Abstract
Extracellular vesicles (EVs) have been implicated in tumorigenesis. Biomolecules which can block EV binding and uptake into recipient cells may be of therapeutic value as well as enhance understanding of EV biology. Here, we show that heparin interacts with uptake of tumor-derived as well as non-tumor-derived EVs into recipient cells. Incubation of glioma cell-derived EVs with heparin resulted in micron-sized structures observed by transmission electron microscopy, with EVs clearly visible within these structures. Inclusion of heparin greatly diminished transfer of labeled EVs from donor to recipient tumor cells. We also show a direct interaction between heparin and EVs using confocal microscopy. We found that the block in EV uptake was at the level of cell binding and not internalization. Finally, incubation of glioma-derived EVs containing EGFRvIII mRNA with heparin reduced transfer of this message to recipient cells. The effect of heparin on EVs uptake may provide a unique tool to study EV function. It may also foster research of heparin or its derivatives as a therapeutic for disease in which EVs play a role.
Keywords: Extracellular vesicles, Exosomes, Glioblastoma, EGFRvIII, Heparin, Tumor
Introduction
The incidence of glioblastoma (GBM) is about 3.5 per 100,000 people per year with a mean overall survival of 1.5 years [1]. Research at the molecular level has begun to unravel some of the characteristics of glioma, which includes production of extracellular vesicles (EVs) [2]. EVs are 30–1000 nm diameter lipid structures produced by normal and tumor cells and they function in cell to cell communication [3]. The content and function of EVs vary depending on the cell of origin. For example, EVs derived from immune cells have potent immuno-stimulatory and antitumor effects in vivo [4]. On the other hand, EVs released from tumor cells can accelerate tumor growth [5, 6]. The observed functional effects of EVs have been attributed to associated proteins, mRNA, miRNA, and DNA [7]. The ability to block transfer of tumor-derived EVs containing oncogenic messages such as EGFRvIII into recipient cells is a potential anti-tumor strategy. We have shown that incubation of EVs from a cell line with heparin blocked their transfer into recipient cells [8]. In the present study we examined whether heparin can be utilized to block EV uptake and transfer of biomolecules from glioma-derived EVs into recipient cells.
Materials and methods
Cell culture
The human GBM cell line U87-MG and the human embryonic kidney cell line, 293T were purchased from the American Type Culture Collection (Manassas, VA). The human glioma cell line, Gli36, was obtained from Dr. Anthony Capanogni (University of California at Los Angeles, Los Angeles, CA). Gli36 were transduced with retroviral vector encoding EGFRvIII [9]. Primary GBM cells GBM11/5 have been described [10]. All of the above cells were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10 % fetal bovine serum (FBS) (Sigma) and penicillin/streptomycin (100 U/ml; 100 μg/ml; Invitrogen, Carlsbad, CA) in a humidified atmosphere supplemented with 5 % CO2 at 37 °C. The primary medulloblastoma cell line D384 was obtained from Dr. S. Pomeroy (Duke University). They were cultured in DMEM containing 10 % FBS, GlutaMAX™ (Invitrogen) and 1 % penicillin/streptomycin. Human umbilical vein endothelial cells (HU-VECs) were provided by Drs. Francis W. Luscinskas and Kay Case (Cell Core Facility, Brigham and Women’s Hospital). HUVECs were cultured in gelatin-coated flasks in endothelial basal medium (Lonza, Allendale, NJ) supplemented with human epidermal growth factor (hEGF), hydrocortisone and GA-1000 (all from Lonza).
Heparin
We used heparin sodium salt, 5,000 USP U/ml, (APP Pharmaceuticals, LLC, Schaumburg, IL) and heparin sodium salt from porcine intestinal mucosa, 180 USP U/mg (Sigma).
Extracellular vesicle isolation
Cells were grown for 48 h in 15 cm cultures plates (~5 million cells plated) in 20 ml DMEM containing 5 % EV-depleted FBS. For each experiment, EVs were purified from 40 to 80 ml of conditioned media by differential centrifugation. Briefly, media was centrifuged 300×g for 10 min at 4 °C followed by 2,000×g for 5 min at 4 °C to pellet dead cells and debris. The supernatant was then filtered through 0.8 μm filter (Thermo Scientific, Lafayette, CO) and ultracentrifuged at 100,000×g for 80 min in a 70Ti rotor. The EV pellet was washed in 12 ml cold 1 × PBS and re-pelleted at 100,000×g for 60 min in a MLA-55 rotor. The resuspended EV pellet was used for experiments.
Transwell system to measure donor to recipient cell EV transfer
Recipient cells were plated (50,000 cells/well) in a 24-well plate. After 24 h, cells were washed and incubated for 30 min at 37 °C in DMEM containing 10 % EV-depleted FBS. Next, heparin was added at the indicated concentration and PKH67-labeled donor cells (50,000 cells/well) were placed in a transwell chamber (1 μm nominal pore size) on top of recipient cells. After 48 h, recipient cells were analyzed for PKH-67 labeling (indicative of EV uptake) using a BD LSRII flow cytometer (Becton–Dickinson, Franklin Lakes, NJ) and analysis software (FlowJo, Ashland, OR).
PKH67 labeling of EVs and direct EV transfer to recipient cells
Purified EVs from 40 ml conditioned media of cells were incubated with the PKH67 green-fluorescent labeling dye (Sigma-Aldrich) at room temperature (RT) for 3 min, as described [10] and washed 2 times to remove unbound dye. Next labeled EVs were incubated in control buffer (PBS) or PBS with 20 μg/ml of heparin for 30 min at room temperature. Then these mixtures were added to wells of recipient cells plated on glass coverslips in 12 well plates. After a 1 h incubation at 37 °C cells were washed in PBS and then fixed in 4 % formaldehyde in PBS before analysis by fluorescence microscopy. Images were acquired using the FITC filter set using the same acquisition settings for all samples. Three images per well of three independent wells were acquired per condition. Images were analyzed for fluorescence intensity using ImageJ. Integrated density was calculated using instructions found on the NIH’s ImageJ website (http://rsbweb.nih.gov/ij/index.html).
Transmission electron microscopy
Purified EVs from 40 ml conditioned media of U87-MG and GBM11/5 cells were resuspended in 1 × PBS. After incubation (30 min) with heparin, freshly prepared 4 % formaldehyde was added to samples before being processed. Fresh carbon-coated grids were placed on top of a drop of the EV suspension. Next, grids were placed directly on top of a drop of 2 % uranyl acetate. The grids were examined with a Technai-12 G2 Spirit Biotwin transmission electron microscope (FEI, Eindhoven, The Netherlands).
Heparin-binding assays
EV/heparin colocalization
For the microscopic visualization of binding of EVs with heparin, 293T cells were plated and labeled with CellTracker™ Red (Life Technologies, Grand Island, NY) according to manufacturer’s recommendations. Briefly, 2 × 106 293T cells plated in 100 mm dish were incubated with CellTracker™ Red in plain media in 37 °C for 30 min followed by a change to normal culture media. Culture media containing EV-depleted FBS was added after 24 h and 293T-derived red EVs were isolated after 48 h according to the ultracentrifugation steps described above.
Next, 10 μg of EVs were mixed with 100 μg/ml of FITC-heparin overnight at 4 °C. FITC-heparin incubated with 1 × PBS without EVs served as negative control. The following day, EVs were pelleted by ultracentrifugation at 100,000×g for 2 h in an Optima MAX-XP ultracentrifuge (Beckman Coulter; MLS-50 rotor). Pellets from each sample were resuspended in 150 μl 1 × PBS. Ten micro-liters of each sample were analyzed in duplicate with confocal imaging using a Zeiss LSM 5 Pascal laser-scanning confocal microscope (Zeiss, Oberkochen, Germany). Images were acquired using a 10, 40 or 63 PlanApo (NA 1.4) differential interference contrast (DIC) objective on an inverted microscope (Axiovert 200 M, Zeiss) equipped with an LSM 510META scan head (Zeiss). Argon ion (488 nm) and HeNe (543 nm) lasers were used for excitation. Green and red fluorescence emissions were detected through BP 505–530 and 560–615 filters, respectively.
EV/heparin pelleting assay
U87-derived EVs were first labeled with PKH67 dye. After removing unbound dye by ultracentrifugation wash steps, EVs were mixed with PBS, or 100 μg/ml of either heparin or streptavidin for 30 min at room temperature. Next samples were centrifuged for 2 h at 100,000×g. EV pellets were resuspended in equal volumes of PBS and fluorescence measured in a FlexStation 3 microplate reader (Molecular Devices, Sunnyvale, CA) using SoftMax Pro software (Molecular Devices).
Heparin/cell preincubation
U87 cells were incubated on ice with 200 μg/ml of heparin or PBS as control for 30 min. Next wells were washed three times with PBS to remove unbound heparin. PKH67-labeled U87-derived EVs were next added to wells and cells incubated at 37 °C for 1 h before fixing in 4 % formaldehyde and visualization by fluorescence microscopy.
EGFRvIII mRNA-uptake assay
EV quantitation
Purified EVs were analyzed for their protein content using Quick Start™ Bradford assay (Bio-Rad, Hercules, CA) and particle concentration using the Nanosight LM10 nanoparticle characterization system (NanoSight, Wiltshire, UK).
RNA isolation from donor cell EVs
Gli36-EGFRvIII-derived EVs obtained from 40 ml conditioned media were purified by differential centrifugation (as described above). For RNA isolation, a total of 1.0 × 1010 EVs and a total of 2.5 × 107 cells were used. The EVs were treated with DNAse (DNA-free kit: Ambion®, Life Technologies, Grand Island, NY) and RNAse inhibitors (Fermentas, Thermo Scientific). Next, the EVs were lysed in 700 μl of Qiazol reagent (Qiagen Valencia, CA). Nucleic acid was extracted using the miRNeasy kit (Qiagen). Total RNA was eluted in RNAse-free water. The quantity and size range of the nucleic acids were evaluated using the 2100 Bioanalyzer (Agilent, Santa Clara, CA) using the RNA 6000 pico chip (Agilent) [10].
EGFRvIII mRNA uptake
To detect transfer of mRNA by EVs into recipient cells, 50,000 cells/well were plated. Purified EVs (3.3 × 109) from 40 ml conditioned media of Gli36-EGFRvIII cells were incubated with or without 100 μg/ml heparin in 1 × PBS at RT for 30 min. This mixture was then added to cells. After 3 h, cells were washed with 1 × PBS. Next cells were lysed in 700 μl of Qiazol reagent (Qiagen). In some experiments, cells were trypsinized and washed before lysing. Nucleic acid was extracted using the miRNeasy kit (Qiagen). Quantity of the nucleic acid was evaluated using the Nanodrop ND-1000 (Thermo Fisher Scientific Inc.).
cDNA reaction
Five hundred ng of cellular RNA was used as input for cDNA reaction using the SuperScript® VILO™ (Invitrogen) in a total of 20 μl. The cDNA synthesis program consisted of 1 cycle at 25 °C for 10 min, 1 cycle at 60 °C for 60 min, 1 cycle at 85 °C for 5 min.
RT-qPCR
1 μl of cDNA was used for each RT-qPCR reaction. Primers and TaqMan MGB probes (Life Technologies) were used to detect human EGFRvIII and GAPDH RNA from cells or EVs. All RT-qPCR reactions were performed in 25 μl reactions using the fast TaqMan MasterMix (Applied Bio-systems). Amplification conditions consisted of 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min, 40 cycles at 95 °C, for 15 s, 1 cycle at 60 °C for 1 min on standard mode and were performed using ABI PRISM 7500 (Applied Bio-systems). The EGFRvIII primers were Forw: CTGCTGGC TGCGCTCTG, Reverse: CGTGATCTGTCACCACATAA TTACC and the probe TTCCTCCAGAGCCCGACT. The GAPDH probe and primer kit was purchased from Life Technologies (Hs03929097_g1, Cat. # 4331182).
Statistics
Statistical analysis was performed using Graphpad Prism® software (version 5.01; La Jolla, CA). Multiple comparisons between groups were performed by a two-way ANOVA test. Statistical significance between two groups was determined using an unpaired t test with statistical significance set to a value of 0.05.
Results
Heparin interferes with tumor derived-EV uptake by recipient cells
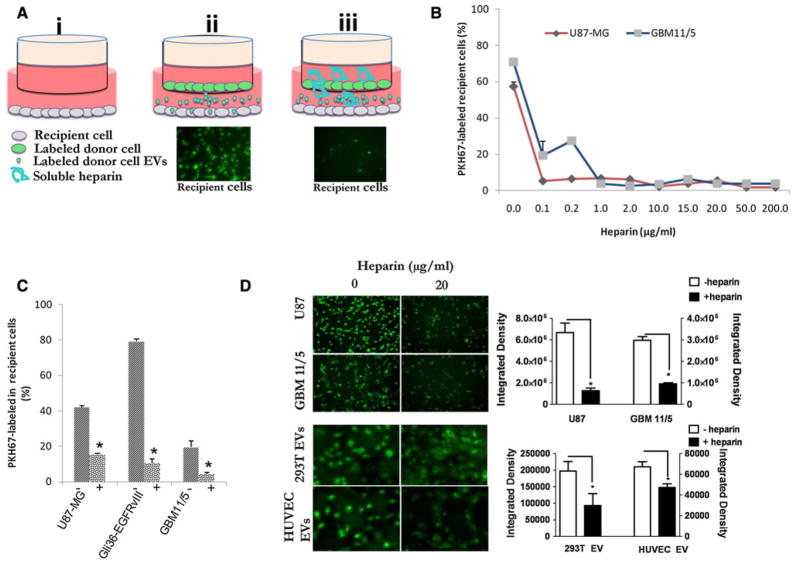
To test the ability of heparin to inhibit transfer of tumor-derived EVs, we used a co-culture system (Fig. 1a). Donor cells were PKH67-labeled (Supplementary Fig. 1) and plated in the top chamber of a transwell dish and the unlabeled recipient cells were plated in the bottom chamber (with different concentrations of heparin added to the media). This allows PKH67-labeled EVs produced by donor cells to pass to the lower chamber while keeping cells separated (Fig. 1a and [11]). After 48 h, using flow cytometry, we measured transfer of EVs from PKH67-labeled D384 donor cells to recipient cells (GBM11/5 or U87-MG) at various concentrations of heparin (Fig. 1b). The lowest heparin concentration tested, 0.1 μg/ml, achieved a 90 % reduction in EV uptake in U87-MG cells, while for GBM11/5 similar block was achieved at a 10-fold higher dose (1 μg/ml) with a reduction of 95 % (Fig 1b, p <0.05). Next, we used PKH67-labeleld GBM11/5, Gli36-EGFRvIII, and U87-MG donor cells in the transwell system on unlabeled recipient cells of the same type at 20 μg/ml of heparin. Heparin caused uptake reduction ranging from 62 to 86 % (Fig. 1c, p <0.05). We also performed controls to rule out that our observed blocking effect of labeled recipient cells was due to a heparin/free PKH67 dye interaction. First we measured the fluorescence intensity of PKH67 incubated with PBS or PBS containing different concentrations of heparin and we observed no significant reduction even at high heparin concentrations (Supplementary Fig. 2). Next we examined whether heparin would block the labeling of cells with free dye in the transwell system and we found no such effect, indicating the block we observed in Fig. 1a–c was due to a heparin/EV interaction and not free dye (Supplementary Fig. 3). Finally, we observed the phenotype of the labeled vesicles in the cells and the staining pattern gave the characterstic punctate dot structures within the cells, indicated dye was associated with EVs (Supplementary Fig. 4). Another possible explanation for the observed lower labeling of recipient cells is that heparin may have effected biogenesis or release of labeled EVs from the donor cell. To test this hypothesis, we incubated GBM 11/5, U87, and D384 with or without heparin at 100 μg/ml for 24 h. Next EVs were harvested by ultracentrifugation and the pellets resuspended in PBS and counted by nanoparticle tracking analysis (NTA). We found no significant differences in the level of vesicles produced by any tested cell in the presence of heparin compared to the cell incubated in control media (Supplementary Fig. 5). To examine whether EV-uptake block is specific for tumor cells, we compared uptake of PKH67-labeled tumor EVs into tumor recipient cells (both U87 and GBM 11/5) with uptake of PKH67-labeled 293T-derived EVs or HUVEC EVs to unlabeled HUVEC recipient cells. For U87 and GBM11/5 EVs/cells 20 μg/ml heparin reduce the uptake by 81 and 68 %, respectively (Fig. 1d, p <0.05). A 53 and 30 % reduction (Fig. 1d, p <0.05) occurred in HUVEC cell uptake of labeled 293T or HUVEC-derived EVs, respectively, in the presence of 20 μg/ml heparin indicating that the blocking effect is universal for both tumor cells and HUVECs, although tumor EVs/cells seem to be more sensitive to the blocking effects.
Fig. 1.
Heparin blocks extracellular vesicle (EV) uptake in recipient cells. a Schematic of experiment of recipient cells uptake of donor cell EVs. (i). Recipient cells are plated in a well. PKH67-labeled donor cells are then added in the upper chamber of a transwell system in the absence (ii) or presence of heparin (iii). 48 h later, recipient cells are then examined for PKH67-labeled EV uptake by fluorescence microscopy (fluorescence images) and flow cytometry. b Flow cytometric quantitation of PKH67-labeled D384 donor cell-derived EVs uptake by recipient glioma cells in the presence of various concentrations of heparin. c Flow cytometric detection of PKH67-labeled U87-MG, Gli36-EGFRvIII, and GBM11/5 donor cell-derived EV uptake in presence or absence of 20 μg/ml heparin into unlabeled U87-MG, Gli36-EGFRvIII and GBM11/5 recipient cells, respectively. d Uptake of purified PKH67-labeled U87 and GBM 11/5-derived EVs into their respective unlabeled recipients (top panels and graph) or 293T-derived or HUVEC-derived EVs into recipient unlabeled HUVEC cells (bottom panels and graph) in presence or absence of 20 μg/ml heparin. *p <0.05
Heparin binds to and causes aggregation of EVs
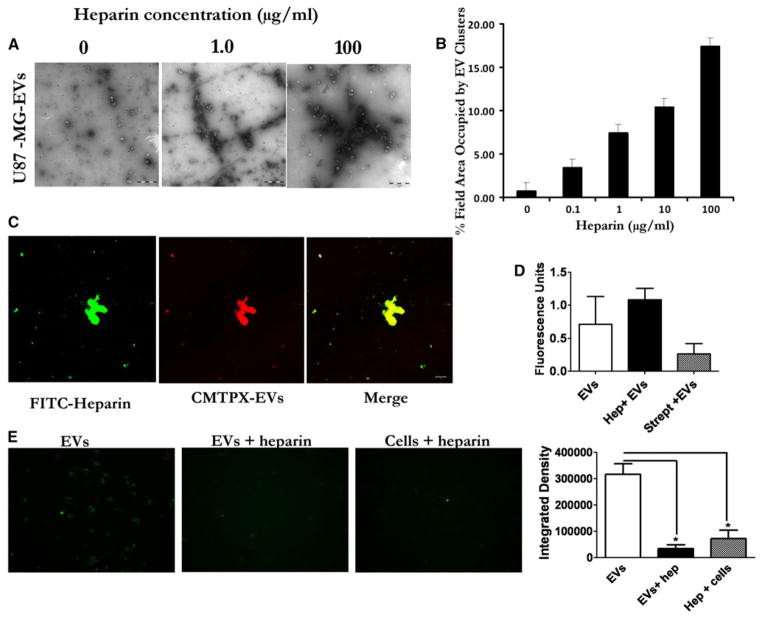
TEM was performed on U87-MG and GBM11/5-derived EVs in the presence of 0, 0.1, 1.0, 10, and 100 μg/ml heparin. Interestingly, we observed increasingly larger clusters/networks of EVs as the heparin concentration increased (Fig. 2a, Supplementary Figs. 6, 7). These aggregates were rarely observed in the absence of heparin (Fig. 2a, 0 μg/ml). Quantification showed an increase in cluster formation and increased area occupied in viewing fields by the EV network as the concentration of heparin increased (Fig. 2b). The TEM analysis suggests direct binding of EVs by heparin, causing aggregation of EVs. Zeta potential predicts the likelihood that suspensions of nanoparticles will aggregate. The zeta potential of U87-MG-derived EVs was determined using electrophoretic light scattering to be −39.1. Addition of 100 μg/ml heparin reduced the zeta potential to −32.6 (Supplementary Fig. 8 p = 0.0033). This supports the TEM data that heparin mediates EV aggregation by binding.
Fig. 2.
Heparin causes EV aggregation and binds EVs. a Suspensions of EVs derived from U87-MG cells in PBS were incubated with 0–100 μg/ml of heparin for 30 min at RT and then imaged by transmission EM (TEM). Scale bar = 1 μm. b Quantification of the TEM images show increased area occupied by the EV clusters as heparin concentration increases. (C) Confocal imaging of complexed FITC-heparin (green) and U87-MG -derived EVs (CMTPX-red). Merging of the images of FITC-heparin and EVs shows colocalization of heparin and EVs in yellow (Scale bar = 10 μm). d Incubating heparin with EVs increases their pelleting efficiency. PKH67 labeled U87 EVs were mixed with PBS, heparin, or streptavidin and pelleted 2 h at 100,000×g. Pelleted EVs were resuspended and fluorescence activity measured using a plate reader. e Binding heparin to cells reduces EV uptake. U87 glioma cells were incubated on ice with PBS or heparin (200 μg/ml) before adding PKH67 labeled U87-derived EVs. For control we incubated heparin and EVs at room temperature (EVs + heparin) before adding to cells. Magnification × 20
To attempt to colocalize FITC-heparin with EVs, we labeled EVs derived from U87-MG, with the red-fluorescent membrane dye, red CMTPX. The EVs collected from these cells were added to FITC-heparin and incubated overnight at 4 °C. Next, EVs were visualized by confocal microscopy at different magnifications. We observed varying sized aggregates and the red-labeled EVs colocalized with FITC-heparin in U87 derived EVs (Fig. 2c) as well as GBM11/5 and 293T cells derived EVs (Supplementary Fig. 9). We hypothesized that the large micron-sized structures observed with heparin and EVs by TEM would allow a more efficient pelleting efficiency when centrifuged. We mixed PKH67 labeled U87 EVs with (a) PBS, (b) heparin, or (c) streptavidin (as a negative control). Next EVs were pelleted for 2 h at 100,000×g and then resuspended in PBS and read using a fluorescence microplate reader. Incubating EVs with heparin lead to the highest recover of EVs (measured by PKH67 fluorescence) over EVs alone and EVs incubated with streptavidin (Fig. 2d). We also considered the possibility that heparin could also bind the cell surface and block EVs from interaction with receptors on the cell surface. To test this, U87 recipient cells were coated with heparin (or PBS alone for control) on ice and free heparin removed by washing with PBS. Next PKH67-labeled U87 EVs were added and the cells switched to 37 °C to allow internalization. We observed a large reduction (4-fold) in EV uptake compared to EVs alone (Fig. 2e). We also incubated EVs and heparin in solution before adding to cells as done in Fig. 1d as control.
Heparin partially blocks transfer of EGFRvIII mRNA in EVs into recipient cells
We next investigated whether heparin can be utilized as a blocking agent to prevent EV-mediated transfer of cellular genetic information into recipient cells. We selected to assay transfer of unique message from the oncogene EGFRvIII. Gli36 cells stably expressing EGFRvIII were used as donor cells. High levels of EGFRvIII mRNA (normalized to GAPDH mRNA) were detected by reverse-transcriptase qPCR (RT-qPCR) in both cellular RNA as well as EV RNA from these cells (Fig. 3a). We next attempted to see if we could detect transfer of EGFRvIII mRNA into U87-MG cells which do not express the mutant form of EGFR. 3.3 × 109 Gli36-EGFRvIII-derived EVs were added to each well of U87-MG cells and incubated for 3 h at 37 °C. RNA was extracted from U87-MG cells and RT-qPCR performed for EGFRvIII and GAPDH mRNA levels. EGFRvIII mRNA was detected in U87-MG cells exposed to EVs (average Ct value, 31.2) while it was not detectable after 40 cycles in U87-MG cells not exposed to EVs (data not shown). We next compared EGFRvIII mRNA values normalized to GAPDH mRNA values for cells exposed to EVs with those of the input vesicles. We found detectable EGFRvIII mRNA in recipient cells (Fig. 3b). To confirm that the detected EGFRvIII was on the inside of recipient cells and not just EVs that bound to the cell surface, after the 3 h incubation step with EGFRvIII-containing EVs, we washed cells and incubated with trypsin (or PBS as control). RNA was isolated and RT-qPCR performed as before. No statistically significant difference in GAPDH-normalized EGFRvIII mRNA levels was detected with or without trypsin treatment indicating the message we are detecting is protected on the inside of the recipient cells (Fig. 3c). We confirmed that the mRNA signal was on the cell interior by incubating U87 cells for 3 h with PKH67-labeled Gli36-EGFRvIII-derived EVs. It was clear from this experiment that the signal was coming from vesicular structures within the cell interior and not the cell surface (Supplementary Fig. 10a, b). To see if heparin could block the uptake of EV-cargoed EGFRvIII mRNA, we mixed EVs with PBS or 100 μg/ml heparin for 30 min at RT before adding to cells for 3 h of incubation at 37 °C. Next, the cells were washed, lysed for RNA extraction and followed by RT-qPCR for EGFRvIII and GAPDH messages in recipient cells. We observed a significant 49 % reduction in the level of oncogene EGFRvIII cDNA in the presence of heparin indicating that heparin can be utilized to block uptake of EV contents into recipient cells (Fig. 3d, p = 0.014).
Fig. 3.
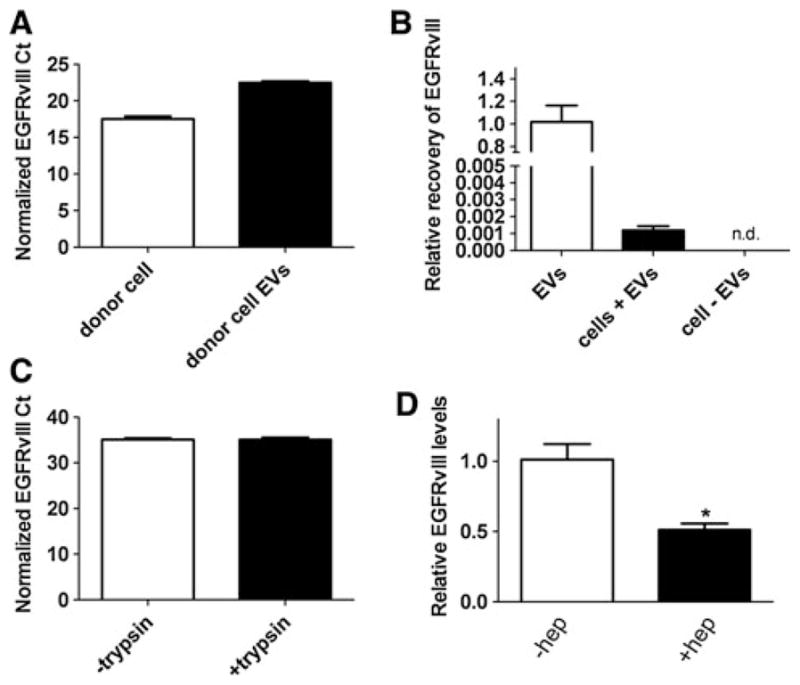
Heparin partially blocks oncogenic EGFRvIII mRNA transfer. a Relative levels of EGFRvIII mRNA in Gli36-EGFRvIII donor cells and their EVs. RNA was extracted from cells or EVs and 1 μg of RNA was used as template for a cDNA reaction. EGFRvIII Ct values were normalized to GAPDH Ct values for each sample. b Transfer of EGFRvIII mRNA inside EVs to recipient U87-MG cells (which lack endogenous EGFRvIII). 3.3 × 109 Gli36-EGFRvIII derived EVs were added to cells and after 3 h at 37 °C RNA was isolated and a RT-qPCR performed to detect EGFRvIII mRNA. After normalization to GAPDH the levels of EGFRvIII were compared to the input levels from donor cell EVs. n.d. = not detectable. c Detected EGFRvIII message is on the inside of recipient cells. After incubation for 3 h at 37 °C, cells were washed and then incubated with trypsin to remove any EVs bound to the cell surface. Control samples were treated with PBS. RNA was isolated and EGFRvIII cDNA detected with RT-qPCR. d Gli36-EGFRvIII derived EVs were incubated with or without heparin (100 μg/ml) for 30 min at RT and next were added directly to recipient U87-MG cells. After 3 h at 37 °C, the total RNA of recipient cells was extracted and used for detection of EGFRvIII mRNA with RT-qPCR. Values were normalized to GAPDH and then compared to the levels in the absence of heparin which was arbitrarily set to 1.0. The depicted graph is representative of one of four independent experiments
Heparin interferes with EV binding to cell surface
To understand whether heparin was blocking the binding of EVs to the cell surface or blocking internalization, we incubated PKH67-labeled Gli36-EGFRvIII-derived EVs with or without 100 μg/ml heparin for 30 min at RT. Next, mixtures were transferred to plated recipient U87-MG cells incubated on ice to prevent cellular internalization. After binding for 30 min, any unbound EVs were rinsed off with PBS and the cells fixed before analysis by confocal fluorescence microscopy. Interestingly, heparin greatly blocked binding of EVs to the recipient cell (Supplementary Fig. 11a). In another set of wells, we rinsed cells after the 30 min binding on ice and incubated the cells at 37 °C for 30 min and visualized internalization of PKH67-labeled EVs by z-stack analysis. Fluorescent signal was highest inside recipient cells in EV samples incubated without heparin (Supplementary Fig. 11b) but some internalized vesicles were also visualized in samples incubated with heparin (Supplementary Fig. 11b) indicating that heparin is most likely blocking ligand binding of the EVs and not internalization per se.
Discussion
We recently observed that heparin blocks uptake of 293T-derived EVs into recipient 293T cells [8]. In the present study, we sought to test the hypothesis that heparin could be used to block uptake of EVs with the potential that this could be used as a tool to study EV function, as well as to develop a therapy for diseases in which EVs have a role in pathogenesis, including progression of cancer such as glioma.
We found heparin efficiently blocked transfer of brain tumor cell-derived EVs into recipient cells (Fig. 1). Heparin interference with EV uptake may occur via more than one process. We detected direct interaction between EVs and heparin co-localization by microscopy (Fig. 2c), as well as aggregation of EVs in the presence of heparin by TEM (Fig. 2a, b). One report exists using artificial giant phospholipid vesicles that heparin caused vesicle adhesion [12] which may support our observations by TEM. Although our binding assay suggest an association of heparin and EVs, it does not indicate the binding affinity and strength of the interaction, which remains to be determined. We also show that binding to the cell surface is blocked by heparin (Supplementary Fig. 11a). This may indicate that EVs contain ligands which bind directly with heparin and that heparin is acting as a decoy for the bona fide receptor on recipient cells (e.g. heparan sulfate proteoglycans, HSPGs). However, since the assays for the blocking experiments (Fig. 1) used a molar excess of heparin, it is possible that heparin may also bind to and block a cell surface receptor utilized by EVs for binding. HSPGs have also been reported to be on the surface of EVs [13, 14], suggesting that HSPGs on the surface of EVs may bind to an EV-binding ligand on the cell surface. In fact when we bound heparin to cells on ice and then added, labeled EVs, uptake was dramatically reduced (Fig. 2e). This may suggest that heparin can bind to both the EV and cell surface to prevent EV internalization.
In addition to the block in uptake of tumor-derived EVs, we explored whether heparin could block the uptake of EVs into primary HUVECs (Fig. 1d). This block was less than the inhibition of EV transfer by heparin observed between glioma cell donor and recipient cells (Fig. 1d). This may indicate differences in receptor/ligand interplay at the cell and EV surfaces in different donor and recipient cell types. The apparent difference in blocking ability between normal and tumor-derived EVs may be important for therapeutic based applications where lower doses of heparin may preferentially block tumor-EV uptake while not interfering greatly with normal cell EVs.
Interestingly, several case reports suggest that in certain cases, heparin has anti-cancer effects in humans [15–24]. Animal studies have observed a decrease in metastasis with injection of heparin [25–27]. The mechanism for heparin’s anti-metastatic effect in an animal model of cancer has been proposed to be a block in tumor cell/platelet interactions which is an interaction known to be important for metastasis [25]. In addition to this mechanism, it is possible that heparin’s anti-cancer role may involve regulating uptake of EVs into recipient cells.
In conclusion, we show that heparin interacts directly with tumor-derived EVs and blocks binding of EVs by recipient cells. Blocking was observed at concentrations as low as 0.1 μg/ml, which is in the range of clinically acceptable heparin concentrations in plasma [28]. Although our results are preliminary, heparin or a more tumor-derived EV specific derivative may have clinical applications in the future to reduce effects of glioma-derived EVs containing environment-modifying cargo. In addition, the ability of heparin to block EV binding to cells provides a tool in assessing functional effect of EV cargo on different cell types.
Supplementary Material
Acknowledgments
This work was supported by NIH/NINDS 1 R21 NS081374-01 (C.A.M.) NIH/NCI grants CA069246 (X.O.B), CA141226 (X.O.B.); CA156009 (X.O.B.) CA141150 (X.O.B.), an American Brain Tumor Association Fellowship (C.A.M.), and a 2010 AMC Scholarship, University of Amsterdam (N.A.A.). We gratefully acknowledge the Neuroscience Center PCR Core facility, which is funded by PHS grant P30NS045776. We acknowledge the MGH Neuroscience Center Microscopy and Image Analysis Core, which is funded by NIH grant P30NS045776. We thank the van Leeuwenhoek Center for Advanced Microscopy (LCAM) at the Academic Medical Center (University of Amsterdam, the Netherlands) for performing the transmission electron microscopy sample processing and imaging. We thank Lori LoGuidice and Ard Jonker (University of Amsterdam) for technical help. C.A.M. has a financial interest in Exosome Diagnostics, Inc. C.A.M.’s interests were reviewed and are managed by the Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.S. is an inventor on the exosome/EVs technology used in this study which has been licensed to Exosome Diagnostics, Inc. He holds equity in, and is now an employee of that company. X.O.B. is on the Scientific Advisory Board of Exosome Diagnostics, Inc.
Abbreviations
- DIC
Differential interference contrast
- DMEM
Dulbecco’s modified eagle’s medium
- EV
Extracellular vesicle
- EGFR
Epidermal growth factor receptor
- FBS
Fetal bovine serum
- FITC
Fluorescein isothiocyanate
- GAG
Glycosaminoglycan
- GBM
Glioblastoma
- hEGF
Human epidermal growth factor
- HSPG
Heparan sulfate proteoglycans
- HUVECs
Human umbilical vein endothelial cells
- RT
Room temperature
- TEM
Transmission electron microscopy
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s11060-013-1235-y) contains supplementary material, which is available to authorized users.
Contributor Information
Nadia A. Atai, Department of Neurology and Program in Neuroscience, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA. Department of Cell Biology and Histology, Academic Medical Center (AMC), University of Amsterdam, 1105 AZ Amsterdam, The Netherlands
Leonora Balaj, Department of Neurology and Program in Neuroscience, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA.
Henk van Veen, Department of Cell Biology and Histology, Academic Medical Center (AMC), University of Amsterdam, 1105 AZ Amsterdam, The Netherlands.
Xandra O. Breakefield, Departments of Neurology and Radiology, Massachusetts General Hospital and Neuroscience Program, Harvard Medical School, Boston, USA
Peter A. Jarzyna, Molecular Imaging Laboratory, MGH/MIT/HMS Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Building 149, 13th Street, Room 6.404A, Charlestown, MA 02129, USA. Harvard Medical School, Boston, MA 02129, USA
Cornelis J. F. Van Noorden, Department of Cell Biology and Histology, Academic Medical Center (AMC), University of Amsterdam, 1105 AZ Amsterdam, The Netherlands
Johan Skog, Exosome Diagnostic, Inc., New York, NY 10032, USA.
Casey A. Maguire, Email: cmaguire@partners.org, Department of Neurology and Program in Neuroscience, Massachusetts General Hospital and Harvard Medical School, Boston, MA 02114, USA
References
- 1.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-Oncol. 2012;14 (suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Nedawi K, Meehan B, Rak J. Microvesicles: messengers and mediators of tumor progression. Cell Cycle. 2009;8(13):2014–2018. doi: 10.4161/cc.8.13.8988. [DOI] [PubMed] [Google Scholar]
- 3.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 4.Chaput N, Taieb J, Andre F, Zitvogel L. The potential of exosomes in immunotherapy. Expert Opin Biol Ther. 2005;5:737–747. doi: 10.1517/14712598.5.6.737. [DOI] [PubMed] [Google Scholar]
- 5.Wieckowski E, Whiteside TL. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36(1–3):247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 6.Clayton A, Mitchell JP, Court J, Mason MD, Tabi Z. Human tumor-derived exosomes selectively impair lymphocyte responses to interleukin-2. Cancer Res. 2007;67(15):7458–7466. doi: 10.1158/0008-5472.CAN-06-3456. [DOI] [PubMed] [Google Scholar]
- 7.Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maguire CA, Balaj L, Sivaraman S, Crommentuijn M, Ericsson M, Mincheva-Nilsson L, et al. Microvesicle-associated AAV vector as a novel gene delivery system. Mol Ther. 2012;20:960–971. doi: 10.1038/mt.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maguire CA, Meijer DH, LeRoy SG, Tierney LA, Broekman ML, Costa FF, et al. Preventing growth of brain tumors by creating a zone of resistance. Mol Ther. 2008;16(10):1695–1702. doi: 10.1038/mt.2008.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skog J, Würdinger T, van Rijn S, Meijer D, Gainche L, Curry WTJ, et al. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. PNAS. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sustar V, Jansa R, Frank M, Hagerstrand H, Krzan M, Iglic A, Kralj-Iglic V. Suppression of membrane microvesiculation–a possible anticoagulant and anti-tumor progression effect of heparin. Blood Cells Mol Dis. 2009;42(3):223–227. doi: 10.1016/j.bcmd.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Nagai A, Sato T, Akimoto N, Ito A, Sumida M. Isolation and identification of histone H3 protein enriched in microvesicles secreted from cultured sebocytes. Endocrinology. 2005;146:2593–2601. doi: 10.1210/en.2004-1478. [DOI] [PubMed] [Google Scholar]
- 14.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 15.Smorenburg SM, Hettiarachchi RJ, Vink R, Büller HR. The effects of unfractionated heparin on survival in patients with malignancy-a systematic review. Thromb Haemost. 1999;82:1600–1604. [PubMed] [Google Scholar]
- 16.Van Noorden CJ, van Sluis GL, Spek CA. Experimental and clinical effects of anticoagulants on cancer progression. Thromb Res. 2010;125(Suppl 2):S77–S79. doi: 10.1016/S0049-3848(10)70019-0. [DOI] [PubMed] [Google Scholar]
- 17.Lebeau B, Chastang C, Brechot JM, Capron F, Dautzenberg B, Delaisements C, et al. Subcutaneous heparin treatment increases survival in small cell lung cancer. “Petites Cellules” Group. Cancer. 1994;74:38–45. doi: 10.1002/1097-0142(19940701)74:1<38::aid-cncr2820740108>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Nitti D, Wils J, Sahmoud T, Curran D, Couvreur ML, Lise M, et al. Final results of a phase III clinical trial on adjuvant intraportal infusion with heparin and 5-fluorouracil (5-FU) in resectable colon cancer (EORTC GITCCG 1983–1987). European Organization for Research and Treatment of Cancer. Gastrointestinal Tract Cancer Cooperative Group. Eur J Cancer. 1997;33:1209–1215. doi: 10.1016/s0959-8049(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 19.Ornstein DL, Zacharski LR. The use of heparin for treating human malignancies. Haemostasis. 1999;S1:48–60. doi: 10.1159/000054112. [DOI] [PubMed] [Google Scholar]
- 20.Gerotziafas GT, Papageorgiou C, Hatmi M, Samama MM, Elalamy I. Clinical studies with anticoagulants to improve survival in cancer patients. Pathophysiol Haemost Thromb. 2008;36:204–211. doi: 10.1159/000175158. [DOI] [PubMed] [Google Scholar]
- 21.Zacharski LR, Ornstein DL. Heparin and cancer. Thromb Haemost. 1998;80:10–23. [PubMed] [Google Scholar]
- 22.Engelberg H. Actions of heparin that may affect the malignant process. Cancer. 1999;85:257–272. doi: 10.1002/(sici)1097-0142(19990115)85:2<257::aid-cncr1>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Hejna M, Raderer M, Zielinski CC. Inhibition of metastases by anticoagulants. J Natl Cancer Inst. 1999;91:22–36. doi: 10.1093/jnci/91.1.22. [DOI] [PubMed] [Google Scholar]
- 24.Hettiarachchi RJ, Smorenburg SM, Ginsberg J, Levine M, Prins MH, Büller HR. Do heparins do more than just treat thrombosis? The influence of heparins on cancer spread. Thromb Haemost. 1999;82:947–952. [PubMed] [Google Scholar]
- 25.Borsig L, Wong R, Feramisco J, Nadeau DR, Varki NM, Varki A. Heparin and cancer revisited: mechanistic connections involving platelets, P-selectin, carcinoma mucins, and tumor metastasis. Proc Natl Acad Sci USA. 2001;98:3352–3357. doi: 10.1073/pnas.061615598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niers TM, Klerk CP, DiNisio M, Van Noorden CJ, Büller HR, Reitsma PH, et al. Mechanisms of heparin induced anti-cancer activity in experimental cancer models. Crit Rev Oncol Hematol. 2007;61:195–207. doi: 10.1016/j.critrevonc.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Nader HB, Chavante SF, dos-Santos EA, Oliveira TW, de-Paiva JF, Jerônimo SM, et al. Heparan sulfates and heparins: similar compounds performing the same functions in vertebrates and invertebrates? Braz J Med Biol Res. 1999;32:529–538. doi: 10.1590/s0100-879x1999000500005. [DOI] [PubMed] [Google Scholar]
- 28.Ginsberg JS. Management of venous thromboembolism. N Engl J Med. 1996;335:1816–1828. doi: 10.1056/NEJM199612123352407. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.