Abstract
A growing body of evidence indicates that G-protein-coupled receptors undergo complex conformational changes upon agonist activation. It is likely that the extracellular region, including the N terminus, undergoes activation-dependent conformational changes. We examined this by generating antibodies to regions within the N terminus of μ-opioid receptors. We find that antibodies to the midportion of the N-terminal tail exhibit enhanced recognition of activated receptors, whereas those to the distal regions do not. The enhanced recognition is abolished upon treatment with agents that block G-protein coupling or deglycosylate the receptor. This suggests that the N-terminal region of μ receptors undergoes conformational changes following receptor activation that can be selectively detected by these region-specific antibodies. We used these antibodies to characterize μ receptor type-specific ligands and find that the antibodies accurately differentiate ligands with varying efficacies. Next, we examined if these antibodies can be used to investigate the extent and duration of activation of endogenous receptors. We find that peripheral morphine administration leads to a time-dependent increase in antibody binding in the striatum and prefrontal cortex with a peak at about 30 min, indicating that these antibodies can be used to probe the spatio-temporal dynamics of native μ receptors. Finally, we show that this strategy of targeting the N-terminal region to generate receptor conformation-specific antisera can be applied to other Gαi-coupled (δ-opioid, CB1 cannabinoid, α2A-adrenergic) as well as Gαs- (β2-adrenergic) and Gαq-coupled (AT1 angiotensin) receptors. Taken together, these studies describe antisera as tools that allow, for the first time, studies probing differential conformation states of G-protein-coupled receptors, which could be used to identify molecules of therapeutic interest.
Family A GPCRs2 play a critical role in normal cell function and are the focus of intense studies and targets for drug development. A tremendous effort has been put toward understanding the mechanism of activation of family A GPCRs at a molecular level. Spin label studies with rhodopsin have shown that exposure to light leads to a movement of helices relative to one another that is important for activation of transducin (1–4). Studies using a variety of techniques suggest that small agonists bind to a pocket formed by the surrounding transmembrane helices, and in addition peptide ligands contact additional determinants in extracellular loops and possibly the N-terminal tail (5–10). Binding of agonists, but not antagonists, leads to the stabilization of the helical bundle into a conformation, which, in turn, leads to the uncovering of molecular determinants at the bottom of the core required for G-protein binding and activation (11). Although a comprehensive mechanism for receptor activation, including the N- and C-terminal regions, is not yet available, accumulating evidence suggests that these regions undergo strong structural perturbations upon receptor activation. Studies with receptors with a large N-terminal tail, such as glycoprotein hormone receptors (12), as well as those with a smaller N-terminal tail, such as opioid and other family A receptors, have provided support for the participation of the N-terminal region in receptor activation (13–15). However, relatively little is known about agonist-mediated change in conformation of the N terminus of native receptors.
Opioid receptors belong to family A GPCRs, and their activation induces systemic responses, such as analgesia, euphoria, and decreased intestinal motility (16–18). These receptors are activated by opioid peptides as well as opiate alkaloids. The alkaloid, morphine, is widely used as an analgesic in the treatment of chronic pain; however, its long term use leads to the development of tolerance and addiction. Therefore, a major research focus has been toward the understanding of the spatio-temporal events that are critical to the development of these side effects. Understanding the mechanisms of receptor activation would allow the design of new drugs that are as effective as morphine in the treatment of chronic pain but with fewer side effects.
Antibodies have been useful in exploring the mechanisms of activation by delineating the domains involved in activity-mediated conformational changes in the case of a variety of signaling proteins and ligand-receptor interactions (19, 20). In the case of GPCRs, although antibodies have been generated to different regions of the receptor, the involvement of the N-terminal region in activity-dependent conformational changes has not been well explored. In a few instances, antibodies to the N terminus appear to discriminate between naive and activated receptors (21, 22). These results suggested that we should be able to target a region of the N terminus for the generation of antibodies that could detect conformational changes in this region following receptor activation. We tested this possibility by generating antibodies against a region in the N terminus of μ-opioid receptors. We find that these antibodies differentially recognize inactive and agonist-activated receptors. Next, we examined if a similar strategy could be used to generate antibodies that detect conformational changes in the N-terminal region a variety of family A GPCRs (differing in the nature of endogenous ligand and G-protein coupling). We find that these antisera can distinguish between inactive and agonist-activated receptors.
Next, we focused on the characterization of μ antibodies because of their high clinical relevance. We show that these antibodies can be used to characterize and screen ligands by whole cell ELISA or flow cytometry. Finally, we show that these antibodies recognize native receptors and that we can quantitate the spatio-temporal dynamics of receptor activation in the brain following peripheral drug administration.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
CHO cells stably expressing FLAG-tagged mouse μ receptors were grown in F-12 medium (23). COS and SKNSH cells were grown in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. COS cells were transfected with FLAG-tagged wild type μ, δ, α2A, CB1, β2, or AT1 receptors using Lipofectamine as per the manufacturer’s protocol (Invitrogen).
Generation of Antibodies
The following peptides (Fig. 1A and supplemental Fig. 1) were used to generate antisera: μ, 14SDPLAPASCSPA25 for SA25 and 5G8 and 52GSHSLCPQTGSPS64 for NT1 and 3D6; δ, 3LVPSARAELQSSPLV17 for LV17; α2A, 15GTEAPGGGTRATPYS29 for GS29; CB1, 27DIQYEDIKGDMASKL41 for DL41; β2, 17SRAPDHDVTQE27 for SE27; AT1, 3SSTEDGIKRI12 for LK12 antibodies. These regions were chosen with the help of ExPASy software so as to exclude regions containing predicted N-glycosylation and phosphorylation sites. The peptides were subjected to a Blast search of the mouse NCBI data base to ensure that they represent unique sequences (<10% overlap). These peptides were synthesized, on a polylysine backbone, as multiple antigenic peptides (MAPs) by Research Genetics (Huntsville, AL). Antisera to μ (SA25 and NT1), δ (LV17), α2A (GS29), or CB1 (DL41) MAPs were generated in rats and to β2 (SE27) or AT1 (LK12) MAPs in rabbits using a standard protocol (24). Monoclonal Abs to μ (5G8 and 3D6) MAPs were generated in mice as described previously (25). These antibodies are highly receptor-specific, since they exhibit low cross-reactivity against other closely related receptors, as examined using a whole cell ELISA (described below) with COS cells expressing the various receptors indicated above. Specificity of the antisera was also examined using an antigen depletion assay, where a 1 mM concentration of the specific MAP or an unrelated MAP (CB1 receptor MAP was used as a nonspecific peptide for SA25, LV17, GS29, SE27, and LK12 antibodies, whereas α2A receptor MAP was used as a non-specific peptide for DL41 antibody) was incubated with 10 μg of Abs for 24 h (4 °C) prior to incubation with cells.
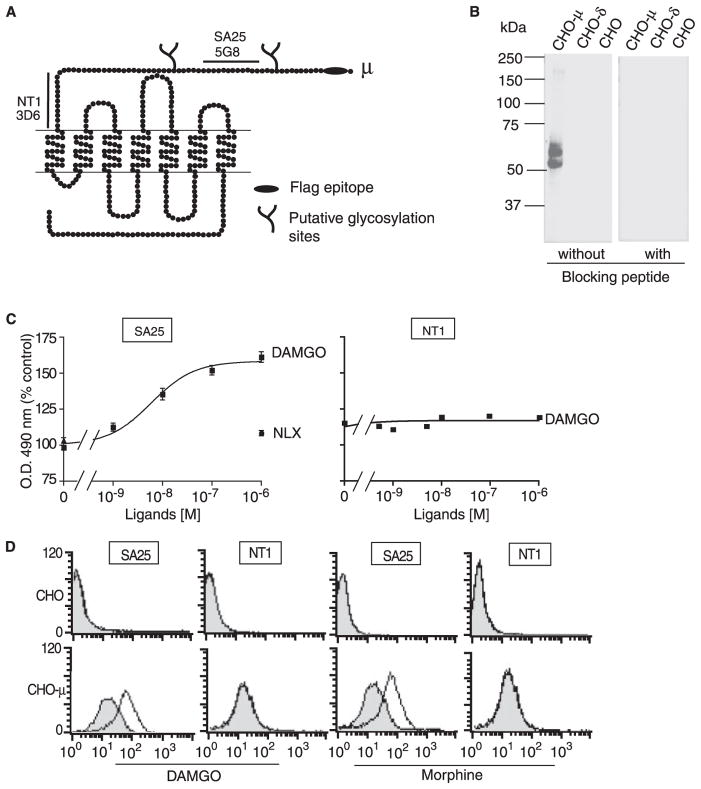
FIGURE 1. Agonist-induced conformational changes in the N terminus of μ receptors.
A, schematic representation of μ-opioid receptors. The FLAG epitope (black ellipse), putative N-linked glycosylation site (branch), and amino acid sequences used for the generation of multiple antigenic peptides (black lines) are indicated. B, Western blot analysis of membranes prepared from CHO cells alone or CHO cells stably expressing μ(CHO-μ) or δ (CHO-δ) receptors with SA25 antiserum preabsorbed without or with the peptide used to generate the antiserum (blocking peptide, 10 mg/ml). C, CHO cells stably expressing μ receptors were treated with the indicated doses of agonists or antagonists and probed with SA25 or NT1 antibodies by ELISA as described under “Experimental Procedures.” Data from vehicle-treated cells were taken as 100%. Treatment of cells with nonselective ligand (1 μM either Delt II, Hu-210, clonidine, isoproterenol, or angiotensin II and probed with SA25 antibody) gave values of 98 ± 5%. Results are the mean ± S.E. (n = 4). D, CHO cells alone (top) or cells stably expressing μ receptors were incubated without (shaded) or with (open) DAMGO or morphine, stained with SA25 or NT1 antibodies followed by Alexa-488-labeled secondary antibodies, and analyzed by flow cytometry. Data are shown from a representative experiment of three.
Western Blot Analysis
Membranes were prepared from CHO cells or from those stably expressing μ- or δ-opioid receptors (26). Membranes (15 μg) were subjected to Western blot analysis using a 1:1000 dilution of SA25 antiserum or SA25 antiserum preabsorbed (10 mg/ml) with the specific peptide used to generate this antiserum and a 1:15,000 dilution of IR Dye 800CW anti-rat IgG. Blots were visualized using the Odyssey Imaging system (Li-Cor Biosciences, Lincoln, NE).
Effect of Ligand Treatment on Receptor Recognition by Abs
CHO cells stably expressing FLAG-tagged μ receptors, COS cells transiently expressing FLAG-tagged μ- or δ-opioid, α2A-or β2-adrenergic, CB1 cannabinoid, or AT1 angiotensin receptors, or SKNSH cells (endogenously expressing these receptors) were plated on poly-L-lysine-treated 24-well plates (2 × 105 cells/well). The next day, cells were washed with phosphate-buffered saline (PBS) and incubated without or with 8–10 doses of ligands in 50 mM Tris-Cl, pH 7.5, or in isotonic HEPES buffer (10 mM HEPES containing 300 mM sucrose and 0.2 mM EDTA, pH 7.4) for 30 min at 37 °C (in the presence of a protease inhibitor mixture; Sigma). There is no significant receptor internalization under these conditions3 due to a lack of K+ and Ca+2 ions, which are required for receptor internalization. Cells were quickly rinsed three times (within 5 min) with cold PBS (washing with 20 μM antagonist in PBS produced similar results) and fixed with ice-cold methanol for 10 min at −20 °C. This treatment was included to help reduce cell loss during multiple washings as determined by protein estimation or recognition by FLAG Ab. We do not observe any significant differences in receptor recognition by SA25 antiserum or FLAG monoclonal Ab in CHO-μ cells that were subjected or not to methanol fixation (0.29 ± 0.01 without and 0.35 ± 0.04 with methanol fixation for SA25 Ab and 0.22 ± 0.01 without and 0.23 ± 0.01 with methanol fixation for FLAG Ab). ELISA was carried out by incubating cells with 3% BSA in PBS for 1 h at 37 °C, followed by overnight incubation at 4 °C with a 1:500 dilution of primary antisera in 1% BSA in PBS. The wells were then washed three times with 1% BSA in PBS (5 min each wash) followed by a 1-h incubation at 37 °C with 1:500 dilution (in 1% BSA in PBS) of secondary antibody coupled to horseradish peroxidase. The wells were washed three times with 1% BSA in PBS (5 min each wash), and color was developed by the addition of the substrate, o-phenylenediamine (5 mg/10 ml in 0.15 M citrate buffer, pH 5, containing 15 μl of H2O2). Absorbance at 490 nm was measured with a Bio-Rad ELISA reader. We do not see a difference in the levels of protein at the end of the assay (197 ± 8 and 197 ± 7 μg/well, respectively, for SA25 and FLAG Ab).
For screening compounds (Table 2), CHO-μ cells (1 × 105) were plated on 96-well Nunc-Immuno™ plates (Nalge Nunc International, Rochester, NY), air-dried at room temperature. The wells were washed with PBS, incubated without or with ligands for 30 min at 37 °C. The extent of receptor recognition by the SA25 Ab was assayed by ELISA as described above.
TABLE 2. Screening of μ ligands using anti-μ (SA25) antibody.
CHO cells expressing μ receptors were treated with 8–10 doses of ligands and probed with μ receptor antibody (SA25) as described under “Experimental Procedures.” Data from untreated cells were taken as 100%. The agonist-mediated increase in [35S]GTPγS binding was measured under the same conditions of antibody binding as described under “Experimental Procedures.” Basal values obtained in the absence of agonist were taken as 100%. Results represent mean ± S.E. (n = 3–5). Statistically significant differences are indicated. ND, not determined. DALDA, H-Tyr-D-Arg-Phe-Lys-NH2; CTAP, D-Phe-c[Cys-Tyr-D-Trp-Arg-Thr-Pen]-Thr-NH2; CTOP, D-Phe-c[Cys-Tyr-D-Trp-Orn-Thr-Pen]-Thr-NH2.
| μ ligands | Ab binding
|
GTPγS binding12
|
||
|---|---|---|---|---|
| EC50 | Emax | EC50 | Emax | |
| M | % control | M | % control | |
| Endo-1 | 2.35 × 10−8 | 230 ± 11a | ND | ND |
| Endo-2 | 8.00 × 10−10 | 196 ± 25a | ND | ND |
| DAMGO | 1.49 × 10−9 | 161 ± 8.2a | 13 × 10−9 | 176 ± 2.1a |
| Methadone | 2.00 × 10−9 | 167 ± 1a | 42 × 10−9 | 169 ± 2.2b |
| Biphalin | 6.85 × 10−8 | 165 ± 5b | ND | ND |
| Fentanyl | 1.64 × 10−8 | 158 ± 5b | 19 × 10−9 | 160 ± 6.8b |
| Super DALDA | 3.06 × 10−9 | 153 ± 1c | ND | ND |
| Met-enk | 1.72 × 10−9 | 151 ± 1c | ND | ND |
| Meperidine | 9.50 × 10−9 | 149 ± 3c | ND | ND |
| β-Endorphin | 7.93 × 10−9 | 145 ± 1c | ND | ND |
| Morphine | 3.45 × 10−8 | 141 ± 3c | 52 × 10−9 | 149 ± 4.6c |
| Naltrexone | >10−6 | 120 ± 1 | ND | ND |
| CTOP | >10−6 | 111 ± 6 | ND | ND |
| CTAP | >10−6 | 108 ± 4 | ND | ND |
| Naloxone | >10−6 | 107 ± 9 | ND | ND |
p < 0.0001 (n = 3–5).
p < 0.001 (n = 3–5).
p < 0.01 (n = 3–5).
Flow Cytometry
Cells (3 × 105/well) were plated onto a 24-well plate. After 48 h, the wells were treated with or without 1 μM [D-Ala2,NMe-Phe4,Gly-ol5]enkephalin (DAMGO) in 50 mM Tris-Cl buffer, pH 7.5, or 1 μM morphine in Dulbecco’s modified Eagle’s medium for 30 min at 37 °C. The extent of cell surface receptor recognition was determined using receptor-specific antiserum and Alexa 488-conjugated goat anti-mouse/anti-rat IgG (1:400 in 50% fetal bovine serum in PBS for 2 h at 4 °C) as described previously (27) using a FACScan flow cytometer (BD Biosciences). Cells remaining in the tubes following fluorescence-activated cell sorting analysis were routinely examined under the microscope to check for cell lysis.
Effect of Peptide:N-glycanase on Antibody Recognition
CHO cells stably expressing μ-opioid receptors (2 × 105) plated on 24-well plates were incubated with or without 1 μM DAMGO in the absence or presence of peptide:N-glycanase F (40 units) for 3 h at 37 °C in 50 mM Tris-Cl buffer, pH 7.5. Cells were quickly washed with ice-cold PBS and fixed with ice-cold methanol for 10 min at −20 °C. The extent of antibody recognition was monitored by ELISA using either SA25, 5G8, NT1, or 3D6 antibodies as described earlier.
Studies with Endogenous Receptors
Membranes (1 μg/well) from C57BL/6 mice brain cortex were coated overnight on a high binding 96-well ELISA plate (Fisher). The next day, cells were washed with PBS and incubated without or with 8–10 doses of ligands in 50 mM Tris-Cl, pH 7.5. Membranes were quickly rinsed three times (within 5 min) with cold PBS; washing with 20 μM antagonist in PBS produced similar results. Wells were treated with ice-cold methanol for 10 min at −20 °C, and the extent of receptor recognition by Abs was assayed by ELISA as described above. Specificity of the antisera in endogenous tissue was determined using the antigen-depleted antisera; these gave a signal that was 10–20% that of sera undepleted or treated with unrelated peptide (supplemental Table 4). To examine the effect of modulators of G-protein activity, cortical membranes (10 μg) were pretreated with a 100 μM concentration of either GTPγS, GPP(NH)p, AlF3, or NaF for 30 min at 37 °C in the presence of a protease inhibitor mixture (Sigma). This was followed by a 30-min treatment with 1 μM DAMGO, and the extent of Ab recognition was assayed by ELISA as described above. The pertussis toxin (50 ng/ml) pre-treatment was carried out overnight at 37 °C; the protease inhibitor mixture was used to protect membrane proteins, including receptors, from degradation. The effect of membrane incubation at 37 °C on receptor integrity was measured by ligand binding in the presence of protease inhibitor mixture using [3H]diprenorphine as described (23). The receptor binding following overnight incubation was 97.2 ± 4.8% of that observed following 30 min of incubation at 37 °C, indicating that under these conditions, no significant loss of receptor binding was observed.
In Vivo Receptor Activation Studies on Brain Sections
Four-month-old male C57BL/6 mice (3–5/group) or μ −/− animals and age-matched sex-matched littermate controls (3–5/group) were injected intraperitoneally with either 10 mg/kg morphine, 10 mg/kg morphine plus 10 mg/kg naloxone, or saline and sacrificed 30 min later or at the indicated times. Brains were dissected and frozen at −80 °C until use. The brains were embedded in M-1 embedding matrix (Thermo Electron Corp., Waltham, MA), and 10-μm serial sections were cut using a MICROM HM 560 CryoStar (Richard-Allan Scientific, Kalamazoo, MI). Areas representing the prefrontal cortex (3.0-2.58 mm; Bregma), striatum (1.5-1 mm; Bregma), or cerebellum (−5.6 to −6.5 mm; Bregma) were collected; at least 10 sections from each animal were probed. Two sections from each region were placed on Fisher Superfrost Plus slides and circled immediately with an ImmEdge PAP pen (Vector Laboratories, Burlingame, CA) to form a waterproof barrier; the resulting wells held ~200 μl. ELISA was performed in these wells using a 1:500 dilution of primary antiserum and 1:500 dilution of secondary antibodies. The reaction product was transferred to a 96-well plate, and absorbance at 490 nm was measured with a Bio-Rad ELISA reader.
RESULTS
Detection of Agonist-induced Conformational Changes in the N Terminus of μ-Opioid Receptors
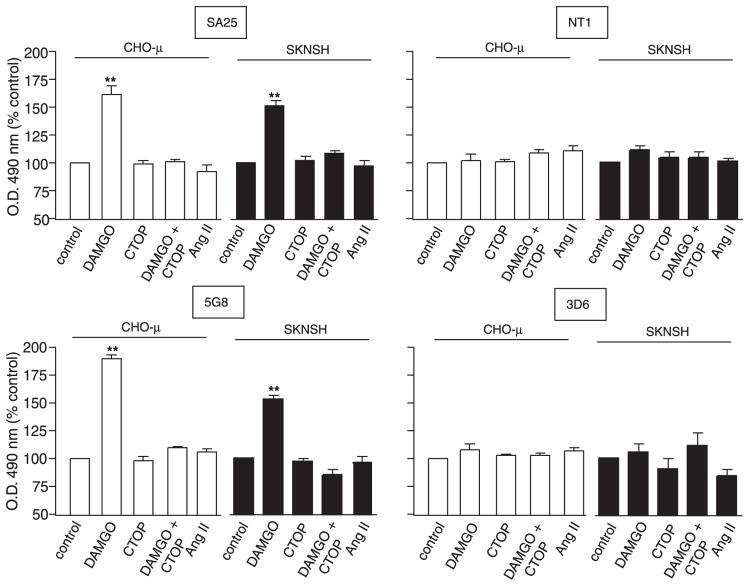
Studies with rhodopsin indicate that the N-terminal region undergoes conformational changes following receptor activation. This suggests that antisera directed against this region of family A GPCRs should be able to distinguish between inactive and activated receptors. In order to see if a similar strategy could be applied to μ-opioid receptors, we targeted the N-terminal domain and generated antibodies to a region proximal (SA25 antibodies) and a region distal to the N terminus (NT1 antibodies) (Fig. 1). These antibodies were characterized in cell lines stably (CHO-μ) or transiently (COS-μ) expressing recombinant μ receptors or expressing native receptors (SKNSH). In all cell lines, these antibodies specifically recognize μ receptors. There was no significant recognition in cells expressing either δ-opioid, CB1 cannabinoid, α2A- or β2-adrenergic, or AT1 angiotensin receptors (Table 1). The specificity of SA25 antisera was confirmed by Western blotting; a specific signal was observed with membranes from CHO cells expressing μ receptors and not from CHO cells alone or from CHO cells expressing δ receptors. Furthermore, when Western blotting was carried out with SA25 antiserum preabsorbed with the specific peptide used to generate the antiserum there was no signal (Fig. 1B), supporting the specificity of the antiserum. This was further supported by studies with membranes from animals lacking μ receptors (supplemental Tables 1 and 2). We examined if these antibodies were able to differentially recognize activated receptors by treating cells with the μ receptor-specific agonist, DAMGO. We find that this leads to a dose-dependent increase in receptor recognition by SA25 but not NT1 antibodies (Fig. 1C). Similar results were obtained upon treatment with morphine (not shown). The increase in recognition by SA25 antibodies does not occur upon treatment with the antagonist, naloxone, or unrelated peptide (Fig. 2). This increase in recognition following receptor activation was further explored by flow cytometry. We find that treatment with either DAMGO or morphine leads to a significant increase in the extent of staining by SA25 but not by NT1 antibodies (Fig. 1D). These results suggest that the region recognized by the SA25 antibodies undergoes conformational changes following receptor activation, whereas the region recognized by the NT1 antibodies does not. To further explore this, we generated monoclonal Abs to peptides directed against these two regions and examined receptor recognition. We find that 5G8, a monoclonal Ab directed against the SA25 epitope, exhibits increased receptor recognition following treatment with agonists, whereas 3D6, a monoclonal Ab directed against the NT1 epitope, does not (Fig. 2). This was seen both in cells expressing recombinant and native receptors (Fig. 2). These results suggest that the increase in recognition by the antibodies is due to a conformational change leading to increased exposure of the region corresponding to the SA25 epitope but not of the region corresponding to the NT1 epitope (proximal to the first transmembrane domain). Taken together, these results suggest that the midregion of the N-terminal tail of μ-opioid receptors undergoes conformational changes following receptor activation.
TABLE 1. Specificity of μ-opioid receptor antibodies.
COS-7 cells were untransfected or transfected with μ- or δ-opioid, CB1 cannabinoid, α2A- or β2-adrenergic, or AT1 angiotensin receptor cDNA and probed with polyclonal antibodies generated to peptides against a region proximal (SA25) or distal to the N terminus (NT1) of μ-opioid receptors (Fig. 1) by ELISA as described under “Experimental Procedures.” A490 nm obtained with untransfected COS-7 cells (0.07 ± 0.01) was subtracted from all groups. OD values obtained with cells expressing μ receptors (0.98 ± 0.01 with SA25 and 0.5 ± 0.01 with NT1) were taken as 100%. Results represent mean ± S.E. (n = 3).
| Cells expressing receptors | Percentage of Ab binding
|
|||||
|---|---|---|---|---|---|---|
| μ-Opioid | δ-Opioid | CB1 cannabinoid | α2A-Adrenergic | β2-Adrenergic | AT1 angiotensin | |
| % | % | % | % | % | % | |
| SA25 | 100 ± 1.0 | 5.1 ± 4.1 | 7.1 ± 1.0 | 4.1 ± 1.0 | 5.1 ± 4.2 | 3.1 ± 2.0 |
| NT1 | 100 ± 2.0 | 2.0 ± 2.0 | 0.0 ± 2.0 | 4.0 ± 2.0 | 6.0 ± 2.0 | 6.0 ± 2.0 |
FIGURE 2. Effect of ligand treatment on μ-opioid receptor recognition by antibodies.
Cells stably (CHO-μ) or endogenously (SKNSH) expressing μ receptors were treated with various ligands (1 μM) and probed with the indicated antibodies as described under “Experimental Procedures.” Angiotensin II was used as a nonselective peptide ligand. Data from untreated “control” were taken as 100%. Statistically significant differences from control are indicated. **, p < 0.001 (n = 3). The OD values with CHO cells not expressing the receptor were <0.005 with or without ligands. The values with CHO-μ cells were 0.4 ± 0.02 for 5G8, 0.38 ± 0.03 for 3D6, 0.42 ± 0.01 for SA25, and 0.28 ± 0.01 for NT1. The values with SKNSH cells were 0.62 ± 0.02 for 5G8, 0.19 ± 0.01 for 3D6, 0.3 ± 0.02 for NT1, and 0.40 ± 0.03 for SA25 antibodies.
Role of G-protein Activity in Ab Recognition
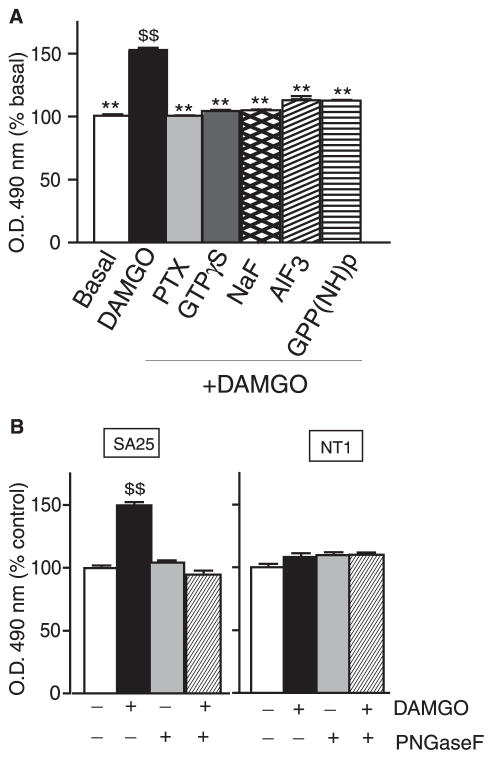
We examined the role of G-protein activity in recognition by SA25 polyclonal or 5G8 monoclonal antibodies by pretreating cortical membranes with agents that uncouple G-proteins (GTPγS, GPP(NH)p, AlF3, NaF, or pertussis toxin) from the receptor. We find that this leads to an impairment of agonist-induced changes in receptor recognition by these antibodies (Fig. 3A), suggesting that the formation of a ternary agonist-receptor G-protein complex by itself is not able to trigger the change in conformation recognized by the antibody. These results suggest that conformational changes induced by postreceptor activation events could be important factors influencing conformational changes at the N-terminal region of μ receptors.
FIGURE 3. Effect of peptide:N-glycanase F treatment and role of G-protein activity.
A, cortical membranes (10 μg) were pretreated with either pertussis toxin (PTX), GTPγS, GPP(NH)p, AlF3, or NaF, followed by treatment with or without 1 μM DAMGO, and probed with a receptor-specific antibody by ELISA as described under “Experimental Procedures.” Data from vehicle-treated cells were taken as 100%. B, CHO cells stably expressing μ receptors were treated with or without 1 μM DAMGO in the absence or presence of peptide: N-glycanase F and probed with μ receptor-specific antibodies by ELISA as described under “Experimental Procedures.” Data from vehicle-treated cells were taken as 100%. Results are the mean ± S.E. (n = 4). **, p < 0.001 versus DAMGO; $$, p < 0.001 versus basal, Dunnett’s test.
Next, we characterized the increased antibody binding by examining the effect of deglycosylation on antibody recognition. We found that treatment with peptide:N-glycanase F abolished the increased recognition following receptor activation by both the SA25 polyclonal and 5G8 monoclonal antibodies (Fig. 3B). This, taken with the fact that peptide:N-glycanase treatment does not affect receptor recognition in the basal state, suggests that receptor glycosylation plays a role in the conformational switch that occurs at the N terminus upon receptor activation. Taken together, these results support the idea that these antibodies recognize changes in conformation at the N terminus following receptor activation.
Screening of Ligands Using Conformation-sensitive Antibodies
Next, we examined if these antibodies can be used as a tool to screen and characterize specific ligands. For this, we set up a simple and rapid screening assay in a 96-well format and probed a series of μ ligands differing in their chemical and pharmacological properties; these included clinically relevant drugs, endogenous peptides, synthetic peptidic and nonpeptidic agonists, and antagonists (Table 2). We found a correlation between the extent of Ab receptor recognition and the reported efficacies of these ligands; morphine, a partial agonist with lower efficacy, exhibits a lower Emax compared with DAMGO, a highly potent full agonist that exhibits a higher Emax (Table 2). In order to further explore this, we carried out dose-response curves and compared the Ab binding properties with the signaling properties. We find a positive correlation between the Emax for Ab binding and that for GTPγS binding (Table 2). These results suggest that our conformation-sensitive Abs can be used to develop rapid and highly sensitive screening assays for the identification of novel small molecule ligands of therapeutic interest.
Detection of Native μ Receptors in the Brain
Next, we examined the extent of recognition of the native receptors using membranes from different brain regions and probing them with SA25 antibodies. The relative levels of Ab binding to various brain regions were measured using a standard curve (generated with cells expressing known levels of μ receptors). Results in Fig. 4 show that there is a good match between the level of antibody binding and the reported level of these receptors; binding is highest in the cortex and lowest in the cerebellum. These results suggest that these antibodies can be used to probe changes in conformation in the N-terminal region following activation of endogenous receptors.
FIGURE 4. Regional distribution of μ receptors examined using SA25 antiserum.

Membranes from various brain regions (~1 μg) were probed with antibodies by ELISA as described under “Experimental Procedures.” In order to quantitate the level of receptors in brain regions by ELISA, a 3-fold serial dilution of CHO cells expressing μ receptors by adding CHO cells not expressing the receptor was carried out (the final cell number was kept constant at 3 × 105 cells/well). Receptor number at each dilution was quantitated by ligand binding as described (27). A parallel set of wells were subjected to ELISA using the SA25 antibody. Results are the mean ± S.E. (n = 3). Triangles, cerebellum; crosses, pons; stars, hippocampus; circles, cortex.
Ab Recognition of Native Receptors in the Brain following Drug Administration
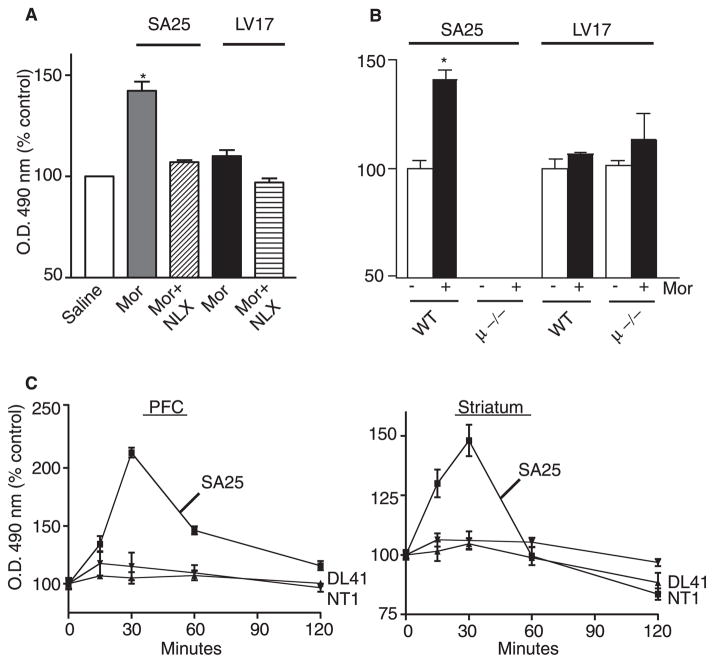
Next we used these antibodies to study the spatio-temporal dynamics of native μ receptors following drug administration. For this, drugs were peripherally administered, and the change in antibody recognition was monitored after 30 min. We find a significant increase in recognition by SA25 antibodies in the brain (striatum) 30 min after peripheral morphine administration (Fig. 5A). This increase could be blocked by co-administration of naloxone with morphine (Fig. 5A). The effect of morphine is selective for μ receptors, since morphine treatment does not lead to significant changes in the recognition of δ receptors by LV17 antibody (Fig. 5A). In order to ensure the specificity of receptor recognition, we used mice lacking μ receptors (μ −/−) and found that there is a significantly decreased recognition that is not affected by morphine administration (Fig. 5B). Under these conditions, the recognition of δ receptors by δ receptor-specific antibodies is not altered (Fig. 5B). Thus, our data show that changes in conformation of native receptors following drug administration can be monitored by N-terminal antibodies.
FIGURE 5. Detection of μ receptors in the brain.
A, animals were injected intraperitoneally with either 10 mg/kg morphine (Mor), 10 mg/kg morphine plus 10 mg/kg naloxone (MOR + NLX), or saline and sacrificed 30 min later. Brains were sectioned, and striatal slices were probed with anti-μ (SA25) or anti-δ (LV17) antibodies. B, mice lacking μ receptors (μ −/−) or wild type (WT) mice were administered 10 mg/kg morphine and sacrificed 30 min later. Brains were sectioned, and antibody recognition on striatal slices was monitored by ELISA using anti-μ (SA25 and NT1) or anti-δ (LV17) antibodies as described under “Experimental Procedures.” Data obtained with saline-injected wild type controls were taken as 100%. C, mice were injected with saline or 10 mg/kg morphine (intraperitoneally) and sacrificed at the indicated times, and brains collected and sectioned as described under “Experimental Procedures.” Prefrontal cortex (PFC) and striatal sections were probed with anti-μ (SA25 and NT1) or anti-CB1 (DL41) receptor antibodies. Data obtained with saline-injected controls were taken as 100%. Data represent mean ± S.E. (n = 3 mice/group). *, p < 0.01, Dunnett’s test.
In order to study the time course of drug-induced change in receptor conformation, we collected brains at various times following peripheral administration of morphine and evaluated slices from prefrontal cortex and striatum for the change in antiserum recognition. We find that the Ab recognition was maximal at 30 min and returned to basal levels by 60 min (Fig. 5C). In contrast, recognition by the NT1 antibody or the DL41 antibody (that recognizes CB1 receptors) was not altered at any time point following morphine administration. Cerebellar slices did not show significant recognition by the SA25 antiserum, and morphine administration had no effect at any of the times tested (not shown). These results are consistent with the notion that morphine administered intraperitoneally reaches the brain and activates the receptor within 30 min; the receptor returns to the basal state of activity by 60 min. This correlates well with the reported time course of behavioral effects of peripherally administered morphine (28).
Detection of Agonist-induced Conformational Changes in the N Terminus of Family A GPCRs
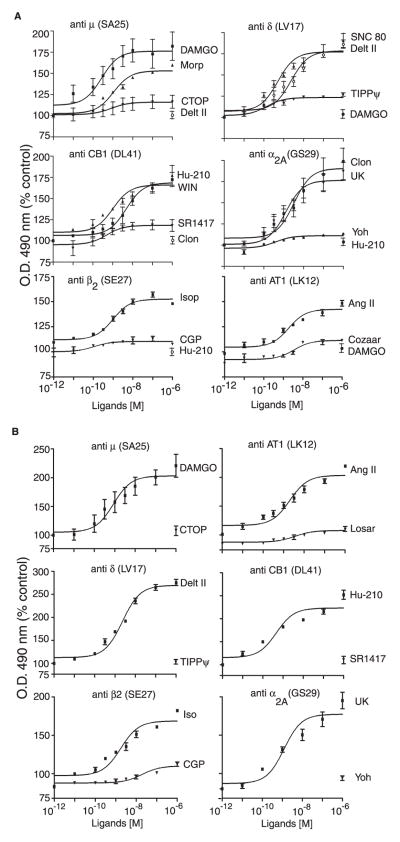
We next examined if the strategy of targeting the N terminus can be used to generate similar conformation-sensitive antibodies to other GPCR family members. We chose receptors that couple to different G-proteins and are activated by different types of ligands (i.e. Gαi-coupled δ-opioid (peptide), CB1 cannabinoid (lipid), α2A-adrenergic (catecholamine), and Gαs-coupled β2 and Gαq-coupled AT1 angiotensin receptors). Regions within the midportion of the N-terminal domain were chosen for the generation of these antibodies (supplemental Fig. 1). We find that these antibodies are highly receptor-specific, since they exhibit low cross-reactivity against other closely related receptors (supplemental Tables 3 and 4). As seen in the case of μ receptor antibodies, treatment with agonists but not antagonists leads to significant increases in receptor recognition in all cases (Fig. 6 and supplemental Table 5). Next, we examined the extent of recognition of the native receptors by these Abs using either a cell line, SK-N-SH (a human neuroblastoma cell line that endogenously expresses these receptors) (Fig. 6A), or mouse cortical membranes (Fig. 6B). Treatment with prototypic agonists leads to a dose-dependent increase in recognition by the antiserum, whereas treatment with antagonists does not (Fig. 6, A and B). This increase is not due to conformational changes indirectly caused by activation of other signal transduction pathways, since activation of an unrelated receptor does not influence Ab recognition of the receptor being probed (Fig. 6). These results suggest that a common mechanism involving structural perturbations of this region following receptor activation leads to increased Ab accessibility to the exposed epitope, although the N-terminal region of these GPCRs is quite diverse.
FIGURE 6. Dose-response curves for recognition of native μ, δ, CB1, α2A, β2, and AT1 receptors.
SKNSH cells (A) or membranes (1 μg) from mouse cortices (B) were treated with the indicated doses of each ligand and probed with antisera raised against μ- or δ-opioid, CB1 cannabinoid, α2A- or β2-adrenergic, and AT1 angiotensin receptors by ELISA as described under “Experimental Procedures.” Data from untreated cells were taken as 100%. Results are the mean ± S.E. (n = 4–5). CTOP, D-Phe-c[Cys-Tyr-D-Trp-Orn-Thr-Pen]-Thr-NH2.
DISCUSSION
GPCRs undergo conformational changes following receptor activation. Although the involvement of the transmembrane regions in conformational changes following the binding of the agonist to the receptor has been extensively documented (1–10), not much is known about the involvement of the N-terminal region. Antibodies have been useful tools in exploring the domains involved in activity-mediated conformational changes of signaling proteins, including receptors (19, 20, 29–36). For example, a monoclonal antibody to the N-terminal region of rhodopsin exhibited a higher degree of recognition for bleached (activated) receptors than for unbleached (inactive) receptors even after detergent treatment. This indicates that photoactivation of rhodopsin induces a conformational change at the N terminus that exposes an epitope that is recognized by the monoclonal antibody (22). In this study, we generated antibodies to distinct regions of the N terminus of μ-opioid receptors to show that a region within the N terminus undergoes significant local movement following receptor activation. We find that this could be a general mechanism shared by other members of family A, since we observe that irrespective of the nature of the ligand or G-protein selectivity, receptor activation leads to the exposure of the epitope recognized by antibodies targeted to the midportion of the N terminus.
The role of the N-terminal region in ligand binding and receptor activation has been mainly explored in receptors with long N-terminal regions. For example, for glycoprotein hormone receptors, the long N-terminal tail constitutes the primary high affinity and selective binding site for receptor agonists (12). In the case of receptors of family C, the very large extracellular N terminus is organized into a domain called the Venus flytrap module that contains the ligand binding pocket (37, 38). Interestingly, the smaller N-terminal tail of family A GPCRs, such as opioid receptors as well as other peptide or amine receptors, has also been proposed to participate in receptor activation (39). In the case of δ-opioid receptors, a random mutagenesis study identified 5 amino acids in the N-terminal region that enhanced the spontaneous activity of the receptor (15). In this study, each mutation substantially modified the chemical nature of the amino acid side chain, suggesting that the N terminus of δ receptors is folded as a domain whose structure and spatial orientation affects receptor function (15). This is also suggested by the structure of rhodopsin, in which the N terminus is folded as a β-sheet over the helical bundle covering it like a lid (40). Although a comprehensive molecular mechanism for receptor activation that includes the extracellular loops and the N- and C-terminal tails is not yet available, the accumulating evidence to date is consistent with the notion that these regions of the receptor undergo substantial structural perturbations upon activation.
An observation in this study has been that increased recognition of the receptor by antibodies targeted to the midportion of the N-terminal region of μ-opioid receptors remains for a prolonged period (ever after the removal of the agonist). In addition, the formation of the ternary complex does not appear to be sufficient, since the recognition persists for a prolonged period of time (well beyond the formation of the ternary complex). It can be postulated that the antibody recognizes the activation-mediated post-translationally modified receptor. It should be pointed out that our assays (in whole cells and membranes) were carried out under conditions that do not allow receptor internalization (and dephosphorylation/resensitization). Hence, the changes in receptor conformation would persist for a long time, as seen in our studies with cells, membranes, and animals. Taken together, these results are consistent with the notion that the SA25 antibody recognizes changes (in the N terminus) that are induced by the activation-mediated long lasting changes to the receptor. We are currently exploring these possibilities.
Although a number of studies have investigated the activation state of heterogeneously expressed opioid receptors, a major research focus has been to identify brain regions and molecular partners playing critical roles in the development of side effects, such as tolerance to and physical dependence on opiates. However, no definite model has emerged that could be used to design a new category of drugs as powerful as morphine but with less abuse potential. This is partly due to difficulties in distinguishing the brain regions targeted by a drug, which depend on the route of administration, dose of the drug, and its bioavailability. For example, morphine has been shown to induce the activation of mitogen-activated protein kinase in a set of cells quite distinct from those that express μ receptors (41). Studies have also shown that the pharmacological properties of a ligand in vitro can be different from those seen in vivo. For example, [D-Pen2,D-Pen5]enkephalin and deltorphin II, two highly δ-selective peptide agonists, are thought to induce analgesia through μ receptors (42). The lack of suitable reagents, thus far, has not allowed the direct evaluation of the extent of activation of receptors of interest. Attempts have been made to probe this using indirect markers (mitogen-activated protein kinase and GTPγS) or receptor knock-out mice. Apart from the radiolabeled GTPγS binding performed on slices (43), few techniques are available that allow the investigation of the spatio-temporal dynamics of receptor activation. This emphasizes the dire need for reliable tools for identification of brain regions where GPCRs are activated at the cellular level. Our antibodies represent a useful and direct approach compared with other time-consuming and labor-intensive techniques. This approach seems to be applicable to many family A GPCRs and opens the way to examine the localization of active receptors as well as the extent of modulation of receptor activity by cross-talk between receptors.
The results of this study not only demonstrate the structural mobility exhibited by the small N-terminal region of distinct types of family A GPCRs but also provide a new and powerful technique to examine the duration and extent of activation of endogenous receptors as well as to screen for drugs that are allosteric modulators of family A GPCRs, which would be of potential therapeutic value.
Supplementary Material
Acknowledgments
We thank Drs. Raphael Rozenfeld for critical reading of the manuscript and Toni Shippenberg for providing the brains from μ −/− mice and littermate controls, Phil Portoghese for biphalin, and Peter Schiller for Super DALDA (H-Tyr-D-Arg-Phe-Lys-NH2).
Footnotes
This work was supported by National Institutes of Health Grants DA08863 and DA19521 (to L. A. D.) and by São Paulo State Research Foundation Grants 04/04933-2 (to E. S. F.) and 04/14258-0 (to A. S. H.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1 and Tables 1–5.
The abbreviations used are: GPCR, G-protein-coupled receptor; Ab, antibody; BSA, bovine serum albumin; DAMGO, [D-Ala2,NMe-Phe4,Gly-ol5]enkephalin; ELISA, enzyme-linked immunosorbent assay; MAP, multiple antigenic peptide; PBS, phosphate-buffered saline; CHO, Chinese hamster ovary; GTPγS, guanosine 5′-3-O-(thio)triphosphate; GPP(NH)p, guanosine 5′-(β,γ-imido)triphosphate.
A. Gupta, I. Gomes, and L. A. Devi, unpublished observations.
References
- 1.Altenbach C, Yang K, Farrens DL, Farahbakhsh ZT, Khorana HG, Hubbell WL. Biochemistry. 1996;35:12470–12478. doi: 10.1021/bi960849l. [DOI] [PubMed] [Google Scholar]
- 2.Altenbach C, Klein-Seetharaman J, Hwa J, Khorana HG, Hubbell WL. Biochemistry. 1999;38:7945–7949. doi: 10.1021/bi990014l. [DOI] [PubMed] [Google Scholar]
- 3.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 4.Yang K, Farrens DL, Altenbach C, Farahbakhsh ZT, Hubbell WL, Khorana HG. Biochemistry. 1996;35:14040–14046. doi: 10.1021/bi962113u. [DOI] [PubMed] [Google Scholar]
- 5.Fadhil I, Schmidt R, Walpole C, Carpenter KA. J Biol Chem. 2004;279:21069–21077. doi: 10.1074/jbc.M311468200. [DOI] [PubMed] [Google Scholar]
- 6.Lu ZL, Saldanha JW, Hulme EC. Trends Pharmacol Sci. 2002;3:140–146. doi: 10.1016/S0165-6147(00)01973-8. [DOI] [PubMed] [Google Scholar]
- 7.Xu W, Li J, Chen C, Huang P, Weinstein H, Javitch JA, Shi L, de Riel JK, Liu-Chen LY. Biochemistry. 2001;40:8018–8029. doi: 10.1021/bi002490d. [DOI] [PubMed] [Google Scholar]
- 8.Mouledous L, Topham CM, Mazarguil H, Meunier JC. J Biol Chem. 2000;275:29268–29274. doi: 10.1074/jbc.M004971200. [DOI] [PubMed] [Google Scholar]
- 9.Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., III Proc Natl Acad Sci U S A. 2002;99:12622–12627. doi: 10.1073/pnas.122357199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer E, Maigret B, Escrieut C, Pradayrol L, Fourmy D. Trends Pharmacol Sci. 2003;1:36–40. doi: 10.1016/s0165-6147(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 11.Gether U. Endocr Rev. 2000;21:90–113. doi: 10.1210/edrv.21.1.0390. [DOI] [PubMed] [Google Scholar]
- 12.Vassart G, Pardo L, Costagliola S. Trends Biochem Sci. 2004;29:119–126. doi: 10.1016/j.tibs.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Lecat S, Bucher B, Mely Y, Galzi JL. J Biol Chem. 2002;277:42034–42048. doi: 10.1074/jbc.M203606200. [DOI] [PubMed] [Google Scholar]
- 14.Levin MC, Marullo S, Muntaner O, Andersson B, Magnusson Y. J Biol Chem. 2002;277:30429–30435. doi: 10.1074/jbc.M200681200. [DOI] [PubMed] [Google Scholar]
- 15.Decaillot FM, Befort K, Filliol D, Yue S, Walker P, Kieffer BL. Nat Struct Biol. 2003;10:629–636. doi: 10.1038/nsb950. [DOI] [PubMed] [Google Scholar]
- 16.Dhawan BN, Cesselin F, Raghubir R, Reisine T, Bradley PB, Portoghese PS, Hamon M. Pharmacol Rev. 1996;48:567–592. [PubMed] [Google Scholar]
- 17.Fang F, Proudfit HK. Brain Res. 1996;722:95–108. doi: 10.1016/0006-8993(96)00198-9. [DOI] [PubMed] [Google Scholar]
- 18.Sora I, Takahashi N, Funada M, Ujike H, Revay RS, Donovan DM, Miner LL, Uhl GR. Proc Natl Acad Sci U S A. 1997;94:1544–1549. doi: 10.1073/pnas.94.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt A, MacColl R, Lindau-Shepard B, Buckler DR, Dias JA. J Biol Chem. 2001;276:23373–23381. doi: 10.1074/jbc.M100057200. [DOI] [PubMed] [Google Scholar]
- 20.Vignal E, Blangy A, Martin M, Gauthier-Rouviere C, Fort P. Mol Cell Biol. 2001;21:8022–8034. doi: 10.1128/MCB.21.23.8022-8034.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cha K, Reeves PJ, Khorana HG. Proc Natl Acad Sci U S A. 2000;97:3016–3021. doi: 10.1073/pnas.97.7.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takao M, Iwasa T, Yamamoto H, Takeuchi T, Tokunaga F. Zoolog Sci. 2002;19:651–659. doi: 10.2108/zsj.19.651. [DOI] [PubMed] [Google Scholar]
- 23.Gomes I, Gupta A, Filipovska J, Szeto HH, Pintar JE, Devi LA. Proc Natl Acad Sci U S A. 2004;101:5135–5139. doi: 10.1073/pnas.0307601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujisawa Y, Furukawa Y, Ohta S, Ellis TA, Dembrow NC, Li L, Floyd PD, Sweedler JV, Minakata H, Nakamaru K, Morishita F, Matsushima O, Weiss KR, Vilim FS. J Neurosci. 1999;19:9618–9634. doi: 10.1523/JNEUROSCI.19-21-09618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes I, Gupta A, Singh SP, Sharma SK. FEBS Lett. 1999;456:126–130. doi: 10.1016/s0014-5793(99)00878-9. [DOI] [PubMed] [Google Scholar]
- 26.Gomes I, Filipovska J, Jordan BA, Devi LA. Methods. 2003;27:358–365. doi: 10.1016/s1046-2023(02)00094-4. [DOI] [PubMed] [Google Scholar]
- 27.Trapaidze N, Gomes I, Bansinath M, Devi LA. DNA Cell Biol. 2000;19:195–204. doi: 10.1089/104454900314465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gouarderes C, Sutak M, Zajac JM, Jhamandas K. Eur J Pharmacol. 2000;406:391–401. doi: 10.1016/s0014-2999(00)00716-0. [DOI] [PubMed] [Google Scholar]
- 29.Lu J, Chuntharapai A, Beck J, Bass S, Ow A, De Vos AM, Gibbs V, Kim KJ. J Immunol. 1998;160:1782–1788. [PubMed] [Google Scholar]
- 30.Morgan EL, Ember JA, Sanderson SD, Scholz W, Buchner R, Ye RD, Hugli TE. J Immunol. 1993;151:377–388. [PubMed] [Google Scholar]
- 31.Garzon J, Castro MA, Juarros JL, Sanchez-Blazquez P. Life Sci. 1994;54:PL191–PL196. doi: 10.1016/0024-3205(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 32.Lebesgue D, Wallukat G, Mijares A, Granier C, Argibay J, Hoebeke J. Eur J Pharmacol. 1998;348:123–133. doi: 10.1016/s0014-2999(98)00136-8. [DOI] [PubMed] [Google Scholar]
- 33.Salle L, Eftekhari P, Aupart M, Cosnay P, Hoebeke J, Argibay JA. J Mol Cell Cardiol. 2001;33:405–417. doi: 10.1006/jmcc.2000.1312. [DOI] [PubMed] [Google Scholar]
- 34.Blanpain C, Vanderwinden JM, Cihak J, Wittamer V, Le Poul E, Issafras H, Stangassinger M, Vassart G, Marullo S, Schlndorff D, Parmentier M, Mack M. Mol Biol Cell. 2002;13:723–737. doi: 10.1091/mbc.01-03-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bozon V, Di Scala E, Eftekhari P, Hoebeke J, Lezoualc’h F, Fischmeister R, Argibay J. Receptors Channels. 2002;8:113–121. [PubMed] [Google Scholar]
- 36.Peter JC, Eftekhari P, Billiald P, Wallukat G, Hoebeke J. J Biol Chem. 2003;278:36740–36747. doi: 10.1074/jbc.M306877200. [DOI] [PubMed] [Google Scholar]
- 37.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 38.Silve C, Petrel C, Leroy C, Bruel H, Mallet E, Rognan D, Ruat M. J Biol Chem. 2005;280:37917–37923. doi: 10.1074/jbc.M506263200. [DOI] [PubMed] [Google Scholar]
- 39.Meng F, Hoversten MT, Thompson RC, Taylor L, Watson SJ, Akil H. J Biol Chem. 1995;270:12730–12736. doi: 10.1074/jbc.270.21.12730. [DOI] [PubMed] [Google Scholar]
- 40.Palczewski K, Kumasaka T, Hori T, Behne CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 41.Eitan S, Bryant CD, Saliminejad N, Yang YC, Vojdani E, Keith D, Jr, Polakiewicz R, Evans CJ. J Neurosci. 2003;23:8360–8369. doi: 10.1523/JNEUROSCI.23-23-08360.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scherrer G, Befort K, Contet C, Becker J, Matifas A, Kieffer BL. Eur J Neurosci. 2004;19:2239–2248. doi: 10.1111/j.0953-816X.2004.03339.x. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Raehal KM, Lin ET, Lowery JJ, Kieffer BL, Bilsky EJ, Sadee W. J Pharmacol Exp Ther. 2004;308:512–520. doi: 10.1124/jpet.103.054049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.