanding of how Hsc70 binds to CHIP and provide a model for how the Hsc70-CHIP complex might function in protein quality control.
Experimental Procedures
Plasmids and Protein Purification
Human Hsc70 (HspA8; 1–646), Hsc70 (1–383) and Hsc70 (391–646) DNA sequences were amplified by PCR and inserted into the pMCSG7 expression vector by ligation-independent cloning as previously described.28 The Hsc70 ΔTail mutant was made using partially overlapping site-directed mutagenesis primers based off of a previously described protocol.29 Forward primer: GCCTGGGGGACCTTTTATTGAAGAGGTTGATTAAGGTGGAGCTC and reverse primer GGAGGAGCTCCACCACCTTAATCAACCTCTTCAATAAAAGGTCCCC were used to remove amino acids 623–641 from the ensuing sequence. Mutagenesis was carried out using the QuickChange protocol developed by Stratagene. Human CHIP (STUB1; 1–303) and UbcH5c (UBE2D3; 1–147) were inserted in the pGEX-6P vector. Expression and purification of all Hsc70 proteins was carried out as previously described,30 except the N-terminal His6 purification tag was not removed during the process. Additionally, ATP-agarose was not used to purify Hsc70 (391–646), as this mutant lacks the nucleotide-binding domain. Isotope labeling was accomplished using our previously reported protocol.31 All peptide sequences were a generous gift from Dr. Duxin Sun (University of Michigan).
Following expression in BL21 E. coli cells, full-length human CHIP was purified by batch processing with GST-sepharose affinity resin (GE Healthcare). The GST tag was then removed by incubating the resin overnight with Turbo Human Rhinovirus 3C protease (Excellgen, Rockville MD) at 4 °C. Free CHIP was isolated in the supernatant, concentrated, and further purified over a Superdex S200 10/300 gel-filtration column (GE Healthcare) in buffer containing 10 mM HEPES pH 7.4, 150 mM NaCl, and 0.05% Tween.
Biolayer Interferometry and Kinetic Analysis
For all binding experiments Hsc70 proteins and the C-terminal peptide were labeled at a 1:1 molar ratio with EZ-link NHS-LC-Biotin (Thermo Scientific) according to the manufacturer's instructions. Using an Octet Red96 system (ForteBio) at 25°C, biotinylated molecules were immobilized on streptavidin (SA) biosensors at a well concentration of 0.5 µM in Buffer A (25 mM HEPES pH 7.4, 5 mM MgCl2, 10 mM KCl, 150 mM NaCl). Immobilization was carried out over 10 minutes, followed by a 10 min incubation in 100 µg/ml biocytin to cap any free streptavidin sites on the biosensor surface. Finally, a 10 min equilibration in Binding buffer (Buffer A with 0.1 mM 2-mercaptoethanol and 0.01% Triton-X 100) was used to prepare the biosensors for CHIP binding. All binding measurements were carried out at the indicated CHIP concentrations over the indicated time period. A single biosensor was used to measure binding to each CHIP concentration in parallel and responses in buffer alone were used to control for signal drift. The data analysis software provided by ForteBio was used to normalize all responses to an appropriate baseline, and subtract the buffer only control. Processed association binding curves were exported to Excel (Microsoft) where responses for the biocytin-capped biosensor surface (without protein immobilized) interacting with CHIP was subtracted from the data. These final values were imported into Prism 5.0 (Graphpad, San Diego CA) where they were fit using the equation for one-phase association. The kobs calculated from each fit were used to determine kon and koff rates for the interaction. Saturation binding data was obtained by fitting raw response values from the association curves using non-linear regression in Prism 5.0.
Nuclear Magnetic Resonance
NMR data were collected using an Agilent/Varian NMR system with a triple resonance cold probe, interfaced to a Oxford instruments 18.7 T magnet (1H 800 MHz). Backbone assignments for the spectra of Hsc70 (391–646) were obtained from a 250 uM triple-labeled sample containing an N-terminal purification tag, which was not cleaved (MHHHHHHSSGVDLGTENLYFQNAM). The construct was determined to be dimeric from 15N relaxation measurements. 3D triple-resonance TROSY spectra HNCO (28h), HNCACO(90h), HNCACB (94h), HNCA (32h) and HNCOCA (64h) were acquired at 30 °C, were processed with NMR Pipe and were peak picked in Sparky. Despite these long acquisition times, the data was very incomplete, yielding only 78 HNCO(i-1), 36 HNCO(i), 52 HNCA(i-1), 59 HNCA(i), 13 HNCB(i-1) and 38 HNCB(i) peaks. The spectra were assigned using Saga32. Saga was expressly developed to allow for systematic assignments in very incomplete data. The program found reliable assignments for the C-terminal 30 residues and the expression tag only. Table 1 in supplementary materials lists the assignments obtained. High intensity of the assigned peaks, as well as their random-coil chemical shifts showed that these areas are dynamic random coil.
The experiments for studying the interaction of Hsc70 with CHIP were carried out with full-length Hsc70 (71 kDa) containing the same N-terminal expression tag. The Hsc70 sample was 15N labeled, in 50 mM K2PO4, 110 µM NRLLLTG peptide, 1 mM ADP, 5 mM MgCl2, 10 mM KCl, 0.02% NaN3, pH 7.2, 25 0°C. The TROSY spectrum with 1:0 Hsc70:CHIP was recorded in 12 hours with a 100 µM Hsc70 sample. The spectrum with 1:0.35 Hsc70:CHIP was recorded in 52 hours with a sample 57 µM in Hsc70 and 21 µM CHIP. The spectra with 1:0.75 Hsc70:CHIP was recorded in 52 hours with a sample 40 µM in Hsc70 and 30 µM CHIP.
The experiments for studying the 15N NMR relaxation of Hsc70 with or without CHIP were carried out with full-length Hsc70 (71 kDa) containing the same N-terminal expression tag. The Hsc70 sample was 15N labeled, at 67 µM in 50 mM K2PO4, 200 µM NRLLLTG, 1 mM ADP, 5 mM MgCl2, 10 mM KCl, 0.02% NaN3, pH 7.2, 25°C. 1D versions of the standard 15N R1 (10 h) and 15N R2 (4.5 h) HSQC experiments (without Rex suppression) were carried out. The data was processed in NMR pipe, and exported in ASCII format using the Pipe2txt.tcl routine. The data were imported into Microsoft Excel, and plotted. For each spectrum, the ranges 9.6–8.6 ppm (structured core residues) and 8.6–7.8 ppm (structured core and mobile tail residues) were integrated and fitted to a single exponential using in-house written non-linear least square fit code with jackknife error estimation.33
1D versions of a 15N chemical shift anisotropy / 15N-1H dipolar transverse cross-correlated relaxation experiment were obtained using symmetric reconversion experiments34. Four spectra, Nxy → Nxy, Nxy → 2NxyHz, 2NxyHz → Nxy and 2NxyHz → 2NxyHz measuring the auto- and cross-correlated relaxation rates over a period T of 10.2 ms were collected in 5 hours total, and processed as described above. The cross correlation rates ηxy were obtained from the equation
where R is given by
Fluorescence Polarization
FP experiments were performed in 384-well, black, low volume, round-bottom plates (Corning) using a BioTek Synergy 2 plate reader (Winooski, VT) at 25°C. To each well was added increasing amounts of protein (0 to 50 uM) and the 5-carboxyfluorescein (5-Fam) labeled SSGPTIEEVD tracer (20 nM) to a final volume of 20 uL in the assay buffer (50 mM HEPES, 75 mM NaCl, 0.01% Triton X-100, pH 7.4). The plate was allowed to incubate at room temperature for 5 min to reach equilibrium. The polarization values in millipolarization units (mP) were measured at an excitation wavelength at 485 nm and an emission wavelength at 528 nm. An equilibrium binding isotherm was constructed by plotting the FP reading as a function of the protein concentration at a fixed concentration of tracer (20 nM). All experimental data were analyzed using Prism 5.0 software (Graphpad Software, San Diego CA) and the inhibition constants were determined by nonlinear curve fitting at the concentration of protein at which 50% of the tracer (ligand) was bound.
Ubiquitination assay
Assays were performed essentially as previously reported.35 Briefly, 1µM CHIP, 1µM UbcH5c, 0.1µM E1, 0.5mM ubiquitin, and 5mM ATP/MgCl2 were incubated with 1µM of the Hsc70 variant indicated for the various time periods mentioned. Samples were stopped by the addition of laemmli buffer and boiled prior to SDS-PAGE and western blotting with HSC70 specific antibodies. The antibody used was purchased from either MBL International (cat # JM-3095), or Santa Cruz (cat # sc-7298).
Results
Full-length Hsc70 (wtHsc70) binds tightly to CHIP
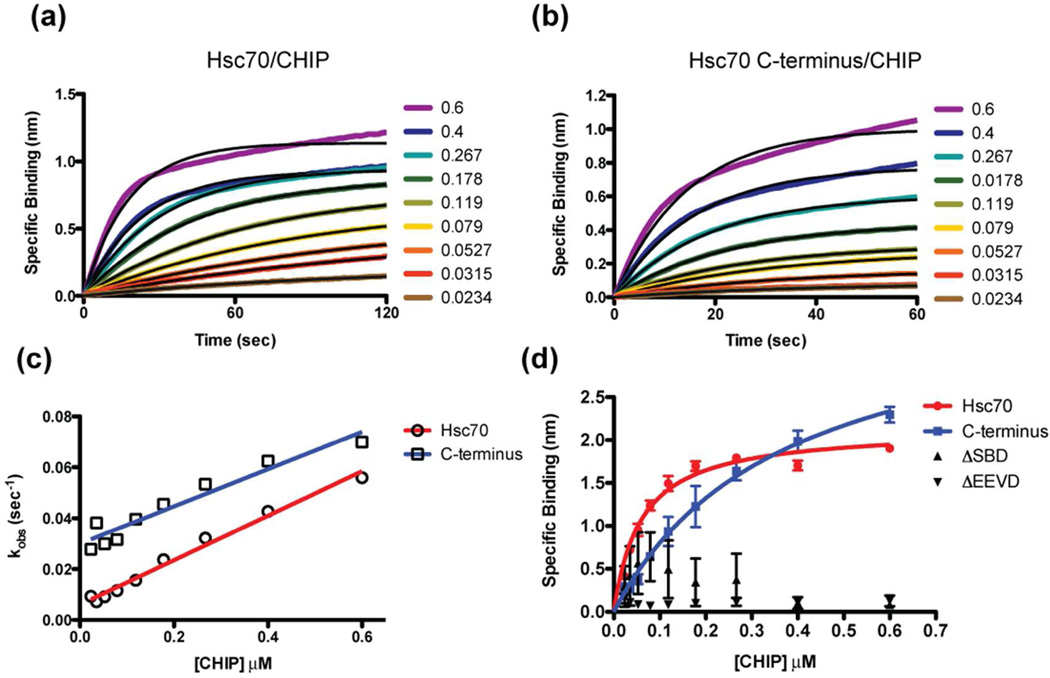
A previous study using surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) determined that a C-terminal peptide of Hsc70 consisting of the last 24 amino acids bound to full-length CHIP with a dissociation constant (Kd) between 1 and 2 µM.27 To probe whether contacts outside of the C-terminus might contribute to the binding affinity, we measured the interaction between immobilized wtHsc70 and full-length CHIP by biolayer interferometry (BLI). BLI association curves for the interaction were first fit using a one-phase exponential equation (Figure 1a). Then, the kobs from these fits were plotted as a function of the CHIP concentration and the kon (slope) and koff (y-intercept) values calculated (Figure 1c and Table 1). By this analysis, the affinity of wtHsc70 for CHIP was calculated to be 0.07 ± 0.01 µM (Table 1). Fitting the equilibrium binding values yielded a similar Kd of 0.060 ± 0.007 µM (Figure 1d and Table 1).
Fig. 1.
Measuring the binding affinity of Hsc70 to CHIP. (a and b) Best-fit data of association binding curves acquired by bio-layer interferometry (BLI). The raw data represent the mean of at least two independent experiments. (a) Biotinylated Hsc70 was immobilized on a streptavidin-coated BLI sensor surface and incubated with increasing µM concentrations of CHIP. After controlling for nonspecific binding of CHIP to the sensor surface and signal drift, specific binding was plotted over time. Black lines represent individual fits of the data using a one-phase exponential equation. (b) Biotinylated Hsc70 C-terminal peptide (SSGPTIEEVD) was immobilized and treated as in (a). (c) Kobs values from the curve-fitting results of (a) and (b) plotted as a function of CHIP concentration gave Kd values of 0.07 ± 0.01 µM and 0.41 ± 0.05 µM for Hsc70 and the C-terminal peptide, respectively. (d) Saturation binding data from the BLI experiments in (a) and (b) gave Kd values of 0.060 ± 0.007 µM and 0.37 ± 0.07 µM for Hsc70 and C-terminal peptide, respectively. Neither the Hsc70ΔSBD, nor the Hsc70ΔEEVD mutant displayed any ability to bind CHIP. Each data point represents the mean of at least two independent experiments, and the error bars represent the standard error (SEM).
Table 1.
Binding values for the Hsc70-CHIP interaction
To compare these values to that of the isolated peptide in the same platform, we immobilized a C-terminal peptide derived from wtHsc70, H2N-SSGPTIEEVD-COOH, and measured its binding to CHIP (Figure 1b). Analysis of the kinetic data revealed a Kd of 0.41 ± 0.05 µM (Figure 1c and Table 1) and the equilibrium values yielded a Kd of 0.37 ± 0.07 µM (Figure 1d and Table 1). In addition, similar results were obtained by fluorescence polarization (FP), using a fluorescent SSGPTIEEVD tracer. In that platform, the Hsc70 C-terminus had a Kd value of 0.61 ± 0.04 µM, agreeing reasonably well with both the literature value and our BLI data 27 (Figure S1). Competition studies showed that the unlabeled peptide had an IC50 value of 2 ± 0.2 µM, while wtHsc70 had an IC50 value of 0.45 ± 0.04 µM (Figure S1).
Together, these results suggest that wtHsc70 binds just ~6-fold tighter to CHIP than the isolated C-terminal peptide. This difference corresponds to an increase in binding free energy of 1.08 kCal/M. This is 3–5 fold less than the energy of a single H-bond and strongly suggests that the additional interaction between CHIP and the core of Hsc70 must be transient and/or non-specific. Examination of the kinetics from the BLI studies shows that this ~ 6-fold advantage in binding affinity is almost entirely afforded by an ~ 5-fold slower dissociation (koff= 0.006 sec−1 vs. 0.03 sec−1, respectively; Figure 1c and Table 1). A marginally faster association (kon= 0.087 µM−1 sec−1 vs. 0.073 µM−1 sec−1, respectively; Table 1) makes up the rest of the discrepancy. So, while the EEVD motif is clearly required for binding, as mutants lacking this sequence (Hsc70ΔEEVD) or the entire C-terminal substrate-binding domain (Hsc70ΔSBD; 1–383) cannot bind CHIP15,16 (Figure 1d), there is only a small difference in affinity between wtHsc70 and its C-terminal peptide, suggesting that contacts outside of the EEVD motif that contribute to the interaction are rather insignificant.
Binding of the Hsc70 EEVD motif to CHIP involves polar interactions
To gain more insight into how EEVD motifs interact with the TPR domain of CHIP, we explored the contributions of the individual residues. Many studies have explored the binding of this region to other TPR domain co-chaperones, especially Hop.16,17 These studies have largely concluded that polar interactions are a common feature of the EEVD-TPR interaction, including a “carboxylate clamp” in the TPR domain that provides selectivity. Using the FP platform, we first confirmed that a fluorescent version of the Hsp90α-derived C-terminus (5FAM-DDTSRMEEVD; residues 723–732) binds to CHIP (Figure 2a). To explore the contribution of Glu730, we generated mutant probes with an Ala or Lys in this position. The Glu730Ala mutant had slightly reduced affinity for CHIP (KD = 1.7 ± 0.24 µM), but switching the charge from anionic to cationic (Glu730Lys) greatly reduced affinity (KD = 6.5 ± 1.5 µM) (Figure 2b). To further test this model, we performed binding studies of the Hsc70 peptide (5FAM-SSGPTIEEVD; residues 637–646) to CHIP at pH 7.4 and 9.4. Consistent with a key role for polar interactions with the glutamic acids, the affinity was reduced more than 10 fold (KD = 0.6 ± 0.05 µM and 11 ± 2.1 µM, respectively) (Figure 2c). Next, we synthesized a shorter fluorescent probe (5FAM-MEEVD) and found that it had a nearly identical affinity to the longer peptide (KD = 1.5 ± 0.076 µM) (Figure 2d). Conversely, the reversed peptide (5FAM-DVEEM) had no affinity for CHIP, consistent with the reported key role for the Val and Asp in the last two positions of the C-terminus.17 Using the MEEVD tracer design, we tested whether replacing the Glu residues with homoserine (Figure 2e), an amino acid that retains electron-accepting character, was compatible with binding. Placing homoserine in either position (5FAM-MHsEVD or 5FAM-MEHsVD) did not disrupt binding (KD = 2.1 ± 0.34 µM and 1.5 ± 0.23 µM, respectively) (Figure 2f). Finally, we generated a point mutation in CHIP that is predicted to disrupt the “carboxylate clamp” (K30A). This mutant had dramatically reduced affinity for the peptides (Figure 2G, note the mP values). Together, these studies suggest that, as has been seen in other EEVD-TPR interactions, polar contacts dominate binding.
Fig 2.
CHIP binding to the C-terminus of Hsc70 involves polar contacts in the EEVD motif. (a) A fluorophore-labelled probe binds to CHIP, but not Hsc70. (b) Switching Glu644 to alanine has a modest effect on affinity for CHIP, but replacement with a cationic lysine greatly reduces affinity. (c) Consistent with the role for a polar contact, the affinity of a fluorescent EEVD probe for CHIP is dependent on pH. There was no change in the fluorescence of the FAM fluorophore under these conditions (not shown). (d) A truncated MEEVD peptide binds with affinity equal to that of the longer probe, but the reversed sequence, DVEEM, had no affinity for CHIP. (e) The chemical structures of glutamic acid and homoserine. (f) Replacing Glu644 for alanine has a modest effect on affinity and replacement with a homoserine restores affinity. Similarly, replacing Glu643 with a homoserine has little effect on affinity. (g) Mutating the proposed “carboxylate clamp” residue in CHIP reduces binding to all of the peptides tested. All results are the average of triplicate experiments performed in triplicate. Error bars represent the standard error of the mean (SEM).
NMR studies identify sharp resonances of residues at the wtHsc70 C-terminus that interact with CHIP
To explore the Hsc70-CHIP complex in more detail, we turned to solution-state NMR. 1H-15N TROSY HSQC spectra were collected for 15N-labeled wtHsc70 (1–646) in the ADP and peptide (NRLLLTG) substrate-bound state. Based on 1D 15N relaxation experiments, the protein is monomeric at 100 µM (see Table 2). The center of the TROSY spectrum was dominated by intense resonances with little chemical shift dispersion (Figure 3b). As identified in our previous study on the highly homologous prokaryotic Hsp70 molecule, DnaK,31 these resonances likely correspond to amino acid stretches that form flexible random coils, such as the linker region between the NBD and SBD and the C-terminal tail (Figure 3a). Conversely, the weaker resonances outside of this central area are indicative of a folded core, including the nucleotide-binding domain (NBD) and the SBD. Some of the resonances of the NBD and SBD are observed in the spectrum of the full-length protein as illustrated by an overlay with the NMR spectra from the individual domains (Figure 3b).
Table 2.
15N relaxation of Hsc70a
| Hsc70:CHIP(1:0) | Hsc70:CHIP(1:1) | |||
|---|---|---|---|---|
| Spec Coverage: | 9.6–8.6 ppm | 8.6-7.8 ppm | 9.6–8.6 ppm | 8.6-7.8 ppm |
| R1 (s−1) | 0.29 ± 0.04 | 0.70 ± 0.04 | 0.37 ± 0.02 | 0.63 ± 0.04 |
| R2 (s−1)b | 135 ± 56 | 17 ± 4 | 65 ± 11 | 22 ± 2 |
| ηxy (s−1)c | 63 ± 4 | 21 ± 1 | 52 ± 3 | 21 ± 1 |
| R2/R1 | 465 ± 188 | 24 ± 5 | 176 ± 27 | 35 ± 4 |
| ηxy/R1 | 217 ± 30 | 30 ± 2 | 140 ± 11 | 33 ± 3 |
| τc (ns)d | 35–42 | 14–15 | 30–33 | 14.5–15.5 |
| MW (kDa)e | 55–65 | 22–23 | 48–52 | 23–24 |
Data obtained from 800 MHz 1D 15N relaxation experiments with 66 µM Hsc70, pH 7.2 at 25 °C. The rates were obtained from integrated data in the ranges indicated. The uncertainties of fit listed were obtained using a jackknife procedure.40
No CPMG (Carr-Purcell-Meiboom-Gill).
N15 chemical shift anisotropy / 15N-1H dipolar transverse cross-correlated relaxation during 10.2 ms obtained using symmetric reconversion.41
The ηxy and R1 rates as a function of τc were computed for a magnetic field of 18.7 T using 1.04 Å for the NH bond length, a rhombic 15N CSA with σ11=230, σ22=87 and σ33=60 ppm, with an angle q between the σ11 axis and the NH bond of 17.5 degrees.43 The non-truncated relaxation equations were used with the ModelFree spectral density function,44 using S2=0.9 and τe=10 ps.
Conversion using equation 42 in Daragan et al.45
Fig. 3.
Sharp resonances of Hsc70 identified by NMR. (a) Graphical representation of the full length Hsc70 construct used for NMR. Areas of flexible, random coil secondary structure are circled, and the corresponding resonances are highlighted in (b). HIS is the un-cleaved 6xHIS tag, NBD is the nucleotide-binding domain, and SBD is the substrate-binding domain of the chaperone. (b) 800MHz TROSY overlay of the spectra from 15N-labeled full length Hsc70 (646 residues, 72kDa) in black, with that of the isolated NBD (residues 1–383, 42kDa) in red, and the isolated SBD (residues 391–646, 28kDa) in green. All spectra were acquired in the presence of excess ADP and model peptide substrate (NRLLLTG).
wtHsc70 is a 71 kDa monomeric protein, while CHIP is a 35 kDa constitutive homodimer of 70 kDa total molecular weight.36,37 Previous data,27 and our own binding model, suggest a 1:1 binding ratio for the Hsc70-CHIP complex. Together, this would make the complex a dimer of dimers containing two molecules of both CHIP and wtHsc70, with an apparent molecular weight of 210 kDa. With such a drastic change in molecular weight, one would expect that the NMR spectrum for wtHsc70 would disappear all together upon binding to CHIP, especially since we are working with pretreated samples of low concentration (~50 µM). However, the spectrum of a 1:1 complex of wtHsc70 and CHIP is barely distinguishable from that of wtHsc70 alone (compare black and red overlays; Figure 4a). The addition of excess CHIP does not further change the spectrum (data not shown). Neither does the addition of the E2 ubiquitin-conjugating enzyme, UbcH5c, which is known to facilitate CHIP-mediated ubiquitination of wtHsc70 (compare red and green overlays; Figure 4a). To reiterate, if a complex with hydrodynamic properties of a 210 kDa globular particle were to exist, one would not be able to observe any NMR spectrum at all. Hence, the small changes seen in the spectrum likely arise from transient and rather non-specific contacts, not stable interactions. However, a closer look reveals that several of the sharp and intense resonances, which were assigned to the unstructured C-terminus of Hsc70, progressively disappear upon complex formation. Only intensity loss, and no line-broadening or shift was observed for these resonances in the spectra with less than stoichiometric amounts of CHIP (Fig S3). These observations are diagnostic of an extremely slow-exchange kinetic regime that is fully consistent with the koff of 0.006 sec−1 obtained from the BLI experiments (see above).
Fig. 4.
C-terminal residues of 15N-labeled wtHsc70 disappear upon binding to CHIP. (a) Full 800 MHz TROSY overlay of the spectra from 15N-labeled wtHsc70 at 100 µM (black) with that of a 67 µM 1:1 mixture of 15N Hsc70 and CHIP (red), and a 60 µM 1:1:1 mixture of 15N Hsc70, CHIP and UbcH5 (green). (b) A 10X high-contour view of detailed area showing Hsc70’s dynamic random coil residues including the ~40 amino acid C-terminal tail; resonance assignments are included. Residues from Hsc70 that interact with CHIP do not appear in the corresponding spectra (red). (c) Depiction of the unstructured C-terminal tail of wtHsc70. Resonances that were assigned are underlined, while those that shifted upon complexation with CHIP are italicized and resonances that disappeared altogether are outlined in red.
Specifically, we found that binding to CHIP caused the resonances for residues 639-GPTIEEVD-646 to disappear from the wtHsc70 NMR spectrum, while the assigned resonances for the “upstream” residues 616–636 of the unstructured tail remained visible and relatively unaffected by the presence of CHIP (Figure 4b and c). These results suggest that the 70 kDa CHIP dimer sequesters the EEVD motif and a few more residues into a large molecular weight complex, broadening the corresponding NMR resonances beyond detection. The results also suggest that the region between the GPTIEEVD motif and the end of the Hsc70 SBD remains a dynamic random coil, even in the presence of bound CHIP. Taken together, these data and the FP studies support a model in which CHIP binding is largely localized to the GPTIEEVD motif.
15N relaxation shows that the folded core of Hsc70 tumbles independently of CHIP in solution
As mentioned above, if CHIP were to stably interact with the Hsc70 core (the well folded regions of the NBD and SBD), the core resonances (i.e. those outside of the central overlapped region in the TROSY spectrum) are expected to disappear. As this is not the case, we conclude that there is no stable interaction between CHIP and the Hsc70 core. But we wondered whether CHIP might still affect the hydrodynamic properties of the core. If so, we would expect that the apparent molecular weight of wtHsc70, as reflected by the rotational correlation time (τc) of the protein, should increase. The following experiments are based on a 1D 15N-1H HSCQ scheme involving variable 15N relaxation rates that affect the intensities of the detected amide-proton signals.38 The logical steps in determining the molecular weight are as follows: 1H intensity changes → 15N relaxation rates → τc → Mr, where Mr is the molecular weight. The first conversion step is a fit to a single exponential, the second step is rooted in NMR relaxation theory, while the third step is governed by the Stokes-Einstein equation. We used this procedure to measure the effect of 15N relaxation on the intensities of the 1H amide-proton spectrum of wtHsc70 both in the absence and presence of CHIP. From the NMR data, three regions can be identified that correspond to resonances of a structured area: 1H > 8.6 ppm, an unstructured area: 8.6 > 1H > 7.6 ppm, and Gln/Asn sidechains: 1H < 7.6 ppm in the wtHsc70 protein (Figure S4). It is apparent that the R1 relaxation of the structured area is much slower than that of the unstructured area, corresponding to a larger rotational correlation time and a larger apparent molecular weight, as expected. For quantitative analysis, R2 relaxation was measured by two different methods (Figs. S5 and S6). The results of all relaxation experiments are summarized in Table 2. The key finding is that the structured area of the wtHsc70, corresponding to the NBD and SBD, has a similar rotational correlation time with or without CHIP present. This is in stark contrast to the expected tripling of rotational correlation time of wtHsc70 that would result from a stable interaction between the core and CHIP in a 210 kDa complex. The reported apparent molecular weight of ~50 kDa for the structured region corresponds more closely to that of the Hsc70 NBD (44 kDa) than to the entire 70 kDa protein, suggesting that the NBD and SBD of Hsc70 are attached by a flexible linker, just as is the case for the E. coli Hsp70 ortholog DnaK in the same nucleotide- and peptide-bound state.31 The results suggest that, in the complex, wtHsc70 and CHIP move independent of one another, like two beads attached by a string. Therefore, any contacts for binding CHIP outside of the EEVD motif cannot be stable and must be transient and/or delocalized.
The Hsc70 C-terminal EEVD motif is both necessary and sufficient for CHIP binding
The binding studies and NMR results all point to the importance of the EEVD motif in binding to CHIP. We wondered whether placing the EEVD elsewhere on Hsc70 might still enable binding. We also wondered whether the association rate of this interaction would resemble that of wtHsc70, even if the EEVD was not displayed in a fully native orientation. To explore these questions, we cloned an Hsc70 mutant missing amino acids 627–641 in the C-terminal tail and then restored an IEEVD sequence at the end of this truncated construct (Hsc70ΔTail; Figure 5a). We then measured this mutant's ability to bind CHIP by BLI. The kinetic analysis revealed that the Hsc70ΔTail mutant had a much slower rate of association than wtHsc70 (~8-fold; Figure 4b and Table 1), however it also had a slower rate of dissociation (~2-fold; Table 1) leading to a comparable Kd of 0.36 ± 0.04 µM. The equilibrium binding data produced a slightly lower Kd of 0.17 ± 0.02 µM (Figure 5b and Table 1). Therefore, placing the EEVD motif on a truncated C-terminal region of Hsc70 did not fully reproduce the kinetic behavior of the wild type protein, but it did restore near normal affinity.
Fig. 5.
The Hsc70 C-terminal domain is sufficient to facilitate CHIP-mediated ubiquitination. (a) C-terminal sequences for wtHsc70 compared to the Hsc70ΔTail mutant. Residues 627–641 have been removed. (b) Kinetic and equilibrium data collected from BLI experiments show comparable binding between CHIP and Hsc70 (1–646) or the ΔTail mutant (1-626-IEEVD). The Kd for Hsc70, as reported in Fig. 1 is ~0.05 µM while the kinetic and equilibrium Kd values for the ΔTail mutant are ~0.08 µM and ~0.17 µM, respectively. (c) In vitro, CHIP-mediated polyubiquitination of Hsc70 is dependent on the EEVD motif, as the Hsc70ΔEEVD mutant is not modified to the same extent. In contrast, placing the EEVD in a non-native segment of the C-terminal domain (ΔTail mutant) reconstitutes activity at 60 minutes. (d) Shortening the tail of Hsc70 (ΔTail mutant) slows ubiquitination, as shown by stopping the CHIP-dependent ubiquitination reactions at earlier times. Error bars represent the standard error (SEM). *Non-specific bands.
The EEVD sequence is required for CHIP-mediated ubiquitination of Hsc70
To achieve a better understanding of the functional consequences of the EEVD-CHIP interaction in vitro, we employed an ubiquitination assay. A mixture of E1, E2 (UbcH5c), CHIP and ATP was used to ubiquitinate Hsc70, and the reaction was monitored by an increase in the apparent molecular mass of Hsc70 by SDS-PAGE. These studies clearly showed that CHIP could mediate the polyubiquitination of wtHsc70, but it did not have activity against either the Hsc70ΔEEVD or the Hsc70ΔSBD mutants (Figure 5c). Interestingly, the Hsc70ΔTail mutant retained near normal activity as a substrate at 60 minutes, suggesting that the exact position of the EEVD motif is not critical for a functional CHIP-Hsc70 interaction to occur. However, when we examined earlier times in the ubiquitination reaction (10 and 30 minutes), we found that Hsc70ΔTail was considerably slower at performing the reaction (Figure 5d). These results suggest that as long as the EEVD is available as an “anchor” in the Hsc70 C-terminus, CHIP is able to bind Hsc70, and locate lysines on which to transfer ubiquitin. However, the length of the flexible tail is important in the efficiency of the reaction, suggesting that Hsc70-CHIP may use a dynamic search strategy to identify ubiquitination sites.
Discussion
In the present study, we explored the binding mode for wtHsc70 and its co-chaperone, CHIP. This complex has many functions, but appears to be particularly important in the turnover of disease-associated proteins and, thus, there is much interest in understanding its interactions. Previous work measured the affinity of a C-terminal Hsc70 peptide for CHIP to be around 1 µM,27 which is in agreement with what we determined by BLI and FP. We found that the full-length wtHsc70 binds CHIP tighter by a factor of 6, suggesting that only weak contacts outside of the EEVD motif exist (representing 1.08 kCal/M). Our results are therefore somewhat aligned with what has been previously discovered for the Hsp70-Hop interaction,17 namely that the EEVD-TPR binding interface is essential but that it may not fully describe these protein-protein interactions. It should, however, be reiterated that these ancillary contacts in Hsc70 appear to be very weak, transient and potentially non-specific, based on our binding and NMR studies. Indeed, the essential nature of the EEVD motif is again illustrated by the ability of the Hsc70ΔTail truncation to recruit CHIP (see Figure 5c), suggesting that simply appending IEEVD to a protein’s sequence might be sufficient, in some cases, to recruit CHIP and induce ubiquitination. However, the slower rate of the reaction with Hsc70ΔTail suggests that the flexibility and length of the intervening region is important in allowing CHIP to “find” its lysine substrates on the protein.
Our NMR studies on the wtHsc70-CHIP complex in solution showed very little difference between the TROSY spectrum of Hsc70 alone (70 kDa) and that of Hsc70 complexed with CHIP (210 kDa). Most notably, only the C-terminal resonances of Hsc70 are significantly affected by the presence of CHIP. In fact, resonances for residues 639–646 (GPTIEEVD) disappear from the NMR spectrum, which is the expected result for residues involved in a high-molecular weight complex. Resonances assigned to Q612 and M617 are unaffected by CHIP, while resonances in the region 620–637 remain sharp but are shifted. This result suggests that CHIP only directly interacts, at a maximum, with residues 639–646 of Hsc70, a result supported by the FP studies (see Figure 2). Thus, it is likely that the N-terminal residues of GPTIEEVD are not directly involved in the interaction, but are just immobilized because of the interaction. The fact that the signals in the region 620–637 remain sharp must indicate that they are not involved in the interaction at all. The observation that some of these residues are shifted upon CHIP addition may be interpreted as a change in their accessible conformations, which are affecting the dynamically averaged chemical shifts of this floppy coil. Even more dramatically, CHIP had no effect on residues N-terminal to M617, including the Hsc70 “core”. 15N relaxation studies fully corroborate these conclusions. Specifically, they indicate that the rotational correlation time of the Hsc70 core is not increased upon binding to CHIP. Therefore, CHIP and Hsc70 appear to move completely independently in the complex. The mobility between these proteins is afforded by the intervening region between the SBD and the IEEVD motif, which remains highly flexible, as discussed above.
At least forty amino acids separate the IEEVD motif from regions of Hsc70 that have secondary structure, meaning that this fully extended tail could reach up to a maximum of ~140 Å. Thus, in the wtHsc70-CHIP complex, the Ubox domain of CHIP would be able to sample a large area around wtHsc70 and its bound clients. This spatial flexibility is likely essential in allowing CHIP to locate lysine residues on a wide variety of Hsc70 client proteins, as is suggested in other degradation complexes.39,40 This conclusion is consistent with the results of the ubiquitination reactions with the Hsc70ΔTail mutant, which suggest that the length of the tail is important in the rate of CHIP’s ubiquitination. We speculate that this flexible region might also be important for ubiquitination of other substrates. wtHsc70 is thought to bind promiscuously to multiple regions of misfolded proteins,41,42 which would require that CHIP be able to adapt to the topology of many different protein surfaces. We speculate that the ancillary contacts in the binding of wtHsc70 to CHIP might, in fact, be indicative of weak, transient interactions produced by CHIP sampling the surface of wtHsc70. Because it is not currently possible to study Hsc70-dependent CHIP ubiquitination of other client proteins in vitro, this model remains to be tested.
Supplementary Material
Acknowledgments
We would like to thank Duxin Sun (University of Michigan, Ann Arbor) for generously synthesizing our peptides, and H.R. Hill for useful discussions. This work was supported by the NIH (NS059690).
Funding: This work was supported by NS059690 to J.E.G. and E.R.P.Z. A Rackham Merit Fellowship and AG000114 supplied support to M.C.S. K.M.S. was supported by NS073936 and AG034228 to H.L.P. And additional funding was supplied by grants NS073899 and BX001637 to C.A.D
Abbreviations
- NBD
nucleotide-binding domain
- SBD
substrate-binding domain
- BLI
biolayer interferometry
- SPR
surface plasmon resonance
- Hsc70
Heat shock cognate 70
- CHIP
C-terminus of Hsc70 interacting protein
Footnotes
Supporting Information Available
Fluorescence polarization data, NMR assignments, as well as high-resolution enlargements of relevant spectroscopic data are provided as supplementary information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hartl FU, Hlodan R, Langer T. Molecular chaperones in protein-folding - the art of avoiding sticky situations. Trends in Biochemical Sciences. 1994;19:20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 2.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annual Review of Physiology. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 3.Imai J, Yashiroda H, Maruya M, Yahara I, Tanaka K. Proteasomes and molecular chaperones: Cellular machinery responsible for folding and destruction of unfolded proteins. Cell cycle (Georgetown, Tex.) 2003;2:585–590. [PubMed] [Google Scholar]
- 4.Kriegenburg F, Ellgaard L, Hartmann-Petersen R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS Journal. 2012;279:532–542. doi: 10.1111/j.1742-4658.2011.08456.x. [DOI] [PubMed] [Google Scholar]
- 5.Stankiewicz M, Nikolay R, Rybin V, Mayer MP. Chip participates in protein triage decisions by preferentially ubiquitinating hsp70-bound substrates. FEBS Journal. 2010;277:3353–3367. doi: 10.1111/j.1742-4658.2010.07737.x. [DOI] [PubMed] [Google Scholar]
- 6.Meacham G, Patterson C, Zhang W, Younger J, Cyr D. The hsc70 co-chaperone chip targets immature cftr for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 7.Younger J, Ren H, Chen L, Fan C, Fields A, Patterson C, Cyr D. A foldable cftr{delta}f508 biogenic intermediate accumulates upon inhibition of the hsc70-chip e3 ubiquitin ligase. J Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adachi H, Waza M, Tokui K, Katsuno M, Minamiyama M, Tanaka F, Doyu M, Sobue G. Chip overexpression reduces mutant androgen receptor protein and ameliorates phenotypes of the spinal and bulbar muscular atrophy transgenic mouse model. J Neurosci. 2007;27:5115–5126. doi: 10.1523/JNEUROSCI.1242-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimura H, Schwartz D, Gygi S, Kosik K. Chip-hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. The Journal of biological chemistry. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 10.Jiang J, Ballinger C, Wu Y, Dai Q, Cyr D, Hohfeld J, Patterson C. Chip is a u-box-dependent e3 ubiquitin ligase: Identification of hsc70 as a target for ubiquitylation. The Journal of biological chemistry. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 11.Kundrat L, Regan L. Identification of residues on hsp70 and hsp90 ubiquitinated by the cochaperone chip. Journal of Molecular Biology. 2010;395:587–594. doi: 10.1016/j.jmb.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urushitani M, Kurisu J, Tateno M, Hatakeyama S, Nakayama K-I, Kato S, Takahashi R. Chip promotes proteasomal degradation of familial als-linked mutant sod1 by ubiquitinating hsp/hsc70. Journal of Neurochemistry. 2004;90:231–244. doi: 10.1111/j.1471-4159.2004.02486.x. [DOI] [PubMed] [Google Scholar]
- 13.Pridgeon JW, Webber EA, Sha D, Li L, Chin L-S. Proteomic analysis reveals hrs ubiquitin-interacting motif-mediated ubiquitin signaling in multiple cellular processes. FEBS Journal. 2009;276:118–131. doi: 10.1111/j.1742-4658.2008.06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qian S, Mcdonough H, Boellmann F, Cyr D, Patterson C. Chip-mediated stress recovery by sequential ubiquitination of substrates and hsp70. Nature. 2006;440:551–555. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin L-Y, Patterson C. Identification of chip, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Molecular and Cellular Biology. 1999;19:4535–4545. doi: 10.1128/mcb.19.6.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Liu F, Hu S, Wang C. Different combinations of the heat-shock cognate protein 70 (hsc70) c-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem J. 2001;359:419–426. doi: 10.1042/0264-6021:3590419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brinker A, Scheufler C, Von Der Mulbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Hartl FU. Ligand discrimination by tpr domains. Relevance and selectivity of eevd-recognition in hsp70 x hop x hsp90 complexes. The Journal of biological chemistry. 2002;277:19265–19275. doi: 10.1074/jbc.M109002200. [DOI] [PubMed] [Google Scholar]
- 18.Schneider C, Sepp-Lorenzino L, Nimmesgern E, Ouerfelli O, Danishefsky S, Rosen N, Hartl FU. Pharmacologic shifting of a balance between protein refolding and degradation mediated by hsp90. Proceedings of the National Academy of Sciences. 1996;93:14536–14541. doi: 10.1073/pnas.93.25.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goebl M, Yanagida M. The tpr snap helix: A novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 20.Das AK, Cohen PT, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for tpr-mediated protein,äìprotein interactions. The EMBO journal. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allan RK, Ratajczak T. Versatile tpr domains accommodate different modes of target protein recognition and function. Cell Stress & Chaperones. 2011;16:353–367. doi: 10.1007/s12192-010-0248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cortajarena AL, Regan L. Ligand binding by tpr domains. Protein Sci. 2006;15:1193–1198. doi: 10.1110/ps.062092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Liu Y-T, Hao R, Chen L, Chang Z, Wang H-R, Wang Z-X, Wu J-W. Molecular mechanism of the negative regulation of smad1/5 protein by carboxyl terminus of hsc70-interacting protein (chip) Journal of Biological Chemistry. 2011;286:15883–15894. doi: 10.1074/jbc.M110.201814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Moroder L, Bartunik H, Hartl F, Moarefi I. Structure of tpr domain-peptide complexes: Critical elements in the assembly of the hsp70-hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2. [DOI] [PubMed] [Google Scholar]
- 25.Graf C, Stankiewicz M, Nikolay R, Mayer M. Insights into the conformational dynamics of the e3 ubiquitin ligase chip in complex with chaperones and e2 enzymes. Biochemistry. 2010;49:2121–2129. doi: 10.1021/bi901829f. [DOI] [PubMed] [Google Scholar]
- 26.Carrigan PE, Nelson GM, Roberts PJ, Stoffer JN, Riggs DL, Smith DF. Multiple domains of the co-chaperone hop are important for hsp70 binding. Journal of Biological Chemistry. 2004;279:16185–16193. doi: 10.1074/jbc.M314130200. [DOI] [PubMed] [Google Scholar]
- 27.Kundrat L, Regan L. Balance between folding and degradation for hsp90-dependent client proteins: A key role for chip. Biochemistry. 2010;49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expression and Purification. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 29.Zheng L, Baumann U, Reymond J-L. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Research. 2004;32:e115. doi: 10.1093/nar/gnh110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between atpase rate and the chaperone activities of escherichia coli heat shock protein 70 (hsp70/dnak) Journal of Biological Chemistry. 2010;285:21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type e. Coli hsp70 (dnak) chaperone complexed with adp and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crippen G, Rousaki A, Revington M, Zhang Y, Zuiderweg EP. Saga: Rapid automatic mainchain nmr assignment for large proteins. Journal of Biomolecular NMR. 2010;46:281–298. doi: 10.1007/s10858-010-9403-2. [DOI] [PubMed] [Google Scholar]
- 33.Quenouille MH. Notes on bias in estimation. Biometrika. 1956;43:353–360. [Google Scholar]
- 34.Pelupessy P, Espallargas GM, Bodenhausen G. Symmetrical reconversion: Measuring cross-correlation rates with enhanced accuracy. Journal of Magnetic Resonance. 2003;161:258–264. doi: 10.1016/s1090-7807(02)00190-8. [DOI] [PubMed] [Google Scholar]
- 35.Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng JM, Gestwicki JE, Paulson HL. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase chip. Molecular Cell. 2011;43:599–612. doi: 10.1016/j.molcel.2011.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nikolay R, Wiederkehr T, Rist W, Kramer GN, Mayer MP, Bukau B. Dimerization of the human e3 ligase chip via a coiled-coil domain is essential for its activity. Journal of Biological Chemistry. 2004;279:2673–2678. doi: 10.1074/jbc.M311112200. [DOI] [PubMed] [Google Scholar]
- 37.Benaroudj N, Triniolles F, Ladjimi MM. Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone hsc70. Journal of Biological Chemistry. 1996;271:18471–18476. doi: 10.1074/jbc.271.31.18471. [DOI] [PubMed] [Google Scholar]
- 38.Cavanagh J, Fairbrother W, Palmer Agiii RM, Skeleton N. Protein nmr spectroscopy: Principles and practice. 2nd edn. Amerstdam: Academic Press; 2007. chap. 8. [Google Scholar]
- 39.Liu J, Nussinov R. Flexible cullins in cullin-ring e3 ligases allosterically regulate ubiquitination. Journal of Biological Chemistry. 2011;286:40934–40942. doi: 10.1074/jbc.M111.277236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdecia MA, Joazeiro CAP, Wells NJ, Ferrer J-L, Bowman ME, Hunter T, Noel JP. Conformational flexibility underlies ubiquitin ligation mediated by the wwp1 hect domain e3 ligase. Molecular Cell. 2003;11:249–259. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 41.Erbse A, Mayer MP, Bukau B. Mechanism of substrate recognition by hsp70 chaperones. Biochemical Society Transactions. 2004;32:617–621. doi: 10.1042/BST0320617. [DOI] [PubMed] [Google Scholar]
- 42.Wegele H, Muller L, Buchner J. Hsp70 and hsp90--a relay team for protein folding. Rev Physiol Biochem Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- 43.Brender JR, Taylor DM, Ramamoorthy A. Orientation of amide-nitrogen-15 chemical shift tensors in peptides: A quantum chemical study. Journal of the American Chemical Society. 2001;123:914–922. doi: 10.1021/ja001980q. [DOI] [PubMed] [Google Scholar]
- 44.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. Journal of the American Chemical Society. 1982;104:4546–4559. [Google Scholar]
- 45.Daragan VA, Mayo KH. Motional model analyses of protein and peptide dynamics using 13c and 15n nmr relaxation. Progress in Nuclear Magnetic Resonance Spectroscopy. 1997;31:63–105. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.