Abstract
We propose a method for differentiating classes of light scatterers based upon their temporal and polarization properties computed from time series of polarization-sensitive optical coherence tomography (PS-OCT) images. The amplitude (motility) and time scale (autocorrelation decay time) of the speckle fluctuations are combined with the cross-polarization pixel-wise to render Motility-, autocorrelation-, and polarization-sensitive (MAPS) OCT contrast images. This combination of metrics provides high specificity for discriminating diffusive gold nano-rods and mammary epithelial cell spheroids within 3D tissue culture, based on their unique MAPS signature. This has implications toward highly specific contrast in molecular (nanoparticle-based) and functional (cellular activity) imaging using standard PS-OCT hardware.
While optical coherence tomography (OCT) has been widely adopted for biomedical imaging, it often lacks the ability to contrast biomarkers of interest. Many methods for adding functional and molecular contrast to OCT have been proposed, including true-color spectroscopic OCT [1], magnetomotive OCT [2], photothermal OCT [3,4], and pump-probe OCT [5]. Here, we propose to exploit the dimension of time in conjunction with polarization-sensitive OCT (PS-OCT) to provide tissue scatterer discrimination. PS-OCT is sensitive to optically anisotropic structures, and has been widely employed to quantify tissue birefringence [6]. At the same time, temporal speckle statistics have begun to implicate several strategies for OCT contrast, including particle diffusion rates [7], capillary velocimetry [8], intracellular motility [9], apoptosis [10], and ciliary activity [11].
In cell biology, 3D tissue culture models are employed to simulate contextual and micromechanical cues that are absent in 2D cell cultures [12]. OCT is advantageous for monitoring 3D tissue models in situations where confocal microscopy cannot provide sufficient depth penetration or speed to image large volumes, such as in breast cancer tissue spheroid models [13]. However, contrast imaging is needed to provide functional information beyond morphology. Here, we employ gold nanorods (GNRs) as diffusive probes into the extracellular matrix (ECM) surrounding breast cancer spheroids. GNRs have been increasingly used in OCT because their surface plasmon resonance can be tuned to near-infrared wavelengths, providing spectroscopic and photothermal OCT contrast [14-16]. In this study, understanding the diffusion of GNRs into 3D tissue models has implications for drug delivery [17], as well as the potential for passive microrheology of the ECM [18].
We propose MAPS (motility-, autocorrelation-, and polarization-sensitive) OCT as a method for distinguishing different types of light scatterers by their temporal and polarization properties. MAPS signatures should be considered in the context of the spatial and temporal resolution of the OCT system and a priori knowledge of the classes of objects to be found within the imaging sample. In the experiments reported here, we consider human mammary epithelial cells (MECs) in a collagen: Matrigel ECM with topically applied GNRs. We chose a frame rate (1.2 Hz) such that the speckle decorrelation due to the adenosine triphosphate (ATP) driven activity of MECs occurred over several frames (τ ~ 4.8 s), allowing for quantification by autocorrelation analysis. The total imaging time (25 s) was chosen to be sufficiently long to observe several decorrelation events from the MECs, providing high speckle fluctuation power, which we will define as motility, below. For the GNRs, our chosen frame rate was too low to capture speckle decorrelation arising from their thermally driven diffusion (τ ~ 0.4 ms when copolarized) and, thus, they exhibited a short autocorrelation decay time in comparison to MECs (Table 1). At the same time, the line scan camera exposure time (0.095 ms at a line rate of 10 kHz) was sufficiently short to capture a single speckle realization from the GNRs within each A-line, resulting in high apparent motility. To complete the MAPS signature, we note that GNRs also exhibit a high amount of cross-polarized scattering owing to their large optical anisotropy. In this Letter, we describe a method that exploits these unique MAPS signatures to contrast specific classes of objects, and demonstrate the ability to differentiate MECs and GNRs in tissue culture.
Table 1.
MAPS Signature for Each Object Class, where ↑ and ↓ Indicate a High or Low Value, Respectively
| Classes of Objects | M (Motility) [a.u.] | A (Autocorrelation) Decay Time [s] | PS (Cross-Polarization) | MAPS Signature |
|---|---|---|---|---|
| GNRs | 36 ± 4 | <0.8 | 0.62 ± 0.07 | ↑↓↑ |
| MECs | 35 ± 9 | 4.8 ± 2 | 0.23 ± 0.12 | ↑↑↓ |
| ECM | 16 ± 3 | 9.7 ± 3 | 0.24 ± 0.11 | ↓↑↓ |
| Noise | 20 ± 4 | <0.8 | 0.52 ± 0.06 | ↓↓↑ |
B-mode OCT images were collected using a polarization-sensitive, spectral-domain OCT system, described in detail previously [18,19]. This system has a center wavelength of 800 nm and axial and transverse resolutions of 3 and 12 μm, respectively. A linear polarization basis was used such that horizontally (H) polarized light was incident upon the sample, and both the copolarized (HH) and cross-polarized (HV) backscattered light were detected. Images were collected over 1.5 mm × 1.5 mm regions sampled into 1000 × 1024 pixels in x and z, respectively. The HH signal was used for motility and autocorrelation analyses, while both the HH and HV signals were used to measure the cross-polarization.
To prepare tissue cultures, MECs (MCF10DCIS.com) were seeded at 30, 000 cells/cm3 into 1:1 collagen I:Matrigel ECM and cultured for 2 weeks before imaging. Images were collected immediately before and 24 h after introduction of GNRs into the cell culture medium (~3.3 × 1011 GNRs/cm3). GNRs stabilized by cetyltrimethylammonoium bromide were prepared by modifying a commonly used method [20], and were ~(83 ± 7) nm × (22 ± 3) nm in size, with a longitudinal surface plasmon resonance (LSPR) tuned to the 800 nm center wavelength of the OCT system (Fig. 1). GNRs were PEGylated using polyethylene glycol (PEG)-thiol with a molecular weight of 1 kDa to prevent adherence to the ECM and ensure they remained freely diffusing during imaging.
Fig. 1.

Transmission electron microscopy (left) and extinction spectrum (right) of the PEGylated GNRs used in these experiments.
OCT is particularly well-suited for imaging MEC tissue cultures, as we have shown previously [13]. Over 1–2 weeks MECs form polarized, acinar structures (spheroids) similar to the structure of in vivo mammary ducts. This unique morphology can be seen in the left column of Fig. 2, where images averaged over time stacks (N = 30) are displayed. Each spheroid is typically comprised of thousands of MECs and appears as a cluster of high light scattering in comparison to the surrounding ECM. Samples with added GNRs display nearly uniform light scattering, due to the penetration of GNRs into the pores of the ECM. Slight inhomogeneities that appear only in the sample with both MECs and GNRs can be suspected to be spheroids that are masked by the surrounding GNRs. Our treatment will utilize MAPS signatures to individually contrast MECs and GNRs in this challenging imaging environment.
Fig. 2.
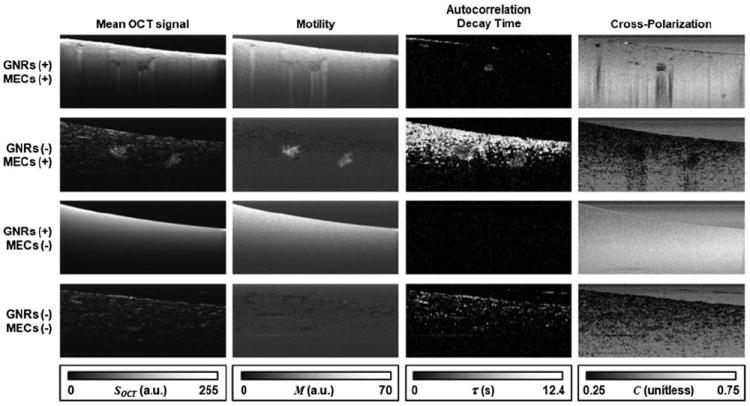
Image stacks of 4 collagen:Matrigel tissue cultures containing both MECs and GNRs (top row), MECs only (second row), GNRs only (third row) and control (bottom row). Each column displays a different way of processing the image stacks, as described in the text. Images represent 1.4 mm × 0.6 mm in x and z.
Motility is typically defined as cellular motion arising from ATP-driven processes, in contrast to other types of biological motion, including diffusion and flow. Here, we employ an experimentally based definition of motility that includes these other types of motion (i.e., the “apparent motility”). We define the motility, M, as the ratio of the measured standard deviation of each pixel to that expected from shot noise,
| (1) |
where SOCT is the OCT signal amplitude at each image position (x, z), sampled at regular intervals ti over N frames. M has a minimum value in the shot-noise limit, and quantifies the relative amount of speckle fluctuation above this limit. Thus, motility is particularly useful for suppressing regions of noise (low scattering) and stationary objects where speckle fluctuations are shot-noise limited, from those exhibiting ATP-driven or thermally driven motion, such as MECs and GNRs, respectively. This is seen in the second column of Fig. 2, where motility is displayed for each tissue culture. Stationary objects such as the ECM are suppressed, while freely diffusing GNRs and live MEC spheroids exhibit high apparent motility.
While motility quantifies the amplitude of speckle fluctuation, the temporal autocorrelation can be used to quantify the time scale of the fluctuation. While recognizing that many types of motion (e.g., diffusion in non Newtonian fluids and ATP-driven motion) do not necessarily follow a simple exponential decay model, we find that a reasonable estimate of the time scale of the fluctuation process can be obtained by fitting log(γ(t)) = −t/τ + c, where γ is the normalized temporal autocorrelation of SOCT subsequently averaged over a local 5 pixel × 5 pixel region, and τ is the 1/e decay time. γ(0) is excluded from the fit, as it contains all of the nondeterministic noise. Regions with no appreciable correlation [defined as γ(t = 1 frame) < e−1 − 0.1] were set to τ = 0. To improve the accuracy of τ, a longer portion of the image stacks (N = 60) was analyzed.
The results of the autocorrelation analysis are shown in the third column of Fig. 2. As expected, GNRs exhibit rapid decorrelation (τ = 0) in comparison to MECs (τ ~ 5 s) and stationary objects such as the ECM (τ > 12 s). For the first time, we begin to see suspected MEC spheroids contrasted within the GNR sample, although the presence of contrast to the ECM within the nonGNR sample suggests that autocorrelation alone is not sufficient to specifically contrast MECs in all samples.
Finally, we employ polarization-sensitive OCT to measure the normalized cross-polarization (C), defined as
| (2) |
where SHV and SHH are the copolarized and cross-polarized OCT signal amplitudes, respectively, obtained under illumination with linearly (H) polarized light. Note that this definition of C is a normalized version of cross-polarization (Q12), according to Bohren and Huffman [21], while remembering that the square of the OCT signal amplitude is proportional to the light intensity scattered from the sample, S2 ∝ |ESample|2 ∝ ISample.
Importantly, cross-polarization is sensitive to the presence of optically anisotropic particles. Our GNRs are highly anisotropic because the LSPR is tuned to the center wavelength of the OCT system (800 nm) and exhibits a longitudinal optical cross section (parallel to the long axis of the rod) ~250× greater than in the transverse directions [18]. This is a regime where the cross-polarization ratio (IHV/IHH) for randomly oriented GNRs can exceed the traditional maximum value of 1/3 [22]. In comparison, other scatterers within the biological medium are not plasmon resonant and are often only weakly optically anisotropic. While many tissues do exhibit cross-polarization (or equivalently depolarization) due to anisotropic scatterers, multiple scattering, or form birefringence, we find that in the absence of GNRs the tissue cultures do not exhibit significant cross-polarization, and thus it is a useful metric here. This is seen in the last column of Fig. 2, where MECs and the ECM have an appreciably lower cross-polarization than regions of GNRs or noise. Interestingly, the cross-polarization within regions of GNRs increases with penetration depth, which we attribute to the onset of multiple scattering.
Now that we have established the MAPS signatures for GNRs and MECs, we can combine the individual metrics to specifically contrast each object class. This can conveniently be computed by linearly mapping each metric onto an unsigned 8-bit integer, where the range from 0 to 255 was chosen to represent the ranges of 20–50 for motility, 0–3 s for τ, and 0.3–0.7 for cross-polarization, in order to optically contrast the MECs and GNRs. Subsequently, each signature is computed by multiplying each metric (for ↑) or its opposite (for ↓) as follows:
| (3) |
Here, SMEC is the ↑↑↓ signature and SGNR is the ↑↓↑ signature, as in Table 1. The results of this operation are displayed in Fig. 3.
Fig. 3.
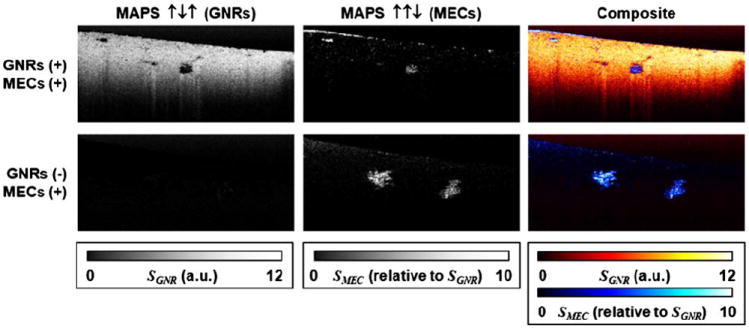
MAPS images for tissue culture samples corresponding to the top two rows of Fig. 2. The GNRs MAPS signature (left column) and MECs MAPS signature (middle) are combined to display a multicontrast image of both GNRs and MECs (right).
In comparison to Fig. 2, where no single metric is sufficient to contrast GNRs or MECs in all sample types, in Fig. 3 we find that MAPS signatures provide specific imaging of both GNRs and MECs, and aid in the identification of MEC spheroids against a highly scattering GNR background. We found that image regions containing GNRs appear spatially well separated from MECs, which we attribute to the expectation that GNRs cannot penetrate the epithelium formed by spheroids while remaining freely diffusing. The sharp delineation between regions of freely diffusing GNRs and MEC spheroids is more apparent in Fig. 4, which displays magnified views of several MEC spheroids. It is also interesting to note that improved contrast to MECs against the ECM background in the nonGNR sample is observed, providing more detailed spheroid morphology. This is significant because we have previously found that spheroid morphology is an important hallmark of premalignancy in breast cancer tissue cocultures [13,23].
Fig. 4.
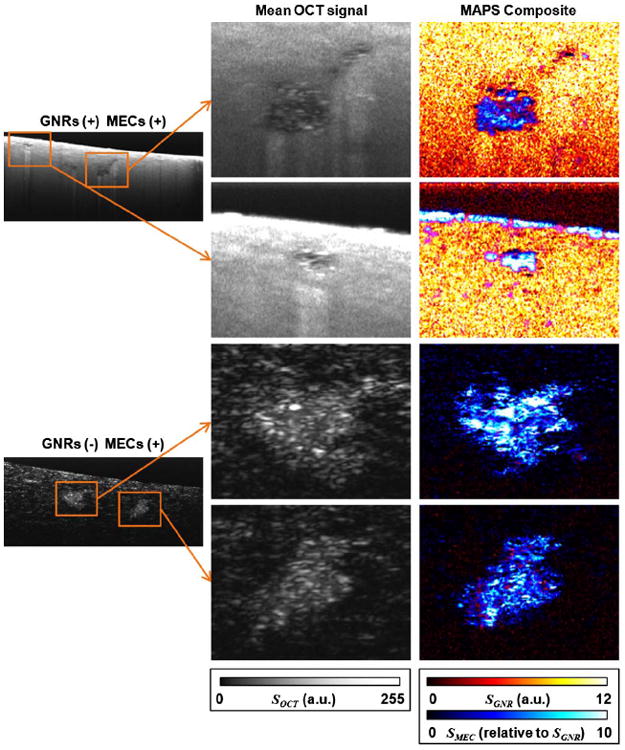
Magnified views (0.27 mm × 0.21 mm, x × z) of MEC spheroids in MEC tissue cultures with GNRs (top half) and without GNRs (bottom half), corresponding to the data of Figs. 2 and 3.
These MAPS images represent an estimate of the presence of GNRs and MECs based solely on their time-dependent and polarization-dependent properties. In the temporal dimension, we described the use of the amplitude (motility) and the time scale (τ) of the speckle fluctuation as easily calculated metrics from a time series of OCT images. Of course, more detailed information can be extracted from temporal speckle statistics, depending on the object classes present in the sample. For example, cluster analysis of motility spectra in coherence imaging has been used to delineate hypoxic versus normoxic tissue cultures in response to drugs [24]. Dynamic light scattering methods have been applied to autocorrelations of OCT data to distinguish between diffusive and deterministic motions [8,25]. In our own studies, we are working toward quantifying rheological properties of the ECM by applying a Stokes–Einstein relation to the statistics of diffusing GNRs [18].
Another finding of this study is that cross-polarized scattering is a good metric for contrasting GNRs in tissue cultures. This may offer utility in molecular contrast OCT applications where GNRs are labeled with specific antibodies [26], or for image guidance in GNR-based drug delivery or photothermal tumor therapy [17]. The added dynamical information in MAPS-OCT could further increase specificity to freely diffusing GNRs versus bound GNRs or endogenous cross-polarizing light scatterers.
MAPS-OCT as demonstrated in this study can also further be generalized to identify a wide range of object classes. We anticipate that these methods will serve as a roadmap for how the richness of temporal and polarization data readily available by standard OCT imaging hardware can be exploited to provide a broad array of functional and molecular contrasts.
Acknowledgments
This work was supported by the NIH via R21HL111968 (A. L. O.), pilot funds from the Carolina Cancer Center of Nanotechnology Excellence (C-CCNE) U54CA119343 (A. L. O.), R01CA138255 (M. A. T.), the Breast Cancer and the Environment Research Program (BCERP) U01ES019472 (M. A. T.), and by the NSF via DMR-1056653 (J. B. T.) We thank Krystian Kozek at NCSU for providing GNR samples for the preliminary experiments.
References
- 1.Robles FE, Wilson C, Grant G, Wax A. Nat Photonics. 2011;5:744. doi: 10.1038/nphoton.2011.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldenburg AL, Wu G, Spivak D, Tsui F, Wolberg AS, Fischer TH. IEEE J Sel Top Quantum Electron. 2012;18:1100. doi: 10.1109/JSTQE.2011.2162580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adler DC, Huang S-W, Huber R, Fujimoto JG. Opt Express. 2008;16:4376. doi: 10.1364/oe.16.004376. [DOI] [PubMed] [Google Scholar]
- 4.Skala MC, Crow MJ, Wax A, Izatt JA. Nano Lett. 2008;8:3461. doi: 10.1021/nl802351p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob D, Shelton RL, Applegate BE. Opt Express. 2010;18:12399. doi: 10.1364/OE.18.012399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer JF, Milner TE, van Gemert MJ, Nelson JS. Opt Lett. 1997;22:934. doi: 10.1364/ol.22.000934. [DOI] [PubMed] [Google Scholar]
- 7.Kalkman J, Sprik R, van Leeuwen TG. Phys Rev Lett. 2010;105:198302. doi: 10.1103/PhysRevLett.105.198302. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan VJ, Radhakrishnan H, Lo EH, Mandeville ET, Jiang JY, Barry S, Cable AE. Biomed Opt Express. 2012;3:612. doi: 10.1364/BOE.3.000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, Radhakrishnan H, Wu W, Daneshmand A, Climov M, Ayata C, Boas DA. J Cereb Blood Flow Metab. 2013;33:819. doi: 10.1038/jcbfm.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farhat G, Mariampillai A, Yang VXD, Czarnota GJ, Kolios MC. J Biomed Opt. 2011;16:070505. doi: 10.1117/1.3600770. [DOI] [PubMed] [Google Scholar]
- 11.Oldenburg AL, Chhetri RK, Hill DB, Button B. Biomed Opt Express. 2012;3:1978. doi: 10.1364/BOE.3.001978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bissell MJ, Radisky D. Nat Rev Cancer. 2001;1:46. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chhetri RK, Phillips ZF, Troester MA, Oldenburg AL. PLoS One. 2012;7:e49148. doi: 10.1371/journal.pone.0049148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldenburg AL, Hansen MN, Ralston TS, Wei A, Boppart SA. J Mater Chem. 2009;19:6407. doi: 10.1039/b823389f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker-Schwartz JM, Meyer TA, Patil CA, Duvall CL, Skala MC. Biomed Opt Express. 2012;3:2881. doi: 10.1364/BOE.3.002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung Y, Reif R, Zeng Y, Wang RK. Nano Lett. 2011;11:2938. doi: 10.1021/nl2014394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alkilany AM, Thompson LB, Boulos SP, Sisco PN, Murphy CJ. Adv Drug Deliv Rev. 2012;64:190. doi: 10.1016/j.addr.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Chhetri RK, Kozek KA, Johnston-Peck AC, Tracy JB, Oldenburg AL. Phys Rev E. 2011;83:040903. doi: 10.1103/PhysRevE.83.040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oldenburg AL, Chhetri RK. Proc SPIE. 2011;7889:78891V. [Google Scholar]
- 20.Nikoobakht B, El-Sayed MA. Chem Mater. 2003;15:1957. [Google Scholar]
- 21.Bohren CF, Huffman DR. Absorption and Scattering of Light by Small Particles. Wiley; 1983. [Google Scholar]
- 22.Khlebtsov N, Mel’nikov A, Bogatyrev V, Alekseeva A, Khlebtsov B. Opt Spectrosc. 2006;100:448. [Google Scholar]
- 23.Casbas-Hernandez P, D’Arcy M, Roman-Perez E, Brauer HA, McNaughton K, Chhetri RK, Oldenburg AL, Troester MA. Breast Cancer Res. 2013 doi: 10.1186/bcr3476. to be published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolte DD, An R, Turek J, Jeong K. Biomed Opt Express. 2012;3:2825. doi: 10.1364/BOE.3.002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J, Wu W, Jiang JY, Zhu B, Boas DA. Opt Express. 2012;20:22262. doi: 10.1364/OE.20.022262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhulkar S, Matthews J, Rawal S, Awdeh RM. Investig Ophthalmol Vis Sci. 2013;54:1192. doi: 10.1167/iovs.12-10794. [DOI] [PMC free article] [PubMed] [Google Scholar]


