Abstract
For several of the proteins in the BioMagResBank larger than 200 residues, 60% or fewer of the backbone resonances were assigned. But how reliable are those assignments? In contrast to complete assignments, where it is possible to check whether every triple-resonance Generalized Spin System (GSS) is assigned once and only once, with incomplete data one should compare all possible assignments and pick the best one. But that is not feasible: For example, for 200 residues and an incomplete set of 100 GSS, there are 1.6*10260 possible assignments. In “EZ-ASSIGN”, the protein sequence is divided in smaller unique fragments. Combined with intelligent search approaches, an exhaustive comparison of all possible assignments is now feasible using a laptop computer. The program was tested with experimental data of a 388-residue domain of the Hsp70 chaperone protein DnaK and for a 351-residue domain of a type III secretion ATPase. EZ-ASSIGN reproduced the hand assignments. It did slightly better than the computer program PINE (Bahrami et al., PLoS Comput Biol. 2009 5 (3): e1000307) and significantly outperformed SAGA (Crippen et al, (2010) J Biomol NMR 46, 281–298), AUTOASSIGN (Zimmerman et al., (1997) J Mol Biol 269:592–610), and IBIS (Hyberts and Wagner (2003) J Biomol NMR 26:335–344). Next, EZ-ASSIGN was used to investigate how well NMR data of decreasing completeness can be assigned. We found that the program could confidently assign fragments in very incomplete data. Here, EZ-ASSIGN dramatically outperformed all the other assignment programs tested.
Introduction
The development of triple resonance NMR methods for assignments of the resonances of protein backbone nuclei in the early 1990’s 1,2 has revolutionized solution protein NMR spectroscopy. Currently, there are more than 5000 proteins with assignments listed in the BioMagResBank. This is a tremendous achievement, but, as Table 1 shows, the vast majority of these assigned proteins are smaller than 25 kDa (200 residues) while the assignments for larger proteins are rather incomplete. These facts are serious obstacles for de-novo NMR structure determination for the majority of proteins: In the human genome the median protein chain length is 423 residues. However, partial assignments for such larger systems are still extremely valuable for the study of protein-protein and protein-ligand interactions, and for studies of conformational/dynamical change and/or allostery as deduced from chemical shift changes, paramagnetic relaxation enhancement, residual dipolar couplings and 15N relaxation (for example, see ref 3).
Table 1.
Completeness of the NMR assignments in the BMRB
| Completeness of assignmenta (%) | ||||||
|---|---|---|---|---|---|---|
| Residues in protein | Occurrence in BMRB | > 90 | 80–90 | 70–80 | 60–70 | < 60 |
| <51 | 440 | 167 | 69 | 79 | 51 | 67 |
| 51–100 | 1628 | 1199 | 278 | 67 | 21 | 53 |
| 101–150 | 2006 | 1469 | 366 | 107 | 22 | 45 |
| 151–200 | 698 | 456 | 151 | 43 | 14 | 30 |
| 201–250 | 205 | 73 | 78 | 31 | 7 | 14 |
| 251–300 | 114 | 53 | 27 | 14 | 7 | 13 |
| 301–350 | 32 | 11 | 8 | 8 | 1 | 4 |
| 351–400 | 24 | 12 | 4 | 1 | 1 | 6 |
| 401–450 | 9 | 0 | 1 | 4 | 0 | 4 |
| 451–500 | 3 | 0 | 1 | 0 | 0 | 2 |
| >500 | 6 | 0 | 2 | 1 | 0 | 3 |
Defined as the number of assignments/number of residues.
How reliable are such partial protein assignments? So far, there has not been a way to assess this question with confidence. In the case of (virtually) complete assignments, it is possible to ensure that (almost) every peak in the triple resonance data is assigned once and only once, that are (almost) no unassigned peaks remain, and that (almost) no unassigned residues remain. In addition, when complete assignments are used for structure determination, one will discover whether they are compatible with a reasonable secondary structure. Regretfully, these tests are not available in cases of partial assignments, especially for proteins of unknown structure. One may use residue-selective labeling to help and/or assess partial assignments and/or use NOESY, but this is not common practice. Systematic mutagenesis is another valid, but labor intensive, approach to guide and/or verify the assignments. Alternatively, one may repeat the assignment process and assess whether a given assignment is reproducible. Such a task is best accomplished with a computer using a fast assignment program. We recently presented such a program, called SAGA 4, which is fast enough to complete a single assignment of a 400-residue protein in about 30 seconds. The program uses a probabilistic branch-and-bound algorithm that automatically repeats from randomly chosen different starting conditions, keeping track of all results. SAGA can produce and evaluate about 4000 different assignments in 24 hours. However, those 4000 independent assignments are nowhere near to astronomical number of possibilities one would need to evaluate to be certain that the correct assignment has been found.
In this report, triple resonance NMR data are referred to as Generalized Spin Systems (GSS), as defined by Montelione and co-workers 5, i.e. a NH “root” with CA(i), CB(i), CO(i) and CA(i-1), CB(i-1), CO(i-1) “rungs”. We do not consider HA(i) and HA(i-1) rungs, because triple resonance data on larger proteins is obtained from perdeuterated systems. To give a sense for the combinatorial barrier to the assignment problem, consider the placement of 100 GSS on 200 residues. One easily perceives that there are 200*199*198*....*100 ~ 1.6×10216 different ways to do this and to exhaustively compare all possible assignments.
Here, we present an approach that dramatically reduces the combinatorial problem by placing GSS on smaller segments. First, we scan the amino acid sequence for stretches of residues that have a unique sequence. This is equivalent to a typical “hand-assignment” approach: one first assigns unique di- or tri-peptides, typically containing amino acids such as alanine, serine, threonine and glycine that can be easily recognized in the NMR data. In the hand assignment, one extends these assignments by adding GSS on both sides. However, in large proteins, there are not many unique di- and tri-peptides to start with. Using a computer, one can easily find and start fitting much larger peptides such as decapeptides which are unique even in large proteins. Unique deca-peptides cover virtually all residues of these proteins. There are just 100*99*…*91 = 6.3×1019 ways of picking 10 GSS out of a collection of 100 to fit a particular unique deca-peptide. As there are at most 190 unique deca-peptides in a 200 residue protein, there are just 1.2×1021 possible assignments. Combined with intelligent search approaches (see below), an exhaustive comparison of all possible assignments is feasible in a minute or less, using a laptop computer.
We have dubbed the computer program “EZ-ASSIGN”. After (several of) the unique deca-peptides have been assigned, the corresponding sequence positions are masked and the assigned GSS are taken out of the GSS pool. Next, the remaining sequence is searched for unique nona-peptides, which are assigned from the remaining GSS, etc., all the way down to mono-peptides. A key virtue of this approach is that more small peptides become unique after larger peptide blocks have been assigned. In EZ-ASSIGN, the user can control the number of required rung matches, the rung match tolerance, the GSS-type classification ranges, search mode, and several other parameters. Thus, a user can design and optimize his/her own protocol, using simple Unix input scripts. EZ-ASSIGN was calibrated using the BMRB data for the 723-residue Malate Synthase (BMRB ID: 5471).
We applied EZ-ASSIGN to the experimental data of two large protein domains: Residues 1–388 of the E. coli Hsp70 chaperone protein DnaK, for which 70% of the backbone assignments were made by hand, 3 and residues 105–456 of a type III secretive ATPase, for which >90% of the backbone assignments were made by hand (P. Rossi, N. K. Khanra, and C. G. Kalodimos, unpublished data). For these proteins, EZ-ASSIGN slightly outperformed the fully automatic computer assignment program PINE6 but dramatically out-performed the programs SAGA4, AUTOASSIGN5 and IBIS7.
We investigated how well the different computer programs performed with NMR data of decreasing completeness, which is common when working with large proteins with limited sample concentration. Because EZ-ASSIGN was expressly written for this situation, it can still confidently assign fragments in severly degraded data, where the other tested programs cannot.
Methods
Data preparation using Sparky
EZ-ASSIGN requires six input files: one for HNCA(i) peaks, one for HNCA(i-1) peaks, one for HNCO(i) peaks, one for HNCO(i-1) peaks, one for HNCB(i) peaks and one for HNCB(i-1) peaks. The files may be empty. In each file, three columns of frequencies, corresponding to the NH “root” and CA, CB, or CO “rungs”, are provided. Optionally, columns listing peak intensities and individual rung tolerances (the frequency range allowed to define a matching rung between adjacent GSS) are accepted. In addition, a sequence file is needed.
EZ-ASSIGN assumes that the numbers in the peak labels (e.g. in Sparky format 8) in each of the six files refer to the same spin system. That is, EZ-ASSIGN assumes that the GSS have been previously constructed. In our experience, the protocol of constructing GSS from crowded spectra of large proteins with limited signal-to-noise requires decisions about noise recognition, the effect of noise and overlap on peak shape and peak position, that are much more reliably made by NMR spectroscopists than computer programs.
Nevertheless, the release package of EZ-ASSIGN comes with an independent program to help assemble the required lists from raw Sparky-style peak pick lists if so desired.
A common problem in spectra of large proteins is the presence of two or more GSS per NH coordinate: for just two NH-overlapping GSS with complete rungs, there are 25= 32 ways to create two complete GSSs. It is best to leave such GSS out of the GSS pool at the beginning of the assignments, and to assign them by hand at the end of the EZ-ASSIGN procedure.
Key to the approach
There are ~ 1.6×10216 different ways to place 100 GSS on 200 residues, so there are an equal number of independent assignments possible. In order to find the correct one, one must evaluate how well the rungs between adjacent GSS match and whether the GSS are compatible with the amino acid sequence. Previously proposed computer assignment algorithms have approached this problem using simulated annealing methods9, genetic algorithms6 and first-best5 approaches. Unfortunately, the best assignment can never be guaranteed by any of these procedures without infinite trial time. However, it is possible to dramatically reduce the number of permutations that must be tested (and thus the trial time) by 1) considering short, unique peptide sequences, 2) making use of the fact that the GSS of seven different amino acid types can be distinguished in triple resonance data (HNCACB) (see Table S1), and 3) building the assignment sequentially from the N-terminus of the unique target peptide. For example, in a 10-residue target peptide beginning with alanine, it is unnecessary to test downstream assignment possibilities for residues 2–10 without first identifying a GSS compatible with alanine at position 1. In Table 2, we show that by considering only appropriate choices for downstream evaluation, there are just 7.8×1012 ways to match 100 GSS to a single deca-peptide, and 1.5×1015 ways to do this for the overlapping deca-peptides in a 200 residue sequence. Hence, with this procedure, one can easily exhaust all possibilities even in larger systems and a fully trustworthy assignment can be obtained.
Table 2.
Permutations of fitting 100 GSSs with known type to a 200-residue protein and shorter peptides, not considering rung-connections
| <Occurrence> in 200 residues | <Occurrence> in 100 GSSs | permutations for 100 GSSs | number | |
|---|---|---|---|---|
| G | 10 | 5 | 10*9*..*6 | 30240 |
| A | 10 | 5 | 10*9*..*6 | 30240 |
| T | 10 | 5 | 10*9*..*6 | 30240 |
| S | 10 | 5 | 10*9*..*6 | 30240 |
| D F I L N Y | 60 | 30 | 60*59*..*31 | 5.8*1047 |
| C E H K M Q R V W | 90 | 45 | 90*89*..*46 | 2.3*1080 |
| Product: 10146 | ||||
| <Occurrence> in 20 residues | <Occurrence> in 100 GSSs | permutations for 100 GSSs | number | |
| G | 1 | 5 | 5 | 5 |
| A | 1 | 5 | 5 | 5 |
| T | 1 | 5 | 5 | 5 |
| S | 1 | 5 | 5 | 5 |
| D F I L N Y | 6 | 30 | 30*29*..*25 | 1.4*109 |
| C E H K M Q R V W | 9 | 45 | 45*44*..*37 | 3.2*1014 |
| 2.8*1026 | ||||
| repeat for 199 20- residue peptides | Product: 5.5*1028 | |||
| <Occurrence> in 10 residues | <Occurrence> in 100 GSSs | permutations for 100 GSSs | number | |
| G | 1 | 5 | 5 | 5 |
| A | 0 | 5 | 1 | 1 |
| T | 1 | 5 | 5 | 5 |
| S | 0 | 5 | 1 | 1 |
| D F I L N Y | 3 | 30 | 30*29*28 | 2.4*104 |
| C E H K M Q R V W | 5 | 45 | 45*44*43*42*41 | 1.3*107 |
| 7.8×1012 | ||||
| repeat for 199 10- residue peptides | Product: 1.5×1015 |
The 200-residue protein would contain 10 A, 10 T, 10 G, 10 S, 60 (D F I L N Y) and 90 (C E H K M Q R V W) types.
100 GSS would contain 5 Ala, 5 Thr, 5 Gly, 5 Ser, 30 (D F I L N Y )and 45 (C E H K M Q R V W) types.
Description of the program
The program first assembles the six input files into a pool of GSSs and selects the possible residue types for both GSS(i) and for GSS(i-1) Residue type is determined using BMRB statistics for CA and CB resonances in diamagnetic proteins (see Table S2). Next, the protein sequence is translated into an “NMR sequence” based on the same statistics. We evaluated three different translations as shown in Table S1. Subsequently, the program starts from the N-terminus and identifies unique sequence stretches (“peptides”) from 1 to 9 residues in length. The program then searches the pool of available GSSs for matches to the unique peptide, starting from the N-terminus. For the N-terminal position, the program checks if the (i-1) rungs of the considered GSS, if available, are compatible with the type of the preceding residue in the complete sequence. If the preceding residue is assigned, the program checks for rung matches and mismatches. Here, and everywhere else in the program, any rung mismatch results in an immediate rejection of that placement, no matter how many other matching rungs are found. When a candidate for the N-terminus of the search peptide is identified, the program searches for a match to the next sequence position in the search peptide. Successful matches for this position must be of the correct amino acid type and must match with the previous position given the rung tolerances and required number of rungs. Rung tolerances may be set globally, but differently, for CA, CB and CO rungs, or individually for each rung. If no match is found for the second position, a new candidate is sought for the N-terminal position, and the search for the second position starts again. If a successful candidate is found for the second position, the search continues to the third position. If none is found, a new candidate for the second place is sought. This process continues until all positions have been filled with candidates or until all possibilities have been exhausted. For the GSS placed on the C-terminus of the peptide, the program checks if it is rung compatible with the N-terminus of a previously assigned stretch, if applicable.
EZ-ASSIGN was not designed to be a fully automatic assignment program. For example, the program does not check whether alternative assignments made for a single peptide use a certain GSS several times or not. Rather, the program lists all possibilities, and lists how many times each GSS was used. It also does not check whether new assignments made for different peptides are compatible with each other, but lists them all. The user must then select among assignments for the best match.
However, if so desired, one may run EZ-ASSIGN in an automatic mode, in which the assignment output is automatically parsed to retain only the last assignment of overlapping assignments. One may then use a script in which that assignment is used a restraint for the following run.
EZ-ASSIGN was written in Fortran90. It was compiled with a GNU compiler on an Apple Macbook Pro 2.4 GHz Intel Core 2 Duo computer, running OSX.6.8. EZ-ASSIGN evaluates all nona-peptides in the NMR data for malate synthase within 10 seconds. With 735 residues and 654 assignments, malate synthase is the largest protein assigned to date; hence there does not appear to be a practical limit to the applicability of EZ-ASSIGN.
Calibration
Calibration of word size, rung tolerance and chemical shift ranges for the residue type identification was accomplished using BMRB data bank data entry 5471 for Malate Synthase. These data are virtually complete with six-rung GSSs for nearly every amino acid. Due to its size (735 residues), the Malate synthase data are a good representation of the statistical variation in resonance positions in all proteins.
1) Chemical Shift ranges
Statistics for CA and CB resonance ranges for different amino acids, as listed in the BMRB, are shown in Table S2. We found no benefit in including CO or N(H) resonance statistics in the definition of amino acid type (not shown). The BMRB also lists standard deviations for the variation of CA and CB resonance positions per residue type. If the BMRB distributions were Gaussian (they are not) one should expect at least 30% of the experimental data at hand to lie outside of the standard deviations. Hence, we used multiplication factors of the BMRB ranges to facilitate a complete assignment. Table S3 shows the performance and reliability of EZ-ASSIGN on Malate synthase as a function of CA and CB range multipliers. The tables show that larger ranges can be used for larger peptide searches than for smaller peptide searches.
2) Word size
Table S4 shows the performance of EZ-ASSIGN on Malate synthase as a function of NMR word size and search length, with other parameters as listed in the table caption. Fewer unique peptides were found when using a 5-letter translation than with a six or seven letter translation. Typically, multiple assignments were found for the unique peptides when word length was large. This was caused by the fact that the same GSS can belong to several different amino acid types, especially when using large multipliers for the CA and CB resonance ranges. When considering peptides longer than approximately 7 residues, there were not large differences between 5-, 6- and 7-letter codes in the number of unique peptides identified and assigned. However, in real data, the bulk of the assignments were obtained when searching for tetra- to hexa-peptides (see below). From Table S4 it is clear that one can obtain many more assignments for tetra- and hexa-peptides using 6 or 7 code without paying too much of a price for multiple assignments.
3) Precision
Next, we addressed the influence of matching tolerance on the assignment performance. EZ-ASSIGN uses a definition that two rungs match if their chemical shifts differ by less than the sum of their tolerances. With the artificial data for malate synthase, identical results were obtained with tolerances between 0.02 and 0.05 ppm (Table S5). Setting the tolerance to 0.1 to 0.15 ppm resulted in greater numbers of incorrect assignments. Experimentally, we found that for a sample of 250 uM triple labeled DnaK(1–388), a C-rung tolerance of 0.05 – 0.08 ppm was optimal for most resonances (see below).
Probability indicator
When searching for assignments requiring only one rung per connectivity, typically many possible assignments are found per trial peptide, especially when the type information is lacking for many GSS. For long search peptides, the number of valid assignments can then run in the thousands per peptide. How to rank these possibilities in the output? One can reasonably assume that assignment with the largest total number of rung matches takes precedent over one with fewer. An assignment which connects to a previously assigned stretch, either front or back or both, is worth considering. If neither of these criteria resolve the issues, one may also evaluate whether the resonance intensities vary wildly over the assignment stretch or not. We have also tested whether 13CA, 13CB and 13CO line width information as provided by the Sparky peak pick function can be used as a criterion to select between different possible assignments. However, we were not able to obtain a meaningful correlation between a line width match and a rung match for the large protein data considered.
In severely incomplete data, there is a whole other issue that needs to be considered: could better GSS have been available in the “missing” data, even if a single assignment was found in the available data? The probability of correctness of the provided assignment in the context of missing data, must be a function of the density of the NMR spectrum of typical protein spectra. For instance, a CA(i) – CA(i-1) rung match at 65 ppm in the available data would likely not occur often in the missing data, while a CA(i) – CA(i-1) rung match at 55 ppm could very well have many alternatives in the missing data.
To illustrate EZ-ASSIGN’s approach to this issue, we will assume a tetra-peptide which has a proposed assignment based on a CA rung match between (i) and (i+1), a CO rung match between (i+1) and (i+2) and a CA+CB rung match between (i+2) and (i+3). We also assume that the protein has 350 theoretical GSS, but that only 340 CO, 300 CA and 250 CB signals are observed. EZ-ASSIGN calculates a probability for that assignment to be correct as follows. For the CA match it consults a database of all 600 CA chemical shifts in the Malate Synthase assignments. The program finds how many corresponding entries exist at that shift with a range as given by the listed tolerances in the experimental data. Suppose that there are 20 entries in the Malate synthase data which fulfill these criteria. Hence, in the 50 missing experimental CA one may expect 50/600*20=1.6 residue with a matching CA within the tolerance range. Now the probability that the found CA GSS connectivity in the observed data is correct, is given by 1/(1+1.6). Similarly, for the next CO rung match, EZ-ASSIGN consults a database of all CO chemical shifts in the Malate Synthase BRMB entry, and finds, for instance, 25. Hence, in the 10 missing experimental CO one may expect 10/600*25=0.4 residue with a matching CO within the tolerance range. Now, the probability that the found CO GSS connectivity in the observed data is correct is given by 1/(1+0.4). If more than one connection is found for a sequential match, a joint statistics, also based on the Malate Synthase data is consulted (how many residues are there with the combined CA and CB frequencies, given the precision ranges). Suppose there are just 2 in Malate synthase; we then expect 100/600*2=0.33 in the missing CA/CB data, and the probability is 1/(1+0.33). The probability for the entire stretch is taken as the product of the probabilities of the individual connectivities. EZ-ASSIGN also has a joint statistics file for CA and CO, and for CB and CO, as well as one for all three. The program computes the probabilities for all pursued assignments. The user can set a probability threshold that will prevent the program from copying low-probability assignments to the output.
Best protocol
We combined the calibrations above to arrive at the assignment protocol as recommended in Tables 3A and 3B. For assignment of malate synthase, tau, DnaK, and type III secretive ATPase, the protocol in Table 3A (searching for unique peptides + adjacent peptides) was consecutively executed three times: once with three required rungs, once with two required rungs and finally with one required rung. The obtained assignments were used as restraints for the next assignments. Next, we executed one or more runs using a search mode in which all remaining peptides were tested, whether they were unique or not (scanning mode 1, see Table 3B). This latter search may pick up assignments that were previously missed because the preceding residue type is not included in the search for unique peptides. Such a scanning mode run can be done with the same ranges as in Table 3A. One may also combine the scanning mode runs with increased CA and CB ranges in order to also capture assignments for residues that lie far outside the BRMB statistics.
Table 3.
| Table 3A. Recommended assignment protocol, search mode 3b | |||
|---|---|---|---|
| Search length | Word length | Alpha Range | Beta Range |
| 9 | 7 | 3.0 | 3.5 |
| 8 | 7 | 2.7 | 3.2 |
| 7 | 7 | 2.6 | 2.8 |
| 6 | 7 | 2.5 | 2.5 |
| 5 | 7 | 2.25 | 2.25 |
| 4 | 7 | 2.0 | 2.0 |
| 3 | 5 | 1.75 | 1.75 |
| 2 | 5 | 1.50 | 1.50 |
| Table 3B. Recommended assignment protocol, search mode 1b | |||
|---|---|---|---|
| Search length | Word length | Alpha Range | Beta Range |
| 5 | 7 | 2.5 | 2.5 |
| 4 | 7 | 2.5 | 2.5 |
| 3 | 5 | 2.5 | 2.5 |
| 2 | 5 | 2.5 | 2.5 |
| 1 | 5 | 2.5 | 2.5 |
| 1 | 5 | 4.0 | 4.0 |
search mode 3: unique peptides and peptides adjacent to previously assigned peptides
search mode 1: all peptides, whether unique or not.
Several variations and extensions of this protocol are possible. One may at the end want to include a run with larger rung tolerances to capture assignments for GSS with low S/N for which the resonance position is not so precisely defined. Or, one may execute the entire protocol of Tables 3A and 3B first with only those GSS for which type-information is available, followed by the entire protocol including all data. Lastly, one may create a protocol which completely simulates a hand assignment: start by assigning unique tri-peptides, and extend them by one GSS at a time.
Use of other software packages
The input lists used for EZ-ASSIGN were used for SAGA4 without change. For PINE6 and AUTOASSIGN5, the HNCA(i) and HNCA(i-1) peak files used for EZ-ASSIGN were combined into a HNCA file, the HNCA(i), HNCA(i-1), HNCB(i) and HNCB(i-1) peak files into a HNCACB file, the HNCA(i-1) and HNCB(i-1) peak files into a HNCOCACB file, and the HNCO(i) peaks, and HNCO(i-1) peaks into a HNCACO file. The HNCA(i-1) peak file served as a HNCOCA file and the HNCO(i-1) peak file as a HNCO file. Peak labels were removed in all files. In these files the N-H coordinates for the corresponding cross peaks are identical. The files do not contain GSS with identical NH coordinates. The files were uploaded on the PINE and AUTOASSIGN webservers. Dr. S. Hyberts (Harvard) was kind enough to assign the PINE files using the program IBIS7.
Availability
EZ-ASSIGN source code, with several utility programs, examples and manuals, is available for download from the University of Michigan Technology Transfer Department (http://inventions.umich.edu/technologies/5729/ez-assign-fortran-source-code-for-nmr-resonance-assignments). The use of the program is unlimited in time and scope, free of charge for academia and non-profits, but bound to rules set forth in a license agreement that can also be found at that website.
Results
Synthetic data
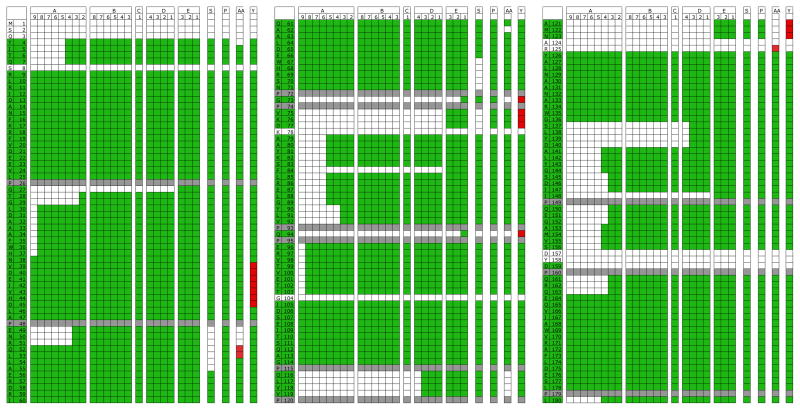
The progress of the assignment of all 723 residues of Malate synthase is illustrated for the first 180 residues in Fig 1. The bulk of the assignments were obtained in the run searching for nona-peptides with GSS connected by three rungs. Nevertheless, runs with decreasing numbers of rung connections and increasing range tolerances were necessary to complete the assignment. The final EZ-ASSIGN assignment, shown in column E1 of Fig1, is equivalent to the literature assignment, except for a missing assignment for D159. It is important to note is that residues not assigned in the literature data were not assigned by EZ-ASSIGN either.
Figure 1.
The progress of the re-assignment of the triple resonance data of Malate Synthase (BMRB 5471) using EZ-ASSIGN. For legibility only the first 180 residues are shown. The available assignments are shown in green on the sequence in the left two columns. Grey fields are Pro residues. The columns A9-A2 report progress using the protocol of Table 3A, requiring 3 rungs connectivities, assigning unique peptides in 7-letter code. Columns B8-B3 show the results of the same protocol requiring 2 rungs, and column C1 the results of the same protocol requiring 1 rungs. Columns D4-1 used the protocol of Table 3B, requiring 1 rung, unique mode. Columns E3-1 used the protocol of Table 3B, requiring 1 rung, scanning mode. Missing columns, such as C9-2, attest to the fact that no new assignments were found for those searches after those obtained by run B3.
Green fields show assignment corresponding to the BRMB file, red ones those that do not.
The column labeled “S” is the assignment obtained with SAGA.
The column labeled “P” is the assignment obtained with PINE. The column labeled “AA” is the assignment obtained with AUTOASSIGN.
The column labeled “Y” shows an “automatic” assignment obtained with EZ-ASSIGN, in which the output of the individual steps A-E was used as restraints for the next steps without human intervention. This run also used unassembled GSS just as in PINE and AUTOASSIGN. Also see Table 4.
The column marked “S” is an assignment of the same GSS data as obtained by using a 30 minute SAGA run. While SAGA makes no errors, the assignment is less complete then the one obtained with EZ-ASSIGN. The column marked “P” is an assignment by using the PINE web server; and the column marked “AA” is an assignment by using the AUTOASSIGN webserver. The column labeled “Y” shows an assignment obtained with EZ-ASSIGN when using the same unassembled GSS just as in PINE and AUTOASSIGN. The GSS were re-assembled using a series of protocols using a series of GSS matching scripts and programs that are part of the EZ-ASSIGN release. Furthermore, in run “Y”, EZ-ASSIGN used the automatic protocol in which the output of the individual steps was used as restraints for the next steps without human intervention, as described above. The result was 614 correct assignments and 38 incorrect assignments, corresponding to a 6 % error rate (see Table 4).
Table 4.
Computer assignments of literature data of Malate Synthase and TAU(11–124)
| MALATE SYNTHASE (BMR5471) | TAU(11–124, BMR17945) | |||
|---|---|---|---|---|
| method | Identical | Different | Identical | Different |
| Literature | 653 | 99 | ||
| EZ-ASSIGN interactive | 652 | 0 | 99 | 0 |
| PINE | 647 | 4 | 91 | 1 |
| SAGA | 569 | 0 | 88 | 2 |
| AUTOASSIGN | 563 | 12 | 31 | 1 |
| EZ-ASSIGN automatic | 614 | 38 | 79 | 9 |
EZ-ASSIGN was used in automatic mode using pre-assembled GSS to address the question as to what type of NMR data are most critical to the assignment of the spectra of large proteins. Removing all CO(i) rungs from the Malate Synthase dataset resulted in 563 correct assignments with 35 errors, while removing all CB(i-1) rungs yielded in 412 correct assignments with 135 errors (results not shown). Comparing these results with assignment using all rungs (614 correct assignments and 38 incorrect) suggests that the HN(CA)CO experiment is almost superfluous, even for proteins of this size. This finding suggests that experimental time is better spent obtaining as many as possible CB(i-1) rungs by running the HNCACB longer.
For those interested in comparison of the performances of fully automatic assignment procedures without human intervention, such as AUTOASSIGN and PINE, with EZ-ASSIGN, column Y is the fair comparison. It appears that PINE is superior to EZ-ASSIGN in the automatic mode, while AUTOASSIGN and EZ-ASSIGN in automatic mode are comparable. However, EZ-ASSIGN was not conceived to be a fully automatic program, and as we will demonstrate below, dramatically outperforms PINE, AUTOASSIGN, SAGA and IBIS on incomplete experimental data. We have not included MARS10 in our comparison because that program’ s webserver does not accept CO rung data. Table 4 lists the final results for Malate Synthase.
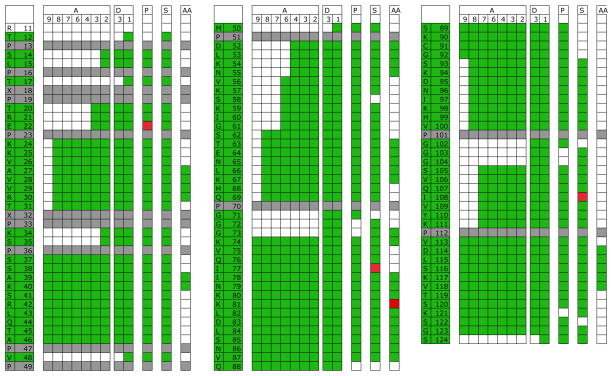
Currently, there is much interest in assigning natively unfolded proteins. The BMRB lists an assignment for 99 of the residues of human tau (11–124), based on HNCACB, HNCOCACB, HNCA and HNCOCA data. The progress of the complete and error free assignment by EZ-ASSIGN is shown in Figure 2. EZ-ASSIGN, when run in the interactive mode, also for this case outperforms all other computer assignment programs tried (see Table 4).
Figure 2.
The progress of the re-assignment of the triple resonance data of human Tau (11–124) (BMRB 17945) using EZ-ASSIGN. The available assignments, based on CA and CB rungs only, are shown in green on the sequence in the left two columns. Grey fields are Pro residues.
The columns A9-A2 report progress using the protocol of Table 3A, requiring 2 rungs connectivities, assigning unique peptides in 7-letter code. Columns D3-1 used the protocol of Table 3B, requiring 1 rung, scanning mode. Missing columns, such as B9-2 and C9-2 attest to the fact that no new assignments were found for those searches after those obtained by run A2.
Green fields show assignment corresponding to the BMRB file, red ones those that do not.
The column labeled “P” is the assignment obtained with PINE.
The column labeled “S” is the assignment obtained with SAGA.
The column labeled “AA” is the assignment obtained with AUTOASSIGN.
Also see Table 4.
Real data
Our group has been working on Hsp70 protein folding chaperones for many years. These 70 kDa monomeric proteins have four domains: a nucleotide-binding domain (NBD; residues 1–390) a substrate-binding domain (SBD residues 400–500), LID (residues 510–610) and TAIL (residues 611–650) (e.g. see 11,12). For the Hsp70 of E. coli, called DnaK, assignments for all individually expressed domains were made by hand using standard suites of six triple resonance experiments, using a samples of ~ 250 μM in protein and using a 800 MHz Varian spectrometer with cryoprobe 3. No NOE data were used. Here we use the spectral peak pick data of triple-labeled NBD(1–388) for tests with EZ-ASSIGN. This NBD has a rotational correlation time of 20 ns at 30 °C 3.
For assignment using EZ-ASSIGN, GSS with overlapping NH coordinates were removed from the DnaK data. Further, the data were idealized for the N and H coordinates; i.e. the NH coordinates of all corresponding cross peaks were made identical, in order to facilitate the GSS assembly by PINE, AUTOASSIGN and IBIS. This did not influence EZ-ASSIGN as that program does not use NH coordinates. Figure 3 shows the progress of the assignment of all 388 residues using the protocols of Tables 3A and 3B as described above. For clarity, only part of the assignment is shown. The progress of the assignment is quite different than that for synthetic data: here most assignments are found while looking for hexa-to-tetra peptides. Also in the figure are the assignments as obtained by SAGA, PINE, AUTOASSIGN and IBIS. As is shown in Table 5 in the column “ALL”, EZ-ASSIGN, in interactive mode, makes the largest number of correct assignments and differs only by one assignment, which turns out to be a mistake in the hand assignment data. PINE is a close second best with many correct assignments and only 9 errors. The other columns in Table 5 will be discussed below.
Figure 3.
The progress of the assignment of the experimental triple resonance data3 of DnaK using EZ-ASSIGN. For legibility only residues 121–300 are shown. The available assignments are shown in green on the sequence in the left two columns. Grey fields are Pro residues.
The columns A report progress using the protocol of Table 3A, requiring 3 rungs connectivities, assigning unique peptides in 7-letter code. The columns B report progress using the protocol of Table 3A, requiring 2-rungs connectivities, assigning unique peptides in 7-letter code. Columns D used the protocol of Table 3B, requiring 1 rung, scanning mode. Missing columns, such as C9-2 attest to the fact that no new assignments were found for those searches after those obtained by run B2.
Green fields show assignment corresponding to the BRMB file, red ones those that do not.
The column labeled “P” is the assignment obtained with PINE.
The column labeled “S” is the assignment obtained with SAGA.
The column labeled “AA” is the assignment obtained with AUTOASSIGN.
The column labeled “I” is the assignment obtained with IBIS.
The shown assignments correspond to the column entry “ALL” in Table 5.
Table 5.
DNAK NBD (1–388) RESULTSa
| Method | Allb | SN>20c | SN>30 | SN>30e | SN>30f | SN>40 | SN>50 | SN>60 |
|---|---|---|---|---|---|---|---|---|
| Hand | 294 | |||||||
| EZ- ASSIGNd | 293/1 | 239/3 | 142/6 | 115/8 | 108/4 | 103/21 | 46/5 | 11/0 |
| Pined | 291/9 | 281/63 | 232/82 | 163/63 | error | error | ||
| SAGAd | 241/6 | 150/3 | 119/14 | 53/5 | 32/7 | 49/25 | ||
| Auto Assign | 202/9 | 55/0 | 39/1 | 14/0 | 12/0 | 5/1 | ||
| IBISd | 261/86 | n.a. | n.a. | n.a. | n.a. | n.a. |
results are listed as total number/number different as compared to the hand assignment
using all data as used by the hand assignment – the actual spectra contained more peaks
taking only data with S/N ratio above listed threshold (see Table S6).
only taking assignments with > 50% probability
NO CO(i) rungs
NO CB(i-1) rungs
To explore whether there is something unique about the data of DnaK that may confuse the previously published assignment programs, we also used experimental data for residues 105–456 of a type III secretive ATPase 13, for which >90% of the backbone assignments were obtained by hand using triple resonance data and NOESY spectra (P. Rossi, N. K. Khanra and C. G. Kalodimos, unpublished data). As Table 6 shows, EZ-ASSIGN is not perfect for this data set, but outperforms the other available programs by an even larger margin. The 17 differences between the EZ and hand assignment are shown in Table S8, and are extensively discussed in the legend to that table. According to the criteria used, the differences are all in favor of the EZ-assignment; it is not that the hand assignments are impossible, but less likely from the triple resonance data alone. Additional data from NOESY spectra have tilted the assignment towards the hand assignment.
Table 6.
Results for type III secretive ATPase(105–456)a
| Method | Allb | Deg1c | Deg2 | Deg3 | Deg4 |
|---|---|---|---|---|---|
| HAND | 317 | ||||
| EZ- ASSIGNd | 298/17 | 216/37 | 134/31 | 66/7 | 28/0 |
| Pined | 304/41 | 265/73 | 251/125 | 174/111 | error |
| SAGAd | 130/20 | 109/34 | 70/19 | 29/10 | 21/7 |
| Autoassign | 123/6 | 64/22 | 18/2 | 7/0 | 0 |
results are listed as total number/number different as compared to the hand assignment
using all data as used by the hand assignment – the actual spectra contained more peaks
Only using data above increasingly higher thresholds (See Table S7).
only taking assignments with > 50% probability
Assignments with degraded experimental data
Every assignment program has been tested with intentionally degraded data. However, performance depends much how one degrades data: for obvious reasons, deleting a series of connected GSS will have quite a different effect than deleting every other GSS. To avoid a subjective degradation protocol, we chose to degrade the DnaK data using actual intensity or S/N information. By deleting individual cross peaks under a certain S/N threshold, this approach simulated data that were recorded in less time, recorded on a less concentrated sample or recorded on a larger protein. By using the same S/N threshold on all spectra, HNCACO cross peaks were lost much earlier than HNCO cross peaks, also in keeping with reality.
Table S6 shows how the selection process affected the data statistics for DnaK. Selecting peaks with SN > 20 caused a substantial change in the number of CB(i-1)-rungs (231 to 166) while the number of other connectivities did not change much. Table 5 shows that this change resulted in a loss of 54 assignments in EZ-ASSIGN, 90 in SAGA and 150 in AUTOASSIGN. PINE lost only 10 assignments, but made 60 additional errors. So even with a small a reduction in data quality, we found that AUTOASSIGN became unusable because it made too few assignments, and that PINE became unreliable because it made too many errors. IBIS was not further pursued because of its less-than-ideal performance on the full data set. The only fully automatic program that remained reliable while still making a large number of assignments was SAGA, but it cannot compete with EZ-ASSIGN when it was run with human intervention. The trend persisted as the data were further degraded.
We again asked the question whether this assignment behavior was peculiar to the data of DnaK. For the Type III ATPase, the first degradation step also mainly caused a loss in CB(i-1) rungs (Table S7), and also resulted in AUTOASSIGN to “drop out” and PINE to make too many errors (Table 6).
Discussion
The results in Tables 5 and 6 demonstrate that EZ-ASSIGN, when run in an interactive way, is a reliable tool to obtain assignments for large proteins.
In case of complete data such as for Malate Synthase, the EZ-assignment is basically finished at the deca-peptide search level. For incomplete data such as for DnaK, only a few of the unique deca-peptides can be assigned because of missing GSS and/or rungs. Here the bulk of the assignment occurs at the hexa-tetra peptide level. For those hexa-tetra peptides for which an assignment is found, the assignment must be robust: the sequence to which the GSS are fitted was chosen to be unique, and matches with all reasonable GSS were surveyed.
If more than one assignment is found for a sequence with well-defined GSS types, it must represent a second conformation in slow exchange. When the GSS types are not well defined, the scientist can possibly distinguish between two or more assignments based on the total number of rung connections, the completeness of type identification, the number of times a GSS was used, whether the peak intensities are roughly equal or not, and the assignment probability factor. If neither of these criteria can resolve the case, all assignments for that peptide must be set aside until more data becomes available (e.g. NOESY, HNCANH14, or HNCAHA/HNCOCAHA).
PINE and EZ-ASSIGN performed about equally well for the reasonably complete data of DnaK (74% of expected HNCACB peaks) and the type III ATPase (85% of expected HNCACB peaks). In those cases, PINE may be the assignment program of choice as it runs in fully automatic mode via a webserver. AUTOASSIGN, IBIS and SAGA did not perform as well on the data of these proteins.
However, when the data were only a little less complete, EZ-ASSIGN clearly outperformed PINE (and all other programs). EZ-ASSIGN obtained assignments in the data with SN > 50 and SN > 60 (Table 5) with a 85% confidence when PINE did not yield a single assignment. Interestingly, in the latter cases AUTOASSIGN and SAGA are better than PINE, but not nearly as good as EZ-ASSIGN. EZ-ASSIGN’s ability to find reliable assignments in very incomplete data can be directly attributed to the strategy of dividing the sequence into smaller unique fragments. In EZ-ASSIGN there is no direct competition from the large fraction of unassigned regions for the same GSS.
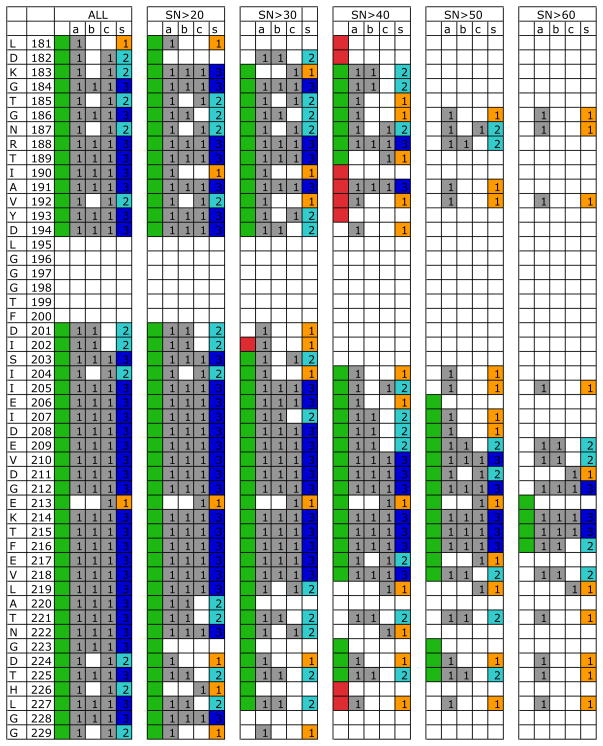
As documented in tables 5 and 6, we show that the assignment deteriorates remarkably rapidly upon slight data degradation. For instance, when the number of HNCACB peaks was only one half of the original, only about 60 out of the earlier 300 assignments for DnaK were found (Table 5). What is the reason for this very non-linear behavior? The results in Table 7 suggest that assignment success was best correlated with the sum of available two-rung and 3-run matches (i.e. the slope of the best fit is closest to unity). Figure 4 shows, at the residue level, how the assignment of a region of DnaK is affected by losses in rungs. The assignment was in most cases lost or became incorrect when the number of matching rungs per GSS pair was less than 2. Since the CA match is almost always present, the success of obtaining an assignment depends on the availability of either a CB or a CO rung.
Table 7.
Parameters for a linear least-square fit of the number of correct assignment vs. the total number of rung matches as identified in left column.
| DnaK(1–388) | ATPase(105–456)a | |||||
|---|---|---|---|---|---|---|
| Rung matches | slope | Y-intercept | R2 | slope | Y-intercept | R2 |
| 1+2+3 | 0.59 | 51 | 0.975 | 0.7 | 113 | 0.93 |
| 2+3 | 0.7 | 34 | 0.993 | 0.86 | 24 | 0.988 |
| 1+3 | 0.47 | 51 | 0.978 | 0.27 | 92 | 0.788 |
| 1+2 | 0.01 | 93 | 0.005 | 0.27 | 110 | 0.518 |
| 3 | 0.58 | −4 | 0.966 | 0.43 | 3 | 0.97 |
| 2 | 0.12 | 38 | 0.587 | 0.43 | 21 | 0.967 |
| 1 | −0.11 | 54 | 0.633 | −0.16 | 89 | 0.36 |
Figure 4.
Graph showing rung statistics and EZ-ASSIGN assignments for degraded experimental NMR data of DnaK. Only residues 181–229 are shown for reasons of legibility.
The obtained assignments are shown in green (correct) or red (wrong). The column labeled “a” indicates presence of CA rung connection, the column labeled “b” indicates presence of CB rung connection, the column labeled “c” indicates presence of CO rung connection. The column labeled “s” gives the total number of rung connections and is color coded yellow for one, cyan for two, and blue for three rung connections. See also tables 5 and S6.
Table 5 shows that the removal of all CO(i) or of all CB(i-1) rungs caused a loss of 30% of the assignments of the data with SN>30. Hence, the experiments are equivalent in context of assignment efficiency, even for real incomplete data in large proteins. This strongly suggests that one may best utilize limited instrument time by collecting a very good HNCACB, rather than spending time collecting a HNCACO spectrum. But that strategy will likely leave many uncertainties in the assignments of glycines which have no CB rung and typically very little CA chemical shift dispersion. Whether that is acceptable depends on the object of the assignment project.
Supplementary Material
Acknowledgments
E.R.P.Z. acknowledges support from National Institutes of Health grant NS059690 (to G.E. Gestwicki, P.I.). I.B. was supported by National Institutes of Health grant HL 102662 (to S. Ragdale, P.I.); P.R. was supported by National Institutes of Health grant AI094623 (to C. G. Kalodimos, P.I.). The authors acknowledge N.K. Khanra (Rutgers) for the type III ATPAse sample preparation and helpful discussions.
Footnotes
Nine tables, sample protocols scripts, and sample output files.
References
- 1.Montelione GT, Wagner G. Conformation-Independent Sequential Nmr Connections in Isotope-Enriched Polypeptides by 1H-13C-15N Triple-Resonance Experiments. J Magn Reson. 1990;87:183–188. [Google Scholar]
- 2.Kay LE, Ikura M, Tschudin R, Bax A. 3-Dimensional Triple-Resonance NMR-Spectroscopy of Isotopically Enriched Proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A. 2009;106:8471–6. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crippen GM, Rousaki A, Revington M, Zhang Y, Zuiderweg ER. SAGA: rapid automatic mainchain NMR assignment for large proteins. J Biomol NMR. 2010;46:281–98. doi: 10.1007/s10858-010-9403-2. [DOI] [PubMed] [Google Scholar]
- 5.Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- 6.Bahrami A, Assadi AH, Markley JL, HRE Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput Biol. 2009:5. doi: 10.1371/journal.pcbi.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hyberts SG, Wagner G. IBIS--a tool for automated sequential assignment of protein spectra from triple resonance experiments. J Biomol NMR. 2003;26:335–44. doi: 10.1023/a:1024078926886. [DOI] [PubMed] [Google Scholar]
- 8.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2000. [Google Scholar]
- 9.Buchler N, Wang H, Zuiderweg ERP, Goldstein RA. Protein Heteronuclear NMR Assignments using Mean-field Simulated Annealing. J Magn Reson. 1997;125:34–42. doi: 10.1006/jmre.1997.1106. [DOI] [PubMed] [Google Scholar]
- 10.Jung YS, Zweckstetter M. Mars – robust automatic backbone assignment of proteins. Journal of Biomolecular NMR. 2004;30:11–23. doi: 10.1023/B:JNMR.0000042954.99056.ad. [DOI] [PubMed] [Google Scholar]
- 11.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuiderweg ER, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imada K, Minamino T, Tahara A, Namba K. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc Natl Acad Sci U S A. 2007;104:485–90. doi: 10.1073/pnas.0608090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frueh DP, Arthanari H, Koglin A, Walsh CT, Wagner G. A double TROSY hNCAnH experiment for efficient assignment of large and challenging proteins. J Am Chem Soc. 2009;131:12880–1. doi: 10.1021/ja9046685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.