Abstract
Human embryonic stem cells (hESCs) are more similar to “primed” mouse epiblast stem cells (mEpiSCs). mEpiSCs, which are derived in Activin A, show an increased propensity to form primordial germ cell (PGC)-like cells in response to bone morphogenic protein 4 (BMP4). Hence, we hypothesized that hESCs derived in the presence of Activin A may be more competent in differentiating towards PGC-like cells after supplementation with BMP4 compared to standard hESC lines. We were able to successfully derive two hESC lines in the presence of Activin A, which were pluripotent and showed higher base levels of STELLA and cKIT compared to standard hESC lines derived without Activin A addition. Furthermore, upon differentiation as embryoid bodies in the presence of BMP4, we observed upregulation of VASA at day 7, both at the transcript and protein level compared to standard hESC lines, which appeared to take longer time for PGC specification. Unlike other hESC lines, nuclear pSMAD2/3 presence confirmed that Activin signalling was switched on in Activin A-derived hESC lines. They were also responsive to BMP4 based on nuclear detection of pSMAD1/5/8 and showed endodermal differentiation as a result of GATA-6 expression. Hence, our results provide novel insights into the impact of hESC derivation in the presence of Activin A and its subsequent influence on germ cell differentiation potential in vitro.
Introduction
The first cells appearing in the mammalian germ cell lineage are the PGCs, a prerequisite to sustain the continuation of species from one generation to the next. Upon spontaneous differentiation of hESCs as embryoid bodies (EBs), several early PGC markers STELLA (Dppa3), FRAGILIS (Ifitm3), cKIT (kit), and late PGC marker VASA (Ddx4) have been detected [1]. Subsequently, several attempts have been made to develop more efficient protocols for deriving germ cells in vitro from ESCs. BMP4 was found to play a prominent role in inducing germ cell gene expression, especially of the postmigratory germ cell marker VASA in in vitro derived PGCs from hESCs [2]. Efforts were also made to develop an efficient step-wise protocol by supplementing hESCs with a growth factor cocktail in a time-dependent manner mimicking the in vivo environment [3–5]. In addition, it was found that coculture of hESCs with Sertoli cells or human fetal gonadal cells boosted germ cell differentiation [6,7] based on the expression of early and late germ cell markers. hESCs were also genetically manipulated by overexpressing DAZL to induce germ cell differentiation or transfected with VASA-pEGFP-1 reporter construct to identify and isolate hESC-derived PGCs in vitro [8,9]. Culture strategies, such as EB culture, adherent culture, colony size, frequency of refreshment of cultures, are all shown to have an effect on directed differentiation of hESCs towards PGCs [6,10].
In all the studies undertaken for hESC differentiation towards PGCs, multiple hESC lines were employed. More evidence suggests that hESC lines differ from each other in their lineage-specific differentiation potential [11,12]. These differences between hESC lines may be due to several factors, such as embryo quality, derivation technique, and inherent genetic identity or used culture conditions during derivation and culture. Therefore, it becomes a necessity to screen for the ideal hESC line before starting differentiation. It is also suggested that derivation of hESC lines in conditions specific towards lineage of interest might help yield valuable results [11,12]; however, this has not been investigated yet.
The TGFβ signaling pathway is composed of two main branches, namely the TGFβ/Activin/Nodal branch involving SMAD2/3 proteins, which maintain hESC pluripotency, and the SMAD1/5/8 branch acting downstream of BMP4 and GDF ligands during differentiation [13,14]. Activin A is one of the important members of the TGFβ superfamily. In situ ligand binding in male Sprague-Dawley rats has shown the ability of Activin A to bind to germ cells [15]. In juvenile mice testis, in vivo progression of germ cell maturation was influenced by Activin A bioactivity during the onset of spermatogenesis [16]. It was found to be the first Sertoli cell by-product that was involved in differentiation of male germ cells in meiotic state [17]. In fetal mouse testis, it keeps a balance between Sertoli and germ cell proliferation [18]. In humans, during the onset of primordial follicle formation, it aids in germ cell proliferation and survival [19] and also regulates follicle formation and growth in vitro [20]. Recently, Activin A was shown to suppress the retinoic acid inhibitor CYP26B1 and to help inducing meiosis [21].
It is becoming increasingly clear that hESCs are in fact similar to mEpiSCs, sharing properties, such as dependence on TGFβ/Activin signaling to maintain their pluripotency and ability to differentiate into PGC-like cells upon exposure to BMP4 [22–25]. In connection to this, our group recently showed that the human ICM outgrowth first undergoes a transient epiblast-like state before hESCs are established, which substantiates their primed pluripotency status [26]. mEpiSCs, which are derived in the presence of Activin A [27], appear to retain characteristics of the original epiblast and show strong potential to generate PGC-like cells in vitro, which are able to further develop into late germ cells [24]. In addition, male and female germ cell specification was induced in mESCs and mouse induced pluripotent stem cells by culturing them in Activin A to obtain early pregastrulating epiblast-like cells (EpiLCs), which gave rise to PGC-like cells in the presence of BMP4 [28,29].
The results that we present here indicate that hESC derivation in the presence of Activin A has a significant impact on the state of the derived hESC lines. We showed that addition of Activin A during hESC derivation induced increased germ cell differentiation potential compared to hESC lines derived in standard conditions without Activin A.
Materials and Methods
Ethical permission
All patients donating embryos signed informed consents before their IVF/ICSI cycle. Approval for the current study was obtained from the Ghent University Hospital Ethical Committee (2009/281) and from the Belgian Federal Ethical Committee on Embryo Research (Adv-030).
hESC derivation and culture
To derive our hESC lines in both Activin A and standard derivation conditions, all human embryos donated for research were randomized and day 6 human blastocysts were scored according to the grading system of Stephenson and colleagues [30]. Blastocysts with grade A and B inner cell mass (large and distinct inner cell mass) [31,32] were exposed to Acidic Tyrode's solution (Cat No. T1788; Sigma) to remove the zona pellucida, washed in hESC media and plated on a feeder layer of CD1 mouse embryonic fibroblasts (MEFs) pretreated with mitomycin-C (Cat No. M4287; Sigma). For standard hESC lines UGent11-5, UGent11-6, and UGent11-7, culturing from blastocyst to hESC stage was carried out in a low-oxygen environment at 37°C, consisting of 6% CO2, and 5% O2, using standard hESC medium composed of Knockout Dulbecco's Modified Eagle Medium (KO-DMEM, Cat No. 10829-018; Invitrogen), 20% Knockout Serum Replacement (KO-SR, Cat No. 10828-028; Invitrogen), 1% nonessential amino acids (NEAA, Cat No. 11140-035; Invitrogen), 0.1 mM L-glutamine (Cat No. 25030-024; Invitrogen), 1% penicillin/streptomycin (Cat No. 15140-122; Invitrogen), 0.1 mM β-mercaptoethanol (Cat No. 31350-010; Invitrogen) and 4 ng/mL basic fibroblast growth factor (bFGF, Cat No. 100-18B; Peprotech). Cultures were refreshed daily and monitored closely for postinner cell mass-intermediates (PICMIs) [26,33]. Within approximately 7 days, hESCs outgrowths were cut mechanically, transferred to freshly inactivated MEFs, and continually mechanically passaged until several colonies appeared. After this, hESCs were maintained and split 1:6 onto freshly inactivated MEFs every 5–6 days using 1 mg/mL collagenase type IV (Cat No. 17104-019; Invitrogen) with daily medium changes to keep hESCs in an undifferentiated state.
For deriving the hESC lines UGent11-4-ActA and UGent12-3-ActA, the medium was supplemented with 20 ng/mL Activin A (Cat No. 338-AC-010; R&D Systems) from the day 6 blastocyst stage onwards until hESC passage 6. After that, the lines were further cultured in standard hESC media without Activin A.
Karyotyping
Early passage hESCs were grown to confluency and exposed to colcemid (1:100 in salt solution, Cat No. 15210-057 Karyomax; Invitrogen) to arrest the cells at metaphase. They were then treated with 0.25% trypsin-EDTA (Cat No. 25200-056; Invitrogen) and exposed to hypotonic HCl solution (Cat No. T-3038; Sigma) followed by fixation in 1:3 methanol/acetic acid solution (Cat No. 1.06009.1000, Merck; Cat No. A6283, Sigma). Karyotype analysis was performed for 10 numerical spreads of metaphase and 3 complete karyograms using G-banding technique as previously described [34].
Alkaline phosphatase live staining
Undifferentiated hESCs were washed twice with DMEM/F-12 (Cat No. 31331-028; Invitrogen) for 2–3 min. They were then incubated in 1×alkaline phosphatase (AP) Live Stain (Cat No.A14353; Molecular Probes, Life Technology) for 30 min. Next, hESCs were washed twice with DMEM/F-12 for 5 min to remove AP Live Stain. Fresh DMEM/F-12 was added to fluorescent-labeled hESC cultures and they were visualized using standard FITC filter by fluorescence microscopy.
Differentiation of hESCs towards PGCs
Activin A-derived hESC lines (UGent11-4-ActA and UGent12-3-ActA) were maintained in hESC conditions similar to standard hESC lines (UGent11-5, UGent11-6 and UGent11-7) after passage 6. All hESC lines were used between passages 20–25 to maintain homogeneity and to understand the impact of derivation conditions on PGC differentiation of hESCs. Also, we performed karyotyping of all hESC lines before starting our experiments. All hESC lines were karyotypically normal. Briefly, hESCs were washed with 1× phosphate buffered saline (PBS), incubated with collagenase for 6 min and subsequently gently scraped from the culture flasks with a flattened needle. Collagenase was neutralized with differentiation medium (80% KO-DMEM, 20% fetal bovine serum, NEAA, L-glutamine, penicillin/streptomycin and β-mercaptoethanol). hESCs were allowed to settle by gravity for 15–20 min and were subsequently resuspended in germ cell differentiation medium consisting of differentiation medium supplemented with 50 ng/mL BMP4 (Cat No. 314-BP-010; R&D Systems). Cells were then transferred onto 24-well Costar Ultra Low Attachment Clear Flat Bottom plates (Cat No. 3473; Corning) and allowed to differentiate for 3, 7 and 14 days in low-oxygen conditions. Germ cell differentiation medium was replaced every alternate day (1 mL per well). For dose-response studies of BMP4, the hESC line UGent11-4-ActA was differentiated as EBs up to day 7 in differentiation medium containing 10 ng/mL BMP4, 50 ng/mL BMP4, or 100 ng/mL BMP4.
Immunocytochemistry
EBs and hESCs were fixed in 4% paraformaldehyde in PBS at 4°C and stored in 100% methanol at −20°C and in 1× PBS at 4°C, respectively. EBs were rehydrated through a series of methanol/water solutions and washed in PBST (PBS+0.1% Tween-20) twice for 10 min. Next, they were rinsed in TBST (Tris-buffered saline+0.1% TritonX-100, pH 7.4) and blocked in 0.5%BSA/TBST blocking solution at 4°C overnight before incubation with primary antibody for 2 days at 4°C (rotating). They were washed in TBST five times for 30 min at room temperature and incubated with secondary antibody overnight at 4°C (rotating). EBs were again washed in TBST (5 times for 30 min), mounted on glass slides using Vectashield with DAPI (Vector Laboratories Ltd.).
hESCs were rinsed in 1× PBS, permeabilized in 0.1%TritonX-100/1× PBS for 8 min, then washed in 1× PBS, and blocked in 0.05% Tween-20/1% BSA/1× PBS blocking solution for 1 h at room temperature. hESCs were then incubated with primary antibody overnight at 4°C, washed three times in 1× PBS, and incubated with secondary antibody for 1 h at room temperature. They were then washed again three times in 1× PBS and subsequently mounted on glass slides using Vectashield with DAPI.
Primary antibodies used were goat anti-OCT4 (1:200, SC-8628; Santa Cruz Biotechnology, Inc.), rabbit anti-NANOG (1:200, AF1997; R&D Systems), rabbit anti-VASA (1:500, ab13840, Abcam; 1:500, AF2030; R&D Systems), rabbit anti-pSMAD2/3 (1:100, No. 9510; Cell Signalling Technologies, Bioké), rabbit anti-pSMAD1/5/8 (1:100, No. 9511, Cell Signalling Technologies, Bioké). Secondary antibodies included donkey anti-goat-FITC (1:200; 705-095-147; Bioconnect) and donkey anti-rabbit-CY3 (1:2000; 711-165-152; Bioconnect). UGent11-4-ActA differentiated as EBs in the presence of BMP4 for 7 days were used as negative controls where only the secondary antibody was applied.
Western blotting
Whole cell lysates were prepared from second trimester fetal testis and BMP4-treated EBs (day 7) derived from UGent11-4-ActA and UGent11-6 hESC lines. Twenty micro gram proteins from each sample were loaded per lane on a 10% SDS-PAGE gel. Next, proteins were transferred to a nitrocellulose membrane. Transferred blots were then blocked in PBT (1× PBS+0.3% Tween) with 1% BSA for 1 h at RT followed by incubation in primary antibody (1:500, rabbit-anti VASA, Cat No. ab13840, Abcam; 1:5000, rabbit-anti β-TUBULIN, Cat No. ab6046, Abcam) overnight at 4°C. Blots were then washed in PBT thrice for 5 min and incubated in secondary antibody (1:20000, goat-anti-rabbit-HRP, Cat No. ab6721, Abcam) for 1 h at RT. Blots were again washed as before, developed using SuperSignal West Dura Chemiluminescent Substrate (Cat No. 34076; Thermoscientific) and were captured using the Bio-Rad VersaDoc imaging system with Bio-Rad Quantity One software.
RNA isolation and reverse transcription quantitative polymerase chain reaction
hESCs and EBs from days 3, 7 and 14 were collected and lysed in 1 mL TRIzol (Cat No. 15596-026; Invitrogen). Total RNA was purified from the inorganic phase using the RNeasy Mini Kit (Cat No. 74104; Qiagen). cDNA was synthesized using the iScript Advanced cDNA Synthesis Kit for RT-qPCR (Cat No. 170-8842; Biorad). cDNA concentration was determined with the Quant-iT Oligreen ssDNA Assay Kit (Cat No. O11492; Invitrogen) on a Tecan fluorescence reader using Magellan software.
Real time polymerase chain reaction (PCR) amplification of cDNA was performed for VASA, OCT4, cKIT, NANOG (Taqman Gene Expression Assays; Applied Biosystems), STELLA, FRAGILIS, SSEA1, SOX2 (Real-time Ready Assays; Roche). For studying the differentiation of day 14 EBs into the three germ layers (differentiated in medium without bFGF), genes analyzed included ectodermal genes NESTIN, PAX6, mesodermal genes HAND1, PECAM1, endodermal genes GATA4, GATA6 and pluripotency gene OCT4. The reference genes were GAPDH, B2M and RPL13A (Table 1). Each quantitative PCR (qPCR) reaction included 20 μl mastermix and 10 ng cDNA. The mastermix consisted of gene specific primers and iTAQ supermix with ROX (Cat No. 172-5855; Biorad) or iTAQ SYBR Green Supermix with ROX (Cat No. 172-5850; Biorad).
Table 1.
Gene Expression Assays Used for Quantitative Real-Time Polymerase Chain Reaction
|
QRT-PCR gene expression assays | |
|---|---|
| Genes of interest | Assay ID |
| VASAa | Hs00987125_m1 |
| cKITa | Hs00174029_m1 |
| OCT4a | Hs01895061_u1 |
| NANOGa | Hs02387400_g1 |
| NESTINa | Hs04187831_g1 |
| PAX6a | Hs00240871_m1 |
| HAND1a | Hs02330376_s1 |
| PECAM1a | Hs00169777_m1 |
| GATA4a | Hs00171403_m1 |
| GATA6a | Hs00232018_m1 |
| STELLAb | 140928 |
| FRAGILISb | 122503 |
| SSEA1b | 118619 |
| SOX2b | 111867 |
| Housekeeping genes | Primer sequences |
|---|---|
| GAPDH | F: AGCCTCAAGATCAGCAATG |
| R: ATGGACTGTGGTCATGAGTCCTT | |
| B2M | F: TGCTGTCTCCATGTTTGATGTATCT |
| R: TCTCTGCTCCCCACCTCTAAGT | |
| RPL13A | F: CCTGGAGGAGAAGAGGAAAGAGA |
| R: TTGAGGACCTCTGTGTATTTGTCAA |
Taqman gene expression assays.
Roche real-time ready assays.
qRT-PCR, reverse transcription quantitative polymerase chain reaction.
The qPCR machine employed was the ABI Prism 7000 Sequence Detection System (Applied Biosystems). The thermocycling conditions were as follows: 2 min at 95°C followed by 45 cycles of 15 s at 95°C followed by 1 min at 60°C. All samples were analysed in biological and technical triplicates. 11-week fetal testis was used as positive control and no cDNA (water) samples or no RT (RNA) samples were used as negative controls. Ct values were normalized to the housekeeping genes and the final values were described as fold change expression values compared to the hESC line UGent11-7. For EB germ layer gene expression analyses, fold change expression values were compared to OCT4 expression in day 14 EBs of each hESC line. Statistical significance was calculated using t-test and ANOVA on means and standard deviations of fold change expression values.
Results
hESC lines derived in the presence of Activin A are pluripotent and show increased propensity towards PGC differentiation
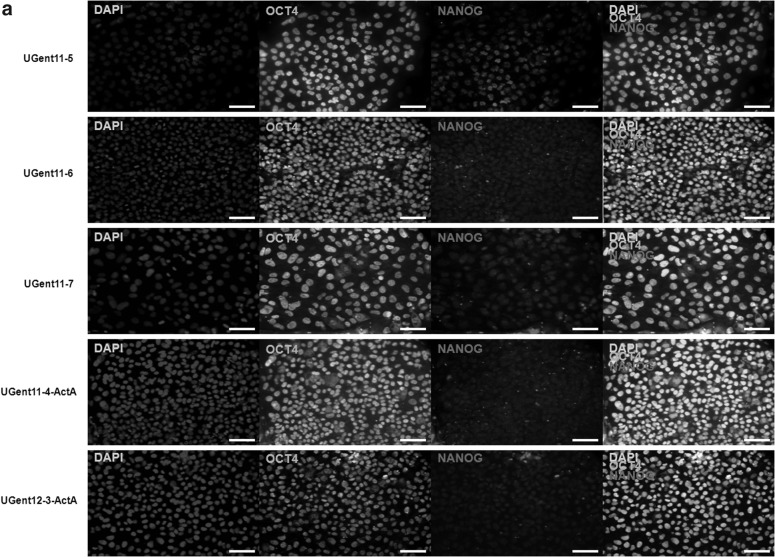
Based on the fact that mEpiSCs are derived in the presence of Activin A [27] and have been previously shown to have high potential to generate PGC-like [24] cells, we also added Activin A at 20 ng/mL to human embryos at the blastocyst stage to determine whether this would influence the in vitro PGC differentiation potential of the derived hESC lines. We were able to successfully derive two hESC lines in the presence of Activin A: UGent11-4-ActA (XX) and UGent12-3-ActA (XY). The other hESC lines UGent11-5 (XX), UGent11-6 (XX) and UGent11-7 (XY) used in this study were all derived conventionally in the absence of Activin A. All hESC lines were pluripotent, expressing OCT4 and NANOG (Fig. 1a), AP positive (Fig. 1b), karyotypically normal (Fig. 1c), and could be differentiated as EBs expressing markers of the three germ layers (Fig. 1d). When SB431542, an inhibitor of Activin A [35], was added at the blastocyst stage, in vitro progression to the PICMI stage was observed; however, no hESC lines could be derived (results not shown). This may likely be due to the fact that SB431542 blocks the TGFβ/Activin/Nodal signaling pathway, which is indispensible to maintain the primed status of pluripotency in hESCs [36]. Hence, this may be the probable cause for the successful derivation of hESCs in the presence of Activin A and failure to generate any hESC lines upon the inhibition of TGFβ/Activin/Nodal signaling pathway by SB431542; thus, further emphasizing the intricate nature of the hESC derivation process.
FIG. 1.
Pluripotency assays on both Activin A-derived and standard human embryonic stem cell (hESC) lines. (a) Immunofluorescence staining for pluripotency markers OCT4 and NANOG, nuclear staining by DAPI and merged images of all hESC lines when maintained in their undifferentiated state. Scale bar, 50 μm. (b) Alkaline phosphatase live staining for all hESC lines in their undifferentiated state. Scale bar, 50 μm. (c) Karyograms for XX hESC lines UGent11-4-ActA, UGent11-5 and UGent11-6 and XY hESC lines UGent12-3-ActA and UGent11-7. All hESC lines were karyotypically normal. (d) Embryoid body (EB) differentiation assay of all hESC lines. (i) Gene expression profile of germ layer markers in day 14 EBs. Fold change values are analyzed compared to OCT4 gene expression in day 14 EBs. For all hESC lines, after 14 days of differentiation as EBs, ectoderm marker NESTIN and endoderm markers GATA4 and GATA6 upregulated significantly compared to OCT4, P<0.05; mesoderm marker HAND1 also showed significant upregulation compared to OCT4, P≤0.001; (ii) Differentiation of all hESCs as EBs on day 3, day 7 and day 14. Scale bar, 100 μm.
Next, we investigated PGC differentiation potential of both Activin A-derived hESC lines and standard hESC lines. At day 0, Activin A-derived hESCs showed significantly higher STELLA expression compared to standard hESCs (7.4 and 9.2-fold increase for UGent11-4-ActA and UGent12-3-ActA, respectively; P≤0.001) (Fig. 2a). The cKIT expression pattern resembled that of STELLA with high expression levels in Activin A-derived hESC at day 0 compared to standard hESCs (Fig. 2b). Hence, these Activin A-derived hESC lines show high inherent expression levels of early PGC markers, which may prone them towards germ cell differentiation. This is further corroborated by the fact that Activin A-derived hESCs showed significant upregulation of VASA compared to standard hESCs on day 7 EBs (10 and 7.5-fold increase in UGent11-4-ActA and UGent12-3-ActA, respectively; P≤0.001). This upregulation of VASA in differentiating Activin A-derived hESCs corresponded to a downregulation in FRAGILIS expression. From day 7 EBs to day 14 EBs, expression levels of both FRAGILIS and VASA were maintained in these differentiating hESCs. In contrast, standard hESCs showed a steady rise in FRAGILIS expression from day 0 to day 14 EBs, while VASA continued to be expressed at a low level (Fig. 2c, d). The pluripotency markers OCT4, SOX2 and NANOG were significantly downregulated in all differentiating hESC lines, while SSEA1 continued to rise from day 0 to day 14 EBs (Fig. 2e–h).
FIG. 2.
Gene expression profiles of all hESC lines upon germ cell-directed differentiation as EBs in the presence of bone morphogenic protein 4 (BMP4) for early primordial germ cell (PGC) markers STELLA, cKIT, FRAGILIS (a–c), late PGC marker VASA (d), pluripotency markers OCT4, SOX2, NANOG (e–g), and the differentiation marker SSEA1 (h). Activin A-derived hESC lines UGent11-4-ActA and UGent12-3-ActA show significantly higher expression for STELLA and cKIT at day 0 and significant upregulation of VASA at day 7, when FRAGILIS is downregulated. For all hESC lines, pluripotency markers OCT4, SOX2, NANOG were significantly downregulated, and SSEA1 expression increased as differentiation proceeded from day 0 to day 14. STELLA, cKIT: UGent11-4-ActA (a′) compared to standard hESC lines at day 0 (a), P≤0.001; UGent12-3-ActA (b′) compared to standard hESC lines at day 0 (b), P≤0.001; FRAGILIS: Day 7 UGent12-3-ActA EBs (a′) compared to day 0 and day 3 EBs (a), P<0.05; VASA: Day 7 UGent 11-4-ActA EBs (a′) compared to day 7 EBs of standard hESC lines (a) P≤0.001; Day 7 UGent12-3-ActA EBs (b′) compared to day 7 EBs of standard hESC lines (b) P≤0.001; OCT4, SOX2, NANOG: for all hESC lines, differentiation as EBs from day 0 to day 14, P≤0.001; SSEA1: P<0.05.
As day 7 marked the critical time point for rise in VASA at the mRNA level in EBs, we immunostained those of the Activin A-derived hESC line UGent11-4-ActA and the standard hESC line UGent11-6 at this time point to check for VASA and OCT4 presence at the protein level. UGent11-4-ActA showed higher expression of VASA compared to UGent11-6 (Fig. 3a and Supplementary Fig. S1 Supplementary Data are available online at www.liebertpub.com/scd). VASA localized to the nucleus and to the cytoplasm of both dividing and nondividing cells. Very little or no OCT4 expression was seen in day 7 UGent11-4-ActA EBs, while UGent11-6 EBs continued to show a few OCT4-positive cells with no or very few cells expressing VASA only in the nucleus (Fig. 3b). It is possible that these OCT4-positive cells of UGent11-6 represent early PGCs. These results were further confirmed by western Blot analysis (Fig. 3c and Supplementary Fig. S2).
FIG. 3.

Representative immunofluorescence staining of Activin A-derived hESC lines and standard hESC lines for the pluripotency marker OCT4 and the late PGC marker VASA together with nuclear stain DAPI on day 7 of germ cell-directed differentiation. (a) Activin A-derived hESC line UGent11-4-ActA showing (i) very low frequency of OCT4-positive cells but a high number of VASA-positive cells and (ii) VASA localizes to both the nucleus and the cytoplasm (*). Scale bar=(i) 50 μm, (ii) 100 μm. (b) Standard hESC line UGent11-6 showing more OCT4-positive cells but no or very low VASA expression in the nucleus and no cytoplasmic localization. Scale bar=(i) 50 μm, (ii) 100 μm. (c) Western blot analysis for VASA on second trimester fetal testis, UGent11-4-ActA and UGent11-6. β-tubulin used as control. (d) Negative control. Scale bar, 50 μm.
50 ng/mL BMP4 is sufficient to direct hESC differentiation towards PGC-like cells
Various reports have studied the dose dependent effect of BMP4 in inducing germ cell gene expression in ESCs [2,3,37,38]. In EBs generated from mESCs, a biphasic response in gene expression was seen upon addition of BMP4 from 20 ng/mL to 500 ng/mL. Here 50 ng/mL, 100 ng/mL and 200 ng/mL showed a gradual increase in early OCT4+ PGC population, whereas 200 ng/mL led to its decrease and induced mesoderm-directed differentiation [38]. PGC-directed differentiation of bone marrow mesenchymal stem cells also showed a BMP4-associated dose dependent increase in VASA expression where 100 ng/mL proved to be the best concentration compared to 25 ng/mL and 50 ng/mL [37]. A cocktail of BMPs, including BMP4, BMP7, and BMP8b supplemented at 100 ng/mL has also proven to be highly beneficial in germ cell differentiation of hESCs compared to 1 ng/mL and 10 ng/mL of BMP4 [2]. Thus, we wanted to determine the optimum concentration of BMP4 for its impact on in vitro PGC differentiation of our Activin A-derived hESC lines. To test this, UGent11-4-ActA was differentiated as EBs in the absence of BMP4, and at 10 ng/mL, 50 ng/mL or 100 ng/mL BMP4 up to day 7. It was observed that 50 ng/mL and 100 ng/mL BMP4 led to significantly higher upregulation of STELLA and VASA expression as compared to 10 ng/mL BMP4 or its absence. However, 50 ng/mL and 100 ng/mL concentrations did not show a significant difference between each other with respect to their effect on STELLA and VASA expression. Hence, 50 ng/mL BMP4 was employed as a standard dosage in our experiments. OCT4 expression was unaffected by different BMP4 concentrations (Fig. 4).
FIG. 4.
BMP4 dose-response study on day 7 EBs derived from the Activin A-derived hESC line UGent11-4-ActA for the early PGC marker STELLA, the late PGC marker VASA and the pluripotency marker OCT4. BMP4 is used at concentrations of 10 ng/mL, 50 ng/mL, and 100 ng/mL. STELLA: 50 ng/mL BMP4 (a′) compared to 10 ng/mL BMP4 and –BMP4 (a), P<0.05; 100 ng/mL BMP4 (b′) compared to 10 ng/mL and –BMP4 (b′), P<0.05. VASA: 50 ng/mL BMP4 (a′) compared to 10 ng/mL BMP4 and –BMP4 (a), P<0.05; 100 ng/mL BMP4 (b′) compared to 10 ng/mL and –BMP4 (b′), P<0.05.
Activin A-derived hESC lines showed high expression of the endodermal marker GATA-6 in the presence of BMP4 and exhibited active TGFβ/Activin signaling
As the Activin A-derived hESC lines were exposed to Activin A during their derivation, we wanted to check whether TGFβ/Activin signaling is active during their differentiation towards PGC-like cells. Translocation of pSMAD2/3, an active component of TGFβ/Activin signaling, from cytoplasm to the nucleus helps in regulating gene expression [13]. Our immunocytochemistry results showed that phosphorylated SMAD2/3 is present in day 7 EBs of UGent11-4-ActA when VASA is upregulated, while it remained absent or at reduced levels in UGent11-6 (Fig. 5a–c). However, this needs to be further investigated.
FIG. 5.
Representative immunofluorescence staining of the TGFβ/Activin signaling pathway mediator pSMAD2/3 and nuclear stain DAPI on day 7 BMP4-induced EBs derived from Activin A-derived hESC lines and standard hESC. (a) The Activin A-derived hESC line UGent11-4-ActA showing phosphorylation and nuclear localization of pSMAD2/3 signifying that Activin signaling is switched on. Scale bar, 50 μm. (b) The standard hESC line UGent11-6 showing low number of pSMAD2/3 positive cells compared to UGent11-4-ActA. Scale bar, 50 μm. (c) Negative control. Scale bar, 50 μm.
As Activin A and BMP4 together are known to induce differentiation towards the endodermal lineage [39], and to further emphasize the fact that derivation in Activin A has an effect on the derived hESC line, we checked whether these hESC lines were also giving rise to endoderm on day 7 EBs, while being differentiated in germ cell differentiation media supplemented with BMP4. We observed GATA-6 positive cells covering the surface of a large majority of EBs in UGent11-4-ActA (Fig. 6a), as well as nuclear pSMAD1/5/8 (Fig. 6b) indicative of their responsiveness to BMP4. Hence, as both pSMAD2/3 and pSMAD1/5/8 are present in differentiating Actvin A-derived hESCs, this further confirms that Activin A-derived hESCs are responsive to both Activin and BMP signaling upon differentiation, which may explain their increased potency to differentiate into PGC-like cells.
FIG. 6.
Immunofluorescence staining for the endodermal marker GATA6 and the BMP signaling mediator pSMAD1/5/8 together with nuclear staining by DAPI on day 7 BMP4-induced EBs derived from Activin A-derived hESC line UGent11-4-ActA. (a) GATA6 is highly expressed in Activin A-derived hESC line as visible by the majority of cells in the embyoid body, which are GATA6-positive. Scale bar, 50 μm. (b) pSMAD1/5/8 is localized to the nucleus showing that cells are responsive to BMP signaling. Scale bar, 50 μm. (c) Negative control. Scale bar, 50 μm.
Discussion
Both mouse EpiLCs [28,29] and mEpiSCs are prone towards germ cell differentiation [24] and are derived in similar culture conditions as mostly used for hESC derivation, except for the extra supplementation of Activin A [27,28]. We successfully showed that supplementing Activin A during derivation of hESC lines can generate pluripotent hESCs, which display predisposition for differentiation towards the PGC-like lineage. Although Activin A has been shown to support pluripotency and self-renewal [40] of hESC culture in feeder-free conditions, derivation of most hESC lines has been performed in the presence of factors, such as bFGF and human recombinant leukemia inhibitory factor [41–50] without Activin A. In 2010, a xeno-free hESC derivation medium called RegES medium was developed, which contained only 5 ng/mL Activin A. However, this medium was solely employed to derive hESC lines in xeno-free conditions for clinical purpose and their lineage-specific differentiation potential was not compared to standard derived hESC lines [51,52].
All hESC lines are known to share similar morphology, growth pattern, and expression of pluripotency markers; however, variations amongst hESC lines exist at the transcriptional level with culture conditions being one of the crucial causative factors [45,46,53,54]. For example, differential expression of 1417 genes was observed upon the culture of HS237 hESC line in medium with or without serum [55]. Hence, variations in hESC lines could be a direct result of the culture environment the embryo was exposed to during the hESC derivation process. In fact, considerable changes at the molecular level have been shown during the process of hESC derivation from blastocyst embryos. These changes have been highlighted in a study by our group [26], where a comparative analysis was performed between ICM, post ICM intermediate (PICMI), and hESCs. It was observed that as the ICM progresses towards hESCs, the intermediate PICMI stage is formed, characterized by the expression of both early and late epiblast markers. Interestingly, germ cell markers, such as cKIT and FRAGILIS also increased along with Nodal/Activin signaling during the PICMI stage. These markers are again downregulated in hESCs although still higher in expression compared to ICM [26]. Thus, exposing hESCs during their derivation to 20 ng/mL Activin A, similar to mouse EpiLCs and mEpiSCs, boosted their germ cell differentiation potential unlike the other hESC lines, which were derived using conventional methods in the absence of Activin A. On the contrary, we observed that addition at the blastocyst stage of SB431542, an inhibitor of Activin A [35], failed to result in hESC lines, further emphasizing the unique significance of the hESC derivation stage.
Germ cell differentiation of hESCs is commonly assessed by determining expression levels of widely used germ cell markers, such as STELLA, FRAGILIS, cKIT, SSEA1, and VASA [56,57]. Our Activin A-derived hESC lines showed significantly higher base levels of STELLA and cKIT as compared to standard derived hESC lines. This may indicate an increased predisposition of these hESC lines towards the germ cell lineage. Moreover, it was recently reported that STELLA plays an active role in inducing germ cell directed differentiation of hESCs [58]. Also, we observed that FRAGILIS continued to rise up to day 7 EBs and as it is subsequently downregulated, there is a concomitant rise in VASA expression. Hence, this expression pattern may indicate the completion of the PGC specification process and that the cells are in a transition between migratory and premeiotic state. For the standard hESC lines, early PGC markers, such as STELLA, FRAGILIS, and cKIT were lowly expressed in the beginning and their expression rose as differentiation progresses after BMP4 supplementation with no or very little expression of VASA. Thus, the conventionally derived hESC lines without Activin A appear to require longer periods for the PGC specification process. We also observed a decrease in the pluripotency markers OCT4, SOX2 and NANOG accompanied by a rise in SSEA1 levels indicating that differentiation has occurred successfully in all hESC lines. Higher expression of VASA was also observed at the protein level in Activin A-derived hESC lines compared to standard hESC lines. Although VASA expression is predominantly localized to the cytoplasm in germ cells, it has also been found in the cell nucleolus of hESC-derived germ cells as described by West and colleagues [59]. Similar to these observations, we also observed both cytoplasmic and nuclear localization of VASA, which may be indicative for the different time points of germ cell differentiation. Standard hESC lines failed to show either of these expression patterns and instead continued to express Oct4; thus, again pointing to the fact that these cell lines may still be in the stage of PGC specification and need extra signals from the environment to continue their lineage progression into late PGCs.
BMPs are known to play an indispensible role for the generation and specification of PGCs. As the in vivo PGC-inducing capacity of BMPs can be extrapolated in vitro, several studies have used BMP4 as a prerequisite factor for differentiation of pluripotent stem cells into germ cells [2,5,8,56]. An important fact is that hESCs respond to BMP4 in a dose-dependent manner. Kee and colleagues observed that adding BMP4 at 1 ng/mL, 10 ng/mL and 100 ng/mL corresponded with a significant rise in expression of VASA [2]. Directed differentiation of bone marrow mesenchymal stem cells towards germ cells was also accompanied by significant upregulation of VASA when the BMP4 concentration was increased from 25 ng/mL to 100 ng/mL [37]. Similarly, we also observed that BMP4 had a dose-dependent effect on PGC differentiation as 50 ng/mL and 100 ng/mL BMP4 significantly upregulated VASA and STELLA expression compared to 10 ng/mL BMP4 or its absence.
It has been observed in mice that BMP4 interacts with the ALK2 receptor in visceral endoderm and conditions the epiblast to form PGCs. In fact, removal of visceral endoderm completely inhibited the formation of PGCs [60]. During gastrulation, extraembryonic visceral endoderm is replaced by definitive endoderm [61] and it has been recently observed that the combination of Activin A and BMP4 results in approximately 20% increase in the formation of definitive endoderm when compared to Activin A alone [61]. Upon differentiation of our Activin A-derived hESCs as EBs in the presence of BMP4, high expression of the endoderm marker GATA-6 was observed at the same time point when VASA was significantly upregulated in our EB cultures (at day 7). BMP4 and TGFβ/Activin signaling were simultaneously active in Activin A-derived hESCs lines due to nuclear localization of pSMAD1/5/8 and pSMAD2/3, respectively. In contrast, no or very low levels of pSMAD2/3 were seen in standard derived hESC lines.
Thus, as TGFβ/Activin signaling plays a role in early cell fate decision, BMP4 and endoderm development are responsible for PGC induction and formation [14,39,60,62,63]; hence, the presence of all these factors in our Activin A-derived hESCs might be responsible for increased differentiation competency towards the germ cell lineage. Collectively, our results further reiterate the notion that Activin A induced hESC derivation helps to generate a pluripotent cell line optimum for studying in vitro differentiation into the germ cell lineage.
Conclusion
The heterogeneity amongst different hESC lines has a significant impact on their differentiation potential. We have successfully shown that the hESC derivation process has profound consequences for the subsequent differentiation potential of the resulting hESC lines. In our study, we have extrapolated the Activin A containing culture conditions used for mEpiSC derivation and generation of mouse EpiLCs to hESC derivation. We were able to show that these Activin A-derived hESC lines have increased propensity towards germ cell differentiation compared to standard hESC lines derived without Activin A. Future research will focus on detailed analysis and comparison of Activin A-derived hESC lines at genome-wide transcriptomics and epigenomic level to standard derived hESC lines and validating the interaction between Activin signalling and germ cell differentiation in vitro. We also propose that a differentiation-oriented derivation strategy of hESC lines may hold a crucial key to success for efficient lineage-specific differentiation.
Supplementary Material
Acknowledgments
G.D., D.D. and this research are supported by the Flemish Foundation for Scientific Research (FWO-Vlaanderen, grant number FWO-3G062910). S.W., D.D. and this research are supported by the Concerted Research Actions funding from Bijzonder Onderzoeksfonds University Ghent (BOF GOA 01G01112). M.V.d.J. holds a PhD grant from the Agency for Innovation by Science and Technology (IWT, grant number SB093128), Belgium. S.C.d.S.L. is supported by the Netherlands Organization for Scientific Research (grant number NWO ASPASIA 016.121.365) and by the Interuniversity Attraction Poles Program (IUAP-07/07). B.H. is supported by a Ghent University grant, Faculty Health and Medical Sciences. P.D.S. is holder of a Fundamental Clinical Research Mandate from the Flemish Foundation of Scientific Research (FWO-Vlaanderen), Belgium. We would like to thank professor Nadine Van Roy and professor Björn Menten of Centre for Medical Genetics for the karyotyping of hESC lines. We would like to thank Ferring Company (Aalst, Belgium) for financial support of this study.
Author Disclosure Statement
No potential conflicts of interest.
References
- 1.Clark AT. Bodnar MS. Fox M. Rodriquez RT. Abeyta MJ. Firpo MT. Pera RA. Spontaneous differentiation of germ cells from human embryonic stem cells in vitro. Hum Mol Genet. 2004;13:727–739. doi: 10.1093/hmg/ddh088. [DOI] [PubMed] [Google Scholar]
- 2.Kee K. Gonsalves JM. Clark AT. Pera RA. Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 2006;15:831–837. doi: 10.1089/scd.2006.15.831. [DOI] [PubMed] [Google Scholar]
- 3.Aflatoonian B. Ruban L. Jones M. Aflatoonian R. Fazeli A. Moore HD. In vitro post-meiotic germ cell development from human embryonic stem cells. Hum Reprod. 2009;24:3150–3159. doi: 10.1093/humrep/dep334. [DOI] [PubMed] [Google Scholar]
- 4.Eguizabal C. Montserrat N. Vassena R. Barragan M. Garreta E. Garcia-Quevedo L. Vidal F. Giorgetti A. Veiga A. Izpisua Belmonte JC. Complete meiosis from human induced pluripotent stem cells. Stem Cells. 2011;29:1186–1195. doi: 10.1002/stem.672. [DOI] [PubMed] [Google Scholar]
- 5.Chuang CY. Lin KI. Hsiao M. Stone L. Chen HF. Huang YH. Lin SP. Ho HN. Kuo HC. Meiotic Competent Human Germ Cell-like Cells Derived from Human Embryonic Stem Cells Induced by BMP4/WNT3A Signaling and OCT4/EpCAM (Epithelial Cell Adhesion Molecule) Selection. J Biol Chem. 2012;287:14389–14401. doi: 10.1074/jbc.M111.338434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bucay N. Yebra M. Cirulli V. Afrikanova I. Kaido T. Hayek A. Montgomery AM. A novel approach for the derivation of putative primordial germ cells and sertoli cells from human embryonic stem cells. Stem Cells. 2009;27:68–77. doi: 10.1634/stemcells.2007-1018. [DOI] [PubMed] [Google Scholar]
- 7.Park TS. Galic Z. Conway AE. Lindgren A. van Handel BJ. Magnusson M. Richter L. Teitell MA. Mikkola HK, et al. Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells. 2009;27:783–795. doi: 10.1002/stem.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kee K. Angeles VT. Flores M. Nguyen HN. Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tilgner K. Atkinson SP. Yung S. Golebiewska A. Stojkovic M. Moreno R. Lako M. Armstrong L. Expression of GFP under the control of the RNA helicase VASA permits fluorescence-activated cell sorting isolation of human primordial germ cells. Stem Cells. 2010;28:84–92. doi: 10.1002/stem.263. [DOI] [PubMed] [Google Scholar]
- 10.Tilgner K. Atkinson SP. Golebiewska A. Stojkovic M. Lako M. Armstrong L. Isolation of primordial germ cells from differentiating human embryonic stem cells. Stem Cells. 2008;26:3075–3085. doi: 10.1634/stemcells.2008-0289. [DOI] [PubMed] [Google Scholar]
- 11.Ramos-Mejía V. Fernández AF. Ayllón V. Real PJ. Bueno C. Anderson P. Martín F. Fraga MF. Menendez P. Maintenance of human embryonic stem cells in mesenchymal stem cell-conditioned media augments hematopoietic specification. Stem Cells Dev. 2012;21:1549–1558. doi: 10.1089/scd.2011.0400. [DOI] [PubMed] [Google Scholar]
- 12.Osafune K. Caron L. Borowiak M. Martinez RJ. Fitz-Gerald CS. Sato Y. Cowan CA. Chien KR. Melton DA. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 13.Shi Y. Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 14.James D. Levine AJ. Besser D. Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- 15.Krummen LA. Moore A. Woodruff TK. Covello R. Taylor R. Working P. Mather JP. Localization of inhibin and activin binding sites in the testis during development by in situ ligand binding. Biol Reprod. 1994;50:734–744. doi: 10.1095/biolreprod50.4.734. [DOI] [PubMed] [Google Scholar]
- 16.Mithraprabhu S. Mendis S. Meachem SJ. Tubino L. Matzuk MM. Brown CW. Loveland KL. Activin bioactivity affects germ cell differentiation in the postnatal mouse testis in vivo. Biol Reprod. 2010;82:980–990. doi: 10.1095/biolreprod.109.079855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meinhardt A. McFarlane JR. Seitz J. de Kretser DM. Activin maintains the condensed type of mitochondria in germ cells. Mol Cell Endocrinol. 2000;168:111–117. doi: 10.1016/s0303-7207(00)00308-7. [DOI] [PubMed] [Google Scholar]
- 18.Mendis SH. Meachem SJ. Sarraj MA. Loveland KL. Activin A balances Sertoli and germ cell proliferation in the fetal mouse testis. Biol Reprod. 2011;84:379–391. doi: 10.1095/biolreprod.110.086231. [DOI] [PubMed] [Google Scholar]
- 19.Coutts SM. Childs AJ. Fulton N. Collins C. Bayne RA. McNeilly AS. Anderson RA. Activin signals via SMAD2/3 between germ and somatic cells in the human fetal ovary and regulates kit ligand expression. Dev Biol. 2008;314:189–199. doi: 10.1016/j.ydbio.2007.11.026. [DOI] [PubMed] [Google Scholar]
- 20.Bristol-Gould SK. Kreeger PK. Selkirk CG. Kilen SM. Cook RW. Kipp JL. Shea LD. Mayo KE. Woodruff TK. Postnatal regulation of germ cells by activin: the establishment of the initial follicle pool. Dev Biol. 2006;298:132–148. doi: 10.1016/j.ydbio.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 21.Kipp JL. Golebiowski A. Rodriguez G. Demczuk M. Kilen SM. Mayo KE. Gene expression profiling reveals Cyp26b1 to be an activin regulated gene involved in ovarian granulosa cell proliferation. Endocrinology. 2011;152:303–312. doi: 10.1210/en.2010-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tesar PJ. Chenoweth JG. Brook FA. Davies TJ. Evans EP. Mack DL. Gardner RL. McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 23.Xu RH. Sampsell-Barron TL. Gu F. Root S. Peck RM. Pan G. Yu J. Antosiewicz-Bourget J. Tian S. Stewart R. Thomson JA. NANOG is a direct target of TGFbeta/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell. 2008;3:196–206. doi: 10.1016/j.stem.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi K. Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development. 2009;136:3549–3556. doi: 10.1242/dev.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna J. Cheng AW. Saha K. Kim J. Lengner CJ. Soldner F. Cassady JP. Muffat J. Carey BW. Jaenisch R. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–9227. doi: 10.1073/pnas.1004584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Leary T. Heindryckx B. Lierman S. van Bruggen D. Goeman JJ. Vandewoestyne M. Deforce D. de Sousa Lopes SM. De Sutter P. Tracking the progression of the human inner cell mass during embryonic stem cell derivation. Nat Biotechnol. 2012;30:278–282. doi: 10.1038/nbt.2135. [DOI] [PubMed] [Google Scholar]
- 27.Brons IG. Smithers LE. Trotter MW. Rugg-Gunn P. Sun B. Chuva de Sousa Lopes SM. Howlett SK. Clarkson A. Ahrlund-Richter L. Pedersen RA. Vallier L. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi K. Ohta H. Kurimoto K. Aramaki S. Saitou M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell. 2011;146:519–532. doi: 10.1016/j.cell.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 29.Hayashi K. Ogushi S. Kurimoto K. Shimamoto S. Ohta H. Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- 30.Stephenson EL. Braude PR. Mason C. Proposal for a universal minimum information convention for the reporting on the derivation of human embryonic stem cell lines. Regen Med. 2006;1:739–750. doi: 10.2217/17460751.1.6.739. [DOI] [PubMed] [Google Scholar]
- 31.O'Leary T. Heindryckx B. Lierman S. Van der Jeught M. Menten B. Deforce D. Cornelissen R. de Sousa Lopes SC. De Sutter P. The influence of early embryo traits on human embryonic stem cell derivation efficiency. Stem Cells Dev. 2011;20:785–793. doi: 10.1089/scd.2010.0338. [DOI] [PubMed] [Google Scholar]
- 32.O'Leary T. Duggal G. Lierman S. Van den Abbeel E. Heindryckx B. De Sutter P. The influence of patient and cohort parameters on the incidence and developmental potential of embryos with poor quality traits for use in human embryonic stem cell derivation. Hum Reprod. 2012;27:1581–1589. doi: 10.1093/humrep/des040. [DOI] [PubMed] [Google Scholar]
- 33.Van der Jeught M. O'Leary T. Ghimire S. Lierman S. Duggal G. Versieren K. Deforce D. Chuva de Sousa Lopes S. Heindryckx B. De Sutter P. The combination of inhibitors of FGF/MEK/Erk and GSK3β signaling increases the number of OCT3/4- and NANOG-positive cells in the human inner cell mass, but does not improve stem cell derivation. Stem Cells Dev. 2013;22:296–306. doi: 10.1089/scd.2012.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisner LF. Johnson JA. Protocols for cytogenetic studies of human embryonic stem cells. Methods. 2008;45:133–141. doi: 10.1016/j.ymeth.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 35.Inman GJ. Nicolás FJ. Callahan JF. Harling JD. Gaster LM. Reith AD. Laping NJ. Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 36.Nichols J. Smith A. Naive and primed pluripotent states. Cell Stem Cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 37.Mazaheri Z. Movahedin M. Rahbarizadeh F. Amanpour S. Different doses of bone morphogenetic protein 4 promote the expression of early germ cell-specific gene in bone marrow mesenchymal stem cells. In Vitro Cell Dev Biol Anim. 2011;47:521–525. doi: 10.1007/s11626-011-9429-0. [DOI] [PubMed] [Google Scholar]
- 38.Young JC. Dias VL. Loveland KL. Defining the window of germline genesis in vitro from murine embryonic stem cells. Biol Reprod. 2010;82:390–401. doi: 10.1095/biolreprod.109.078493. [DOI] [PubMed] [Google Scholar]
- 39.Teo AK. Ali Y. Wong KY. Chipperfield H. Sadasivam A. Poobalan Y. Tan EK. Wang ST. Abraham S, et al. Activin and BMP4 synergistically promote formation of definitive endoderm in human embryonic stem cells. Stem Cells. 2012;30:631–642. doi: 10.1002/stem.1022. [DOI] [PubMed] [Google Scholar]
- 40.Unger C. Skottman H. Blomberg P. Dilber MS. Hovatta O. Good manufacturing practice and clinical-grade human embryonic stem cell lines. Hum Mol Genet. 2008;17:R48–R53. doi: 10.1093/hmg/ddn079. [DOI] [PubMed] [Google Scholar]
- 41.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 42.Reubinoff BE. Pera MF. Fong CY. Trounson A. Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 43.Cowan CA. Klimanskaya I. McMahon J. Atienza J. Witmyer J. Zucker JP. Wang S. Morton CC. McMahon AP. Powers D. Melton DA. Derivation of embryonic stem-cell lines from human blastocysts. N Engl J Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- 44.Andrews PW. Benvenisty N. McKay R. Pera MF. Rossant J. Semb H. Stacey GN SCotISC Initiative. The International Stem Cell Initiative: toward benchmarks for human embryonic stem cell research. Nat Biotechnol. 2005;23:795–797. doi: 10.1038/nbt0705-795. [DOI] [PubMed] [Google Scholar]
- 45.Ware CB. Nelson AM. Blau CA. A comparison of NIH-approved human ESC lines. Stem Cells. 2006;24:2677–2684. doi: 10.1634/stemcells.2005-0452. [DOI] [PubMed] [Google Scholar]
- 46.Adewumi O. Aflatoonian B. Ahrlund-Richter L. Amit M. Andrews PW. Beighton G. Bello PA. Benvenisty N. Berry LS, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 47.Lerou PH. Yabuuchi A. Huo H. Miller JD. Boyer LF. Schlaeger TM. Daley GQ. Derivation and maintenance of human embryonic stem cells from poor-quality in vitro fertilization embryos. Nat Protoc. 2008;3:923–933. doi: 10.1038/nprot.2008.60. [DOI] [PubMed] [Google Scholar]
- 48.Fraga AM. Souza de Araújo É. Stabellini R. Vergani N. Pereira LV. A survey of parameters involved in the establishment of new lines of human embryonic stem cells. Stem Cell Rev. 2011;7:775–781. doi: 10.1007/s12015-011-9250-x. [DOI] [PubMed] [Google Scholar]
- 49.Camarasa MV. Galvez VM. Brison DR. Bachiller D. Optimized protocol for derivation of human embryonic stem cell lines. Stem Cell Rev. 2012;8:1011–1020. doi: 10.1007/s12015-012-9377-4. [DOI] [PubMed] [Google Scholar]
- 50.Tannenbaum SE. Turetsky TT. Singer O. Aizenman E. Kirshberg S. Ilouz N. Gil Y. Berman-Zaken Y. Perlman TS, et al. Derivation of xeno-free and GMP-grade human embryonic stem cells—platforms for future clinical applications. PLoS One. 2012;7:e35325. doi: 10.1371/journal.pone.0035325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rajala K. Lindroos B. Hussein SM. Lappalainen RS. Pekkanen-Mattila M. Inzunza J. Rozell B. Miettinen S. Narkilahti S, et al. A defined and xeno-free culture method enabling the establishment of clinical-grade human embryonic, induced pluripotent and adipose stem cells. PLoS One. 2010;5:e10246. doi: 10.1371/journal.pone.0010246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skottman H. Derivation and characterization of three new human embryonic stem cell lines in Finland. In Vitro Cell Dev Biol Anim. 2010;46:206–209. doi: 10.1007/s11626-010-9286-2. [DOI] [PubMed] [Google Scholar]
- 53.Allegrucci C. Young LE. Differences between human embryonic stem cell lines. Hum Reprod Update. 2007;13:103–120. doi: 10.1093/humupd/dml041. [DOI] [PubMed] [Google Scholar]
- 54.Abeyta MJ. Clark AT. Rodriguez RT. Bodnar MS. Pera RA. Firpo MT. Unique gene expression signatures of independently-derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- 55.Skottman H. Strömberg AM. Matilainen E. Inzunza J. Hovatta O. Lahesmaa R. Unique gene expression signature by human embryonic stem cells cultured under serum-free conditions correlates with their enhanced and prolonged growth in an undifferentiated stage. Stem Cells. 2006;24:151–167. doi: 10.1634/stemcells.2004-0189. [DOI] [PubMed] [Google Scholar]
- 56.Aflatoonian B. Moore H. Germ cells from mouse and human embryonic stem cells. Reproduction. 2006;132:699–707. doi: 10.1530/REP-06-0022. [DOI] [PubMed] [Google Scholar]
- 57.Matsui Y. The molecular mechanisms regulating germ cell development and potential. J Androl. 2010;31:61–65. doi: 10.2164/jandrol.109.008094. [DOI] [PubMed] [Google Scholar]
- 58.Wongtrakoongate P. Jones M. Gokhale PJ. Andrews PW. STELLA facilitates differentiation of germ cell and endodermal lineages of human embryonic stem cells. PLoS One. 2013;8:e56893. doi: 10.1371/journal.pone.0056893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West FD. Machacek DW. Boyd NL. Pandiyan K. Robbins KR. Stice SL. Enrichment and differentiation of human germ-like cells mediated by feeder cells and basic fibroblast growth factor signaling. Stem Cells. 2008;26:2768–2776. doi: 10.1634/stemcells.2008-0124. [DOI] [PubMed] [Google Scholar]
- 60.de Sousa Lopes SM. Roelen BA. Monteiro RM. Emmens R. Lin HY. Li E. Lawson KA. Mummery CL. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Genes Dev. 2004;18:1838–1849. doi: 10.1101/gad.294004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis SL. Tam PP. Definitive endoderm of the mouse embryo: formation, cell fates, and morphogenetic function. Dev Dyn. 2006;235:2315–2329. doi: 10.1002/dvdy.20846. [DOI] [PubMed] [Google Scholar]
- 62.Lawson KA. Dunn NR. Roelen BA. Zeinstra LM. Davis AM. Wright CV. Korving JP. Hogan BL. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Genes Dev. 1999;13:424–436. doi: 10.1101/gad.13.4.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dudley B. Palumbo C. Nalepka J. Molyneaux K. BMP signaling controls formation of a primordial germ cell niche within the early genital ridges. Dev Biol. 2010;343:84–93. doi: 10.1016/j.ydbio.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.