Abstract
Some introduced populations thrive and evolve despite the presumed loss of diversity at introduction. We aimed to quantify the amount of genetic diversity retained at introduction in species that have shown evidence of adaptation to their introduced environments. Samples were taken from native and introduced ranges of Arctotheca populifolia and Petrorhagia nanteuilii. Using microsatellite data, we identified the source for each introduction, estimated genetic diversity in native and introduced populations, and calculated the amount of diversity retained in introduced populations. These values were compared to those from a literature review of diversity in native, confamilial populations and to estimates of genetic diversity retained at introduction. Gene diversity in the native range of both species was significantly lower than for confamilials. We found that, on average, introduced populations showing evidence of adaptation to their new environments retained 81% of the genetic diversity from the native range. Introduced populations of P. nanteuilii had higher genetic diversity than found in the native source populations, whereas introduced populations of A. populifolia retained only 14% of its native diversity in one introduction and 1% in another. Our literature review has shown that most introductions demonstrating adaptive ability have lost diversity upon introduction. The two species studied here had exceptionally low native range genetic diversity. Further, the two introductions of A. populifolia represent the largest percentage loss of genetic diversity in a species showing evidence of substantial morphological change in the introduced range. While high genetic diversity may increase the likelihood of invasion success, the species examined here adapted to their new environments with very little neutral genetic diversity. This finding suggests that even introductions founded by small numbers of individuals have the potential to become invasive.
Keywords: Asteraceae, biological invasions, caryophyllaceae, genetic diversity, microsatellite, rapid evolution
Introduction
Biological invasions present one of the greatest environmental challenges of our time, yet the drivers of successful invasion remain poorly understood. The concept that genetic diversity in the founding population is positively related to the probability of invasion success is one of the oldest hypotheses in invasion biology (e.g., Mayr 1965) and continues to be supported by recent research (Crawford and Whitney 2010; Jones and Gomulkiewicz 2012). However, the debate over the importance of genetic diversity to invasion success continues (Sakai et al. 2001; Kolbe et al. 2004; Roman and Darling 2007; Hufbauer 2008). This hypothesis presents a paradox because genetic bottlenecks are expected to occur at introduction, reducing the potential for introduced populations to adapt to novel environments (Allendorf and Lundquist 2003; Allendorf and Luikart 2007), but despite this, many introduced populations thrive. In some cases, this has been explained by high levels of propagule pressure through multiple introduction events, resulting in introduced populations having greater genetic diversity than is found in the native range (Kolbe et al. 2004; Genton et al. 2005). However, successful invasions are not always accompanied by high genetic diversity and sometimes are depauperate in neutral genetic variation (Ren et al. 2005; Mergeay et al. 2006; Zimmermann et al. 2010).
Many plant and animal populations expanding into novel environments not only thrive but also exhibit rapid evolutionary changes in crucial traits such as dispersal ability, reproductive output, phenotypic plasticity, and size (Blossey and Nötzold 1995; Cody and Overton 1996; Siemann and Rogers 2001; Bossdorf et al. 2005; Phillips et al. 2006; Richards et al. 2006; Cheptou et al. 2008; Ridley and Ellstrand 2009; Buswell et al. 2011). This empirical evidence is supported by simulations, demonstrating that evolution may move at a faster rate when an organism's environment varies (Kashtan et al. 2007) and invasive populations often experience extreme environmental shifts. Further, it appears that rapid evolution in invasive species may be quite common. For example, Buswell et al. (2011) studied herbarium specimens of 23 plant species introduced to Australia and sampled repeatedly across the past ∼150 years to identify evidence of significant morphological change. They concluded that changes had occurred in 70% of these species following their introduction and that this was most likely the result of rapid evolution. Evidence of rapid evolution in novel environments supports the idea that genetic diversity is important to the success of introduced populations because adaptations following introduction are more likely to be derived from standing genetic variation rather than mutation (Barrett and Schluter 2008). Nevertheless, several studies have demonstrated rapid evolution in the presence of low genetic diversity in introduced ranges (Dlugosch and Parker 2008b; Harris et al. 2012), suggesting that the level of standing genetic diversity required for adaptation may, in fact, be quite low.
Quantitative genetic theory predicts that the extent of adaptive genetic change due to pre-existing genetic variation in the initial population in a new environment, as well as adaptation due to new mutations arising in the new environment, will be an increasing function of selection, genetic diversity, genetically effective population size, and number of generations (Robertson 1960; Weber 2004). The extent of adaptation expected for both pre-existing diversity and novel mutation can be predicted (see Appendix S1 for details), and these predictions are supported by empirical evidence (Frankham 1980b, 1983; Weber 2004; Frankham et al. 2010). As these predictions assume that genetic variation is neutral, genetic adaptation should increase with levels of neutral genetic variation, other factors being equal (Frankham et al. 1999, 2010).
In this study, we examine the relationship between neutral genetic diversity and rapid evolution in introduced species, using two species that have exhibited significant morphological change since their introduction to Australia. First, unlike many other studies of rapid evolution after introduction, we aimed to determine the exact source population(s) for the introductions for both species. This information is important for the accurate comparison of the genetic characteristics of the introduced populations from the actual source populations in their native ranges. Second, we characterized genetic diversity in the native and introduced ranges of both species. Finally, we surveyed the literature to determine (1) whether the levels of genetic diversity we found in the native range of both species were typical of other species within those families and (2) whether our species retained similar levels of genetic diversity at introduction compared with other introduced species showing evidence of rapid evolution in their introduced environments. We expected that genetic diversity in the species studied here would not be low relative to their families because they had demonstrated the ability to undergo morphological change in their introduced environments. We also expected the change in diversity between the native and introduced populations in our study species to be similar to that of other introduced species showing evidence of rapid evolution.
Methods
Study species
We aimed to select species that had shown potential for rapid evolution through postintroduction morphological change. We chose species with restricted native and introduced ranges so that we could comprehensively sample across their distributions. Annual or short-lived perennial species with sexual reproduction were selected, because these species have had more generations since introduction, increasing the opportunity for evolution to occur in the introduced range. We avoided selecting crop and pasture species that were likely to have been introduced many times.
Arctotheca populifolia (Fig. 1), chosen based on the findings of rapid morphological change in introduced populations (Buswell et al. 2011), is in the Asteraceae and is a perennial, herbaceous succulent native to South Africa and introduced to Australia (Harden 1992). The first records of this species in Australia occurred in the 1930s on both the east and west coasts (AVH Database, 2012). The Australian distribution encompasses coastal environments from Geraldton (Western Australia) to northern New South Wales (Heyligers 1998). The Global Compendium of Weeds lists A. populifolia as an agricultural and environmental weed (GCW database 2012).
Figure 1.

Australian sample of Arctotheca populifolia (photograph by C. Brandenburger).
The second study species was Petrorhagia nanteuilii, which is an annual, herbaceous plant in the Caryophyllaceae. It is native to western Europe and western North Africa (Ball and Heywood 1964) and introduced to Asia, Australia, North America, South America, and Macaronesia (GRIN database 2012). This species was first recorded in Australia in 1882, and, currently, the Australian distribution is restricted to the southeast, ranging from Brisbane to Adelaide (AVH Database, 2012). Petrorhagia nanteuilii is also listed in the Global Compendium of Weeds as an agricultural and environmental weed (GCW database 2012).
Similar to A. populifolia, P. nanteuilii also showed evidence of morphological change over time since introduction (see Results, below). This was determined using herbarium specimens following methods described by Buswell et al. (2011). We measured height on all available specimens at the National Herbarium of Victoria (MEL) at the Royal Botanic Gardens, Melbourne. This gave data for 184 plants from 56 herbarium sheets ranging in collection date from 1882 to 1998. No leaf traits were measured because leaves do not press well in this species. All plants had grown in the range of the introduction, in Victoria and New South Wales in Australia. We ran a general linear model including region and year as predictors and log10-transformed height as a dependent variable. The term for region was included to prevent the possibility that a population expansion along an environmental gradient would be mistaken for adaption to the native range across time (Buswell et al. 2011). To do this, we recorded the region of origin for each sample. Because most regions were represented by relatively few specimens, we pooled bioregions to construct four broad climate regions: (1) humid coast and hinterlands (including East Gippsland, Victoria, and the New South Wales Central Coast and South Coast), (2) humid highlands (including Eastern Highlands, the Snowfields, and the Southern Tablelands), (3) subhumid slopes (including the Victorian Midlands and Riverina, and the New South Wales South West Plains, South West Slopes, and North West Slopes), and (4) semi-Mediterranean (including the Victorian Volcanic Plain, the Grampians and Wannon). In order to acknowledge the nonindependence of plants from the same herbarium sheet, we weighted individuals according to the number measured on the herbarium sheet such that the weights for all the plants on each sheet sum to one. For example, a single plant on a sheet received a weight of one, while two individuals on the same sheet each received a weight of 0.5. Analyses were performed in JMP, version 5 (SAS Institute, Cary, NC).
For both species, we also measured plant height over time in the native range, using the methods described above. This was done in order to determine whether any changes identified in the introduced range were concurrently occurring in the native range, perhaps as a result of global climate change. For these data, region was not included as a term due to the small number of samples available for each region. In total, 52 herbarium samples from 28 sheets were measured from the native range of A. populifolia and 86 samples from 26 sheets for native range P. nanteuilii.
Genetic sampling
We sampled leaves from 348 A. populifolia plants from 10 sites covering the native range (N = 188; Fig. 2A) and seven sites across the introduced range in Australia (N = 160; Fig. 2B, triangles). For P. nanteuilii, we sampled a total of 345 plants, including those from 12 sites in the native range (N = 282; Fig. 2C) and two sites across the introduced range in Australia (Fig. 2B, squares). Two attempts were made to sample this species in the vicinity of Adelaide, South Australia, at the westernmost reported extreme of the Australian distribution, and in the vicinity of Sydney where P. nanteuilii has also been reported; however, on all occasions, none were present. Leaves were placed in vials containing a solution of 40% sodium chloride, 4% sodium ascorbate, 4% silica, and 3% cetyltrimethylammonium bromide (Thompson 2002) and stored at 4°C. To prepare samples for extraction, leaves were removed from the preservative, washed in Milli-Q water, patted dry, and frozen at −70°C prior to freeze drying. Freeze-dried samples were crushed and DNA was extracted using a NucleoSpin 96 Extraction II Kit (Macherey-Nagel, Düren, Germany).
Figure 2.
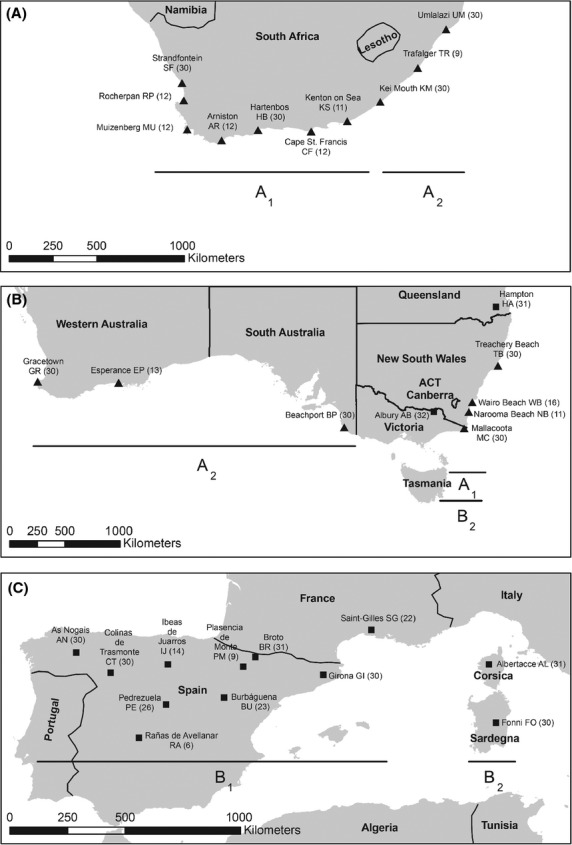
Sampled areas with place name abbreviations and number of individuals sampled in parentheses. (A) native range samples of Arctotheca populifolia. (B) Australian introduced range samples of A. populifolia (triangles) and Petrorhagia nanteuilii (squares). (C) native range samples of P. nanteuilii. Genetic groups are indicated by bars labeled with group name (i.e., A1; see Results). Note that group assignment of P. nanteuilii samples PM and GI is ambiguous (see Figs 3 and 4).
Microsatellites were developed using next-generation sequencing on the GS-FLX 454 platform (Roche, Manheim, Germany) following methods described by Abdelkrim et al. (2009). QDD v 0.9.0.0 Beta (Meglécz et al. 2010) was used to identify microsatellites, and primers were designed using the program Primer 3 (Rozen and Skaletsky 2000). A panel of polymorphic markers was chosen for each species (A. populifolia, seven microsatellite loci; P. nanteuilii, 12 microsatellite loci; Table S1). Using universal primers (Neilan et al. 1997) having four differently colored fluorescent labels, we multiplexed PCRs within label color and multiloaded all loci for each species into a single reaction per individual. The step-down PCR protocol consisted of ten cycles each at the following annealing temperatures: 70°C, 64°C, 58°C, 54°C, 50°C. Samples were genotyped using an ABI 3730 (Applied Biosystems, Foster City, CA) using GS-500 (Liz) in each capillary as a size standard. Allele sizes were estimated on GeneMapper, version 3.7 (Applied Biosystems).
Statistical analyses of genetic data
We tested microsatellite data for departures from Hardy–Weinberg and linkage equilibrium in Arlequin, version 3.5.1.2 (Excoffier et al. 2005), and P-values were Bonferroni corrected. We used Structure, version 2.2 (Pritchard et al. 2000; Falush et al. 2003), to determine whether multiple genetic groups were present across the range of each species and to determine the native source of introduced populations. For this analysis, we used the admixture model with correlated allele frequencies and tested the number of genetic groups (K) for each value of K between one and ten. We ran ten replicates for each value of K, each run having a burn-in period of 100,000 Markov chain Monte Carlo steps followed by 106 iterations. The most likely number of genetic groups was inferred using Evanno et al.'s (2005) ΔK method. We determined group membership assignment of each sample using the highest proportion of membership across all ten runs of Structure. Principal coordinate analysis (PCoA) conducted in GenAlEx v. 6.3 (Peakall and Smouse 2006) was used to visualize genetic distances (Nei 1972) between populations.
Many authors have stressed that a spectrum of diversity measures gives the best summary of diversity (Pielou 1966; Hill 1973). Therefore, we used measures closely related to each of Hill's first three diversity orders: zero (number of alleles, NA; allelic richness, R), unity (Shannon's Index, SH), and two (Hardy–Weinberg expected heterozygosity, HE). To calculate NA, R, and HE for each sample, we used FSTAT, version 2.9.3.2 (Goudet 1995, 2002), and SH was calculated using GenAlEx. For greatest utility in future comparisons, we also convert diversity orders 1 and 2 into their effective number equivalents, which avoid many well-known problems of diversity measures (Jost et al. 2010; Leinster and Cobbold 2011). The respective effective numbers equivalents are 1Dwithin = 2∧SH, and 2D = 1/(1−HE).
We used nonparametric Mann–Whitney U-tests to compare within-population diversity levels between the samples identified as sources for Australian introductions of both species because these data could not be made normal by transformation. Three approaches were used to assess diversity between populations. Pairwise FST values were calculated in Arlequin for comparison with other studies that quote this measure. Pairwise values for Shannon's mutual information index (SHUA) were calculated in GenAlEx. Compared with FST, mutual information is known to be more robust to a wide range of population sizes and dispersal rates (Sherwin et al. 2006; Dewar et al. 2011); additionally, the mutual information index can be converted to a numbers equivalent (1Dbetween), which avoids some serious problems that occur with other between-population measures (Jost et al. 2010).
We surveyed the literature regarding HE measured from polymorphic microsatellite data in species from both families containing our study taxa, Asteraceae and Caryophyllaceae, to determine whether HE estimates generated from native populations in this study were congruent with those from other members of the same family. This search was conducted in Google Scholar using the family name as a search term in conjunction with the terms “microsatellite” and “heterozygosity” in August 2012. Where data were given for multiple populations within a study, a mean value of HE was used. We avoided including estimates generated from introduced ranges, those of populations suspected of hybridization, and those of cultivated populations. Then, we surveyed the literature for examples of species showing evolutionary change in their introduced range, where genetic diversity had been estimated in both the native and introduced ranges. This search was conducted in Google Scholar in October 2012 using the terms “introduced” and “heterozygosity” in conjunction with either “rapid evolution” or “contemporary evolution.” Additionally, we included studies referenced in a review of genetic variation across native and introduced ranges (Dlugosch and Parker 2008a) showing evidence of morphological change in the introduced environment. We calculated the ratio of diversity found in the introduced range to that found in the native range (RHE), which gives an estimate of diversity retained after introduction, assuming no changes in diversity have occurred in the introduced range. For this calculation, we only used estimates of HE generated from microsatellite data because the absolute values of diversity estimates differ according to the marker used, and we wanted to directly compare these results to those generated in the current study.
Results
Evidence of morphological change
Petrorhagia nanteuilii in the introduced range showed a significant increase in log10 height through time that was not driven by region sampled (weighted general linear model including a term for region; R2 = 0.06; Fyear1,176 = 5.06; Pregion = 0.13; Pyear = 0.026; Fig. 3). Although the predictive power of this relationship is low, the magnitude of change is high, with average plant height increasing by almost 30% between 1882 and 1998.
Figure 3.
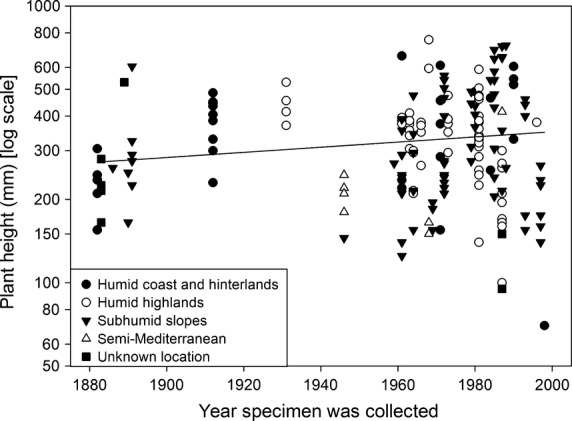
Log10 plant height of Petrorhagia nanteuilii introduced to Australia measured from herbarium specimens sampled from 1880 to 2000, classed by climatic region. Values increased significantly across time (weighted general linear model including a term for region; R2 = 0.06; Fyear1,176 = 5.06; Pregion = 0.13; Pyear = 0.026).
In the native range of A.populifolia, log10 plant height was unchanged across the period of this study (weighted general linear model; R2 = 0.003; Fyear1,52 = 0.15; Pyear = 0.70; Fig. S1a). In the native range of P. nanteuilii, log10 plant height decreased through time (weighted general linear model; R2 = 0.05; Fyear1,85 = 3.98; Pyear = 0.049; Fig. S1b).
Microsatellite markers
We found no evidence for departures from Hardy–Weinberg and linkage equilibrium in the microsatellite data for A. populifolia. Two of the twelve loci developed for P. nanteuilii (Pna06 and Pna16) significantly deviated from Hardy–Weinberg equilibrium and were excluded from downstream analyses. The remaining ten loci showed no significant departures from equilibrium. Petrorhagia nanteuilii has previously been reported to be tetraploid (Thomas and Murray 1981). Although we found no evidence of tetraploidy in the microsatellite data presented here, it is possible that in allotetraploid species, only a single parental genome may be amplified from any pair of primers.
Population structure
The Structure analysis of A. populifolia suggested the presence of two genetic groups (Fig. 4). One group included native samples extending from the western edge of the range in South Africa to Kenton on Sea as well as samples from south-eastern Australia (Fig. 4, group A1). The second group contained native samples from the eastern edge of the range to Kei Mouth and also included samples from Western Australia (Fig. 4, group A2). Two genetic groups were identified in P. nanteuilii: One group consisted of most sampling localities in Spain and France (Fig. 4, group B1) and a second group (Fig. 4, group B2) contained samples from two localities in Spain (Plasencia del Monte, PM; and Girona, GI), Corsica, Sardinia, and Australia. The samples from Corsica, Sardinia, and Australia had membership proportions for group B2 in excess of 0.97, whereas those from Spain were lower (PM, B2 membership proportion = 0.61; GI, B2 membership proportion = 0.80).
Figure 4.
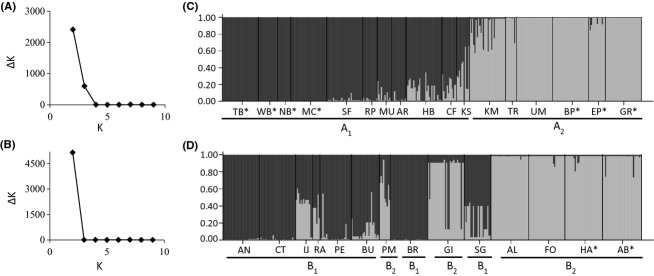
Structure analyses. Evanno et al.'s (2005) ΔK values for each putative number of populations (K) for (A) Arctotheca populifolia and (B) Petrorhagia nanteuilii. Structure Q plots generated using the maximum value of ΔK indicate A. populifolia (C) and P. nanteuilii (D) samples represent two genetic groups each, demarcated by labeled bars (i.e., A1). Samples from the introduced range are denoted by asterisks. Each individual is represented by a vertical line showing degree of admixture. Sample name abbreviations are defined in Figure 1.
Principal coordinate analysis plots were generally concordant with Structure results. Arctotheca populifolia samples from group A1 (Fig. 5A, diamonds) formed two distinct clusters representing native and introduced populations, respectively. A1-introduced samples were more closely related to A1 native samples than any A2 samples (Fig. 5A, circles). Within the A2 group, the Kei Mouth sample was separated from all other samples. The remaining native A2 samples were clustered with introduced A2 samples. Petrorhagia nanteuilii samples from group B1 (Fig. 5B, diamonds) were clustered together. The two Spanish samples that had lower membership proportions for group B2 in the Structure analysis were clustered with samples in group B1. Group B2 included the introduced sample from Hampton, which was clustered with the native sample from Corsica, and the introduced sample from Albury, which clustered with the native sample from Sardinia (Fig. 5B, circles).
Figure 5.
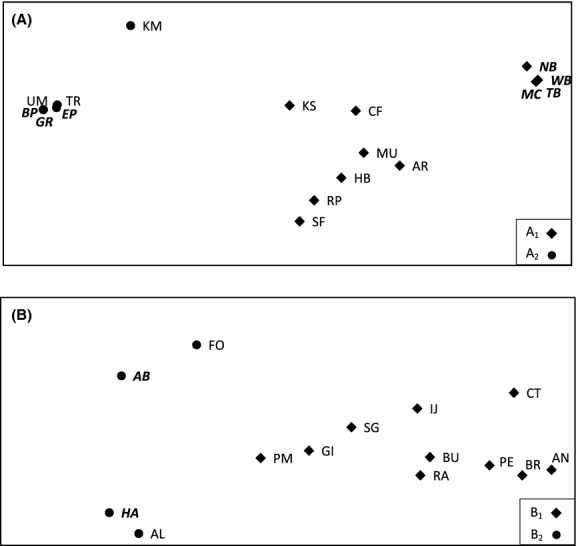
Principal coordinates analysis of genetic distance between Arctotheca populifolia samples (A) and Petrorhagia nanteuilii samples (B). Genetic groups identified in Structure analyses denoted by diamonds (A1 and B1) and circles (A2 and B2). Sample name abbreviations are defined in Figure 1 and those in bold represent introduced samples.
Genetic diversity
In the introduction to eastern Australia, native samples of A. populifolia had significantly higher HE (Table 1) than did introduced samples (A1 introduction: Mann–Whitney U, P < 0.01), while in the introduction to Western Australia, HE was not different between native and introduced samples (A2 introduction: Mann–Whitney U, P = 0.70). Diversity was low within the second introduction: We found a single genotype across all seven loci in one A. populifolia sample from the northeastern extreme of the native range (Umlalazi) and three samples from Australia (Beachport, Narooma and Wairo Beach). Assuming a single introduction of P. nanteuilii to Australia, the estimated HE in the native samples was not different to that of introduced samples (B2 introduction: Mann–Whitney U, P = 0.33). Values of R were similarly low in both species (Table 1). Despite the low values of genetic diversity, we detected that within populations, the total number of alleles detected for each species was not particularly low; 36 alleles were detected across the seven loci used for A. populifolia and 50 alleles across the ten loci used for downstream analysis in P. nanteuilii. This highlights the strong genetic diversity found between samples across the native ranges of both species (Table 1; FST, 0.33–0.56; SHUA, 0.09–0.33).
Table 1.
Estimation of diversity within native and introduced populations of Arctotheca populifolia and Petrorhagia nanteuilii including measures across three diversity orders: zero (allelic richness, R), unity (Shannon index, SH; and the effective numbers equivalent, 1Dwithin), and two (Hardy–Weinberg expected heterozygosity, HE; and the effective numbers equivalent, 2D). For both species, genetic differentiation within the native and introduced ranges was calculated using FST and Shannon's mutual information index (SHUA), and the numerical equivalent of SHUA (1Dbetween)
| Arctotheca populifolia (A1) | Arctotheca populifolia (A2) | Petrorhagia nanteuilii (B2) | ||||
|---|---|---|---|---|---|---|
| Statistic | Native | Introduced | Native | Introduced | Native | Introduced |
| Mean R (range) | 2.2 (2.0–2.6) | 1.0 (1.0–1.1) | 1.3 (1.0–1.8) | 1.1 (1.0–1.2) | 1.11 | 1.4 (1.3–1.4) |
| Mean SH (range) | 0.79 (0.57–0.96) | 0.08 (0.0–0.30) | 0.18 (0.0–0.46) | 0.04 (0.0–0.09) | 0.09 (0.08–0.11) | 0.33 (0.29–0.38) |
| Mean 1Dwithin (range) | 1.73 (1.48–1.94) | 1.06 (1.0–1.23) | 1.14 (1.0–1.38) | 1.03 (1.0–1.06) | 1.07 (1.05–1.08) | 1.26 (1.22–1.30) |
| Mean HE (range) | 0.34 (0.23–0.43) | <0.01 (0.0–0.01) | 0.07 (0.0–0.18) | 0.01 (0.0–0.03) | 0.04 (0.02–0.05) | 0.13 (0.11–0.15) |
| Mean 2D (range) | 1.53 (1.31–1.74) | 1.01 (1.0–1.01) | 1.09 (1.0–1.22) | 1.01 (1.0–1.03) | 1.04 (1.02–1.06) | 1.15 (1.12–1.18) |
| Mean Pairwise FST (range) | 0.33 (0.09–0.56) | <0.01 (0.0–0.01) | 0.40 (0.19–0.59) | 0.05 (0.02–0.08) | 0.56 (0.18–0.77) | 0.522 |
| Mean Pairwise SHUA (range) | 0.23 (0.08–0.41) | <0.01 (0.0–0.01) | 0.09 (0.01–0.16) | 0.01 (0.01–0.002) | 0.33 (0.27–0.41) | 0.262 |
| Mean 1Dbetween (range) | 1.18 (1.06–1.33) | 1.001 | 1.06 (1.01–1.12) | 1.01 (1.00–1.01) | 1.25 (1.21–1.33) | 1.202 |
All measures equal mean.
Denotes a single pairwise comparison.
We found microsatellite estimates of HE in 28 Asteraceae species from 28 different genera ranging from 0.22 to 0.88 (mean HE = 0.58, Fig. 6 and, Table S2). Of the approximately 2200 species in Caryophyllaceae (Schweingruber et al. 2011), microsatellite data exist for only eight species from five genera. We used expected heterozygosity measures from all eight of these species ranging from 0.35 to 0.93 (mean HE = 0.64, Fig. 6 and Table S2). In this study, values of HE across the native ranges of A. populifolia and P. nanteuilii were lower than almost all values we found for confamilials (Fig. 6; A. populifolia HE = 0.26; P. nanteuilii HE = 0.17) and both were significantly lower than the mean of these values for each family (one-sample t-tests: A. populifolia, P < 0.001; P. nanteuilii, P = 0.001).
Figure 6.
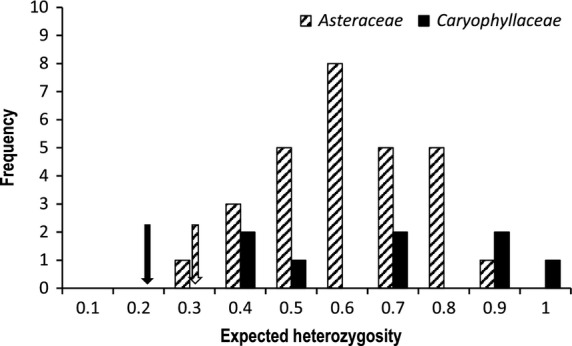
Native range estimates of expected heterozygosity from microsatellite data in Asteraceae (striped) and Caryophyllaceae (solid) families (see Table S1, for details). Arrows indicate the level of heterozygosity found in the native ranges of the species used in the present study (calculated from all sites sampled in the native range of each species).
We found 19 examples in the literature of species that had demonstrated evolutionary change in their introduced environment and where genetic diversity had been estimated in both the native and introduced range. Eight of these studies used allozymes, half of which found higher genetic diversity in the native range (Table 2). Of the eleven studies that used microsatellites, only one estimated a higher genetic diversity in the introduced range as compared to the native range. Ten of the microsatellite studies could be directly compared to our results (i.e., they provided estimates of HE). In these studies, the ratio of diversity in the introduced range to the diversity in the native range was an average of 0.81 (RHE range: 0.30–1.22, Table 2). In the present study, more diversity was found in the introduced range than the native source population of P. nanteuilii (RHE = 3.25). However, RHE for A. populifolia was much lower than estimates found in the literature search (A1 introduction: RHE = 0.01, A2 introduction: RHE = 0.14). In fact, both introductions of A. populifolia had the lowest ratios of any example we found.
Table 2.
Species showing evidence of evolutionary change in introduced environments and for which genetic diversity was measured in native and introduced populations. The ratio of genetic diversity in the introduced range to the native range is given, and the direction of change is given (Trend). The statistics used to calculate diversity included allelic richness (R), expected heterozygosity (HE) and genet richness (GR). Effective number equivalents (E) have been calculated for native/introduced diversity
| Genetic Diversity | |||||||
|---|---|---|---|---|---|---|---|
| Species | Native | Introduced | Ratio | Trend | Statistic | E | Reference |
| Allozymes | |||||||
| Acridotheres tristis | 0.06 | 0.03 | 0.50 | − | HE | 1.06/1.03 | Baker and Moeed (1987), Berthouly-Salazar et al. (2012) |
| Bufo marinus | 0.391 | 0.36 | 0.91 | − | HE | 1.64/1.56 | Estoup et al. (2001), Phillips et al. (2006) |
| Cedrus atlantica | 0.19 | 0.16 | 0.88 | − | HE | 1.23/1.20 | Bariteau and Ferrandes (1992), Lefevre et al. (2004) |
| Clidemia hirta | 0.04 | 0.06 | 1.40 | + | HE | 1.04/1.06 | DeWalt et al. (2004), DeWalt and Hamrick (2004) |
| Fringilla coelebs | 0.05 | 0.07 | 1.40 | + | HE | 1.05/1.07 | Baker (1992) |
| Gambusia affinis | 0.14 | 0.15 | 1.07 | + | HE | 1.16/1.17 | Stearns (1983), Scribner et al. (1992) |
| Passer montanus | 0.10 | 0.08 | 0.77 | − | HE | 1.11/1.08 | Barlow (1980), St. Louis and Barlow (1988) |
| Phalaris arundinacea | 1.89 | 2.27 | 1.20 | + | R | 1.89/2.27 | Lavergne and Molofsky (2007) |
| Mean2 | 0.99 | ||||||
| Microsatellites | |||||||
| Alliaria petiolata | 0.22 | 0.12 | 0.55 | − | HE | 1.28/1.14 | Durka et al. (2005), Bossdorf et al. (2004a,b) |
| Ambrosia artemisifolia | 0.76 | 0.75 | 0.99 | − | HE | 4.10/3.94 | Genton et al. (2005), Hodgins and Rieseberg (2011) |
| Carpodacus mexicanus | 0.81 | 0.77 | 0.95 | − | HE | 5.24/4.37 | Able and Belthoff (1998), Egbert and Belthoff (2003), Hawley et al. (2006) |
| Coregonus albula | 0.60 | 0.73 | 1.22 | + | HE | 2.47/3.65 | Amundsen et al. (2012) |
| Drososphila suboscura | 0.87 | 0.70 | 0.80 | − | HE | 7.94/3.33 | Huey et al. (2000), Pascual et al. (2001) |
| Linepithema humile | 0.64 | 0.20 | 0.31 | − | HE | 2.78/1.25 | Tsutsui et al.(2000) |
| Microstegium vimineum | 0.24 | 0.16 | 0.67 | − | HE | 1.32/1.19 | Novy et al. (2012a,b) |
| Oryctolagus cuniculus | 0.69 | 0.67 | 0.97 | − | HE | 3.23/3.03 | Williams and Moore (1989), Zenger et al.(2003) |
| Phragmites australis | 0.74 | 0.22 | 0.30 | − | GR | 0.74/.022 | Saltonstall and Stevenson (2007), Kettenring and Mock (2012) |
| Rhagoletis completa | 0.52 | 0.50 | 0.96 | − | HE | 2.08/2.00 | Chen et al. (2006) |
| Thymallus thymallus | 0.191 | 0.13 | 0.68 | − | HE | 1.23/1.15 | Koskinen et al. (2002) |
| Mean2 | 0.81 | ||||||
| Arctotheca populifolia | |||||||
| A1 introduction | 0.34 | <0.01 | 0.01 | − | HE | 1.53/1.01 | This study |
| A2 introduction | 0.07 | 0.01 | 0.14 | − | HE | 1.09/1.01 | This study |
| Petrorhagia nanteuilii | |||||||
| B2 introduction | 0.04 | 0.13 | 3.25 | + | HE | 1.04/1.15 | This study |
These estimates are from primary introductions, which were the sources of secondary introductions (“introduced” values for these species).
Mean includes all species having HE estimates.
Discussion
Genetic diversity has been demonstrated to be positively correlated with invasion success (Crawford and Whitney 2010), and standing genetic variation is believed to be important to invasive species' ability to adapt to novel environments (Barrett and Schluter 2008). However, it is becoming clear that introduced populations with very low neutral genetic diversity are sometimes successful invaders (Ren et al. 2005; Mergeay et al. 2006; Zimmermann et al. 2010) and have the ability to adapt to their new environments (Dlugosch and Parker 2008b; Harris et al. 2012). Here, we provide two examples of species that have established, spread, and adapted to the environment in their introduced range in Australia (Buswell et al. 2011 and Fig. 3), yet have significantly lower genetic diversity in their native ranges than do confamilials. Interestingly, for both of these species, the changes we identified in the introduced ranges were not found in the native ranges, ruling out the possibility that global processes are driving these changes.
Despite the fact that the two separate introductions of A. populifolia described here had comparatively low levels of genetic diversity in the native source populations, these two introductions represent a larger percentage loss of genetic diversity than found in any introduction identified in our review of species, showing substantial morphological change in the introduced range. Similarly, introduced populations of invasive Japanese knotweed (Fallopia species complex) harbored very low genetic diversity at Amplified Fragment Length Polymorphism markers despite displaying significantly different phenotypes in a common garden setting (Richards et al. 2008). While greater levels of diversity may increase the likelihood of invasion success (Crawford and Whitney 2010; Jones and Gomulkiewicz 2012), it is clear that some introduced species, such as those discussed here, are able to become invasive and adapt to their new environments with very little neutral genetic diversity. This has important management implications because it demonstrates that even introductions from very small numbers of individuals have the potential to become invasive.
The review we have conducted specifically examines loss of genetic diversity at introduction in species where some evidence of adaptive change has been documented in the introduced environment. It would be useful to compare the associated change in genetic diversity in this group with that of a group of species which has been introduced but has shown no evidence of adaptation to novel environments; we might anticipate that the latter would show more loss if diversity is important to adaptive potential. Unfortunately, there is a bias in reporting which makes this difficult. However, we can compare the results of our review (19% loss of GD in the introduced range) with that of Dlugosch and Parker (2008a), who found 22.6% loss of diversity at introduction irrespective of evidence of adaptive change (Dlugosch and Parker 2008a) (two-sample t-test, P = 0.72). This suggests that neutral genetic diversity is not important to adaptive potential in introduced species.
The ability of populations with low current “neutral” diversity to evolve could be due to (1) retention of greater adaptive than neutral genetic variation due to either chance or balancing selection on adaptive variation (Reed and Frankham 2001), (2) contributions of mutations to selection response, especially when selection lasts for more than twenty generations (Frankham 1980b, 1983; Hill 1982a,b), (3) loss of neutral genetic diversity after much of the adaptation has occurred, or (4) some combination of these. Substantial adaptive genetic changes can still occur in populations subject to bottlenecks, and there is often large variation among replicates (Frankham 1980a). We are unable to distinguish between these hypotheses, but the likelihood of contributions from mutations that arose after introduction increases as the level of neutral genetic diversity in the introduced population decreases.
Recent research suggests that epigenetic modifications (DNA methylation) may also play an important role in invasion success. Using the invasive Japanese knotwood populations discussed above, Richards et al. (2012) demonstrated that although genetic diversity was extremely low, significant epigenetic differentiation occurred between sites, suggesting a possible nongenetic mechanism for adaptation. Theoretical work on epigenetic selection models indicates that increased phenotypic change can occur in populations with no genetic variation as a result of epigenetic changes (Geoghegan and Spencer 2012). In fact, Liebl et al. (2013) found a negative relationship between genetic and epigenetic diversity in introduced populations of sparrows (Passer domesticus) and speculated that epigenetic variation may provide a mechanism for adaptation over the short time scales relevant to invasions.
Source population identification
Our results highlight the importance of determining source populations prior to assessing changes in genetic diversity between native and introduced ranges. Few studies investigating this topic have done this, but levels of genetic diversity can be very different across a species' native range, as we found with A. populifolia. Identifying the source of an invasion assures that observed differences between introduced and native populations are not the result of diversity within the native range and prevents actual differences from becoming obscured (Dlugosch and Parker 2008a). Similarly, for studies attempting to identify contemporary evolution in introduced species, it is vital that the correct source population is identified in the native range.
Our analyses identified two genetic groups of A. populifolia. One group consisted of western South African native samples and eastern Australian introduced samples. PCoA indicated that the latter were most similar to native samples from the south coast of South Africa ranging from Muizenberg to Kenton on Sea. The second group contained samples from the east coast of South Africa and introduced samples ranging from Margaret River in Western Australia to Beachport in South Australia. The two genetic groups identified in South Africa correspond perfectly to the Cape Seashore Vegetation (Group A1) and the Subtropical Seashore Vegetation (Group A2) described by Mucina and Rutherford (2006). A single genotype across seven loci was found in all 30 individuals sampled at Umlalazi (eastern South Africa), and this genotype was found in every individual sampled in Beachport and 39 of the 43 individuals sampled in Western Australia. These results indicate two separate introductions to Australia. This is supported by morphological data indicating that two forms of A. populifolia exist in Australia, both of which are found in South Africa (Heyligers 2007). The Victorian coastline separates the two morphological groups (Heyligers 2007) as well as the genetic groups found in this study.
Samples within native populations of both species were highly differentiated. This is possibly due to our intentional selection of species with restricted ranges so that we could sample comprehensively across the native ranges. Although restricted ranges and high levels of population differentiation can be caused by limited dispersal, there is independent evidence that there may be a small amount of long-distance dispersal in A. populifolia. Heyligers (2007) argued that the distribution of A. populifolia morphs in Australia could be explained by dispersal of achenes via coastal currents and that historical records of first appearance showed an eastward progression of this introduction from Western Australia to South Australia. Given this evidence, one might expect to find a more cosmopolitan distribution of this species, but to our knowledge, A. populifolia is only found in southern Africa and Australia.
The Structure analysis of P. nanteuilii indicated that two genetic groups exist in the native range, but only one of these was represented within Australia. Mediterranean island samples from Corsica and Sardinia were most similar to samples from Australia, supporting a single source for this introduction. However, while the PCoA supported the membership of group B1 determined in Structure (Fig. 5B), the samples contained within group B2 were not well clustered. In fact, the PCoA indicated that the introduced sample from Hampton was closely related to the native sample from Corsica, whereas the introduced sample from Albury was closely related to the native sample from Sardinia. This raises the possibility that two introductions of this species into Australia may have occurred.
Comparisons of genetic diversity between introduced and native ranges
Low genetic diversity in introduced populations can reflect a genetically impoverished source (Voss et al. 2012). Our identification of the source populations for the introductions discussed here allows us to confirm that the sources were genetically impoverished. Within native and introduced populations of A. populifolia and P. nanteuilii, values of HE were considerably lower than found in other species within these families (Table 1; Fig. 6; Table S2). One native sample of A. populifolia displayed no gene diversity (i.e., had a single genotype across seven loci). Variation was even lower in the introduced range of A. populifolia than the already low variation in the native range, suggestive of a small number of founders.
Despite low levels of within-population neutral genetic diversity, both of these species are widespread in their introduced range in Australia. Similarly, Hardesty et al. (2012) found extremely low levels of neutral genetic variation in highly successful introductions of Miconia calvescens (mean HE = 0.07). However, because the ability to evolve in response to a novel environment may depend on the level of adaptive variation present, the relevance of neutral markers variation has been questioned (Reed and Frankham 2001). Although a positive correlation has been reported between quantitative trait (QST) and microsatellite variation (FST), quantitative variation is usually higher (Merilä and Crnokrak 2001; Leinonen et al. 2008) and the ability to predict QST increases with increasing values of FST (Leinonen et al. 2008). Population bottlenecks are predicted to reduce additive genetic variance (Wright 1951; Chakraborty and Nei 1982), but in some circumstances, such populations may experience an increase in additive genetic variance for traits with at least some nonadditive genetic variation (Willis and Orr 1993; Wang et al. 1998; Willi et al. 2006). While this shift in additive genetic variance may not always result in an increased ability to adapt to novel selection pressures (van Heerwaarden et al. 2008), the combination of adaptation with low diversity at neutral markers indicating the presence of a bottleneck has been identified here and in other studies (Koskinen et al. 2002; Yonekura et al. 2007; Dlugosch and Parker 2008b). Frankham et al. (1999) showed that the effects of population bottlenecks on ability to evolve in response to environmental change closely followed neutral expectations. Further, a number of data sets indicate that genetic variation involved in adaptation to new environments is approximately additive (de Oliveira and Cordeiro 1980; Frankham et al. 1999), in contrast to the fitness variation in the environment to which populations have been adapted long-term, where there is usually a predominance of nonadditive variation (i.e., the occurrence of increased additive genetic variation in bottlenecked populations probably does not apply to populations adapting to new environments).
Finally, we speculate on whether intrinsic characteristics of the species might be affecting the diversity. Finding a genetically monomorphic sample in the native range may suggest alternate forms of reproduction across the species' distribution. When Roman and Darling (2007) examined successful introductions having decreased genetic diversity in the introduced range, 63% had reproductive abilities other than those involving sexual recombination. It is possible that A. populifolia may have the ability to reproduce via apomixis, spread vegetatively, or self-fertilize, although none of these reproductive mechanisms have been reported in this species. Arctotheca populifolia has previously been reported as diploid (Norlindh 1967), and our investigations of ploidy in both native and introduced samples support this (see Appendix S2). This suggests that apomixis is an unlikely explanation for the observed genetic pattern because, among Asteraceae, this reproductive mechanism is normally only found in polyploids (Noyes 2007). Vegetative reproduction has been reported in the congener A. calendula (Bossard 2000), but does not explain the biogeographic patterns of A. populifolia described here. Baker's Law (Stebbins 1957) states that self-fertilization should provide an advantage to colonizing populations (Baker 1955) and could explain the genetic patterns we have identified in eastern South Africa and Australian introduced populations. Baker's Law is supported by three findings: i) increased frequency of self-compatible species on islands (Barrett et al. 1996), ii) species capable of autonomous seed production had larger invasive ranges (van Kleunen and Johnson 2007), and iii) species naturalized outside of their native range are more likely to self-fertilize than congeners only found in their native range (van Kleunen et al. 2008).
Conclusion
Considerable effort has been invested in identifying drivers of invasion success, including the importance of genetic diversity to invasiveness. While genetic diversity may be related to invasion success in some species (Crawford and Whitney 2010), increasingly, evidence suggests that genetic diversity is not essential to a species' ability to invade novel environments. Here, we have identified two species with low levels of neutral genetic variation in both their native and introduced ranges, which appear to have adapted and spread in their introduced range. Recent empirical evidence and simulations suggest that a number of factors influence the relationship between genetic diversity and invasion success and that complexities such as competitive interactions and diversity of the native community are likely to be important (Chang and Smith 2012; Hovick et al. 2012; Jones and Gomulkiewicz 2012). Further, it appears that epigenetic modifications may play a role in facilitating invasion success immediately following invasion, although this idea has not yet been rigorously tested. In combination, these results suggest that genetic diversity measures alone are inadequate predictors of invasion success.
Acknowledgments
We thank Carolyn Porter, Maurizio Rossetto, Rod Peakall, Peter Lockhart and Mike Gardner for early discussions regarding the approach to this project, Ray Blick for assistance with sample collection, the Australian Genome Research Facility for assistance with DNA extractions and next-generation sequencing, and the Ramaciotti Centre for Gene Function Analysis for assistance with next-generation sequencing and analysis. This research was funded by an ARC grant to A. T. Moles, R. Frankham and W. B. Sherwin (DP0984222) and a Research Fellowship from Deakin University to L. A. Rollins.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Appendix S1. Theory of adaptive genetic change in novel environments.
Appendix S2. Chromosome investigations of Arctotheca populifolia.
Table S1. Characterization of microsatellite loci in Arctotheca populifolia (N = 348) and Petrorhagia nanteuilii (N = 377) including locus name, GenBank accession number, primer sequences, repeat motif, number of alleles and allele size range.
Table S2. Expected heterozygosity (HE) estimates from microsatellite data for species within the Asteraceae and Caryophyllaceae families. Effective number equivalents (2D) have been calculated. For Asteraceae, estimates were included for one species per genus (N = 28). For Caryophyllaceae, all estimates identified in the literature were included (N = 8). The number of loci (L), samples (S) and total number of individuals (I) are given for each study.
Figure S1. (A,B) Plant height (log10 transformed) of Arctotheca populifolia (A) and Petrorhagia nanteuilii (B) measured from herbarium specimens sampled in the native range between 1891–2003 and 1848–1985, respectively. For A. populifolia, values do not change significantly over this time period (weighted general linear model; R2 = 0.003; Fyear1,52 = 0.15; P = 0.70). For P. nanteuilii, plant height decreased through time (weighted general linear model; R2 = 0.05; Fyear1,85 = 3.98; Pyear = 0.049).
References
- Abdelkrim J, Robertson BC, Stanton JL, Gemmell NJ. Fast, cost-effective development of species-specific microsatellite markers by genomic sequencing. Biotechniques. 2009;46:185–192. doi: 10.2144/000113084. [DOI] [PubMed] [Google Scholar]
- Able KP, Belthoff JR. Rapid ‘evolution’ of migratory behaviour in the introduced house finch of eastern North America. Proceedings of the Royal Society of London. Series B: Biol. Sci. 1998;265:2063–2071. [Google Scholar]
- Allendorf FW, Luikart G. Conservation and the genetics of populations. Malden, Massachusetts: Blackwell; 2007. [Google Scholar]
- Allendorf FW, Lundquist LL. Introduction: Population biology, evolution, and control of invasive species. Conserv. Biol. 2003;17:24–30. [Google Scholar]
- Amundsen P-A, Salonen E, Niva T, Gjelland K, Præbel K, Sandlund O, et al. Invader population speeds up life history during colonization. Biol. Invasions. 2012;14:1501–1513. [Google Scholar]
- AVH Database. Australia's virtual herbarium. 2012. Available at: http://avh.ala.org.au. (accessed 17 August 2012) [Google Scholar]
- Baker HG. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution. 1955;9:347–349. [Google Scholar]
- Baker AJ. Genetic and morphometric divergence in ancestral European and descendent New Zealand populations of chaffinches (Fringilla coelebs. Evolution. 1992;46:1784–1800. doi: 10.1111/j.1558-5646.1992.tb01169.x. [DOI] [PubMed] [Google Scholar]
- Baker AJ, Moeed A. Rapid genetic differentiation and founder effect in colonizing populations of common mynas (Acridotheres tristis. Evolution. 1987;41:525–538. doi: 10.1111/j.1558-5646.1987.tb05823.x. [DOI] [PubMed] [Google Scholar]
- Ball PW, Heywood VH. A revision of the genus Petrorhagia. Bulletin of the British Museum (Natural History) Botany. 1964;3:121–172. [Google Scholar]
- Bariteau M, Ferrandes P. Amelioration des especes vegetales cultivees: objectifs et criteres de selection. In: Gallais A, Bannerot H, editors. Mieux comprendre. Paris: INRA; 1992. pp. 732–743. [Google Scholar]
- Barlow JC. Proc. Int. Ornithol. Congress. 1980;17:1143–1149. Adaptive responses in skeletal characteristics of the New World population of Passer montanus. [Google Scholar]
- Barrett RDH, Schluter D. Adaptation from standing genetic variation. Trends Ecol. Evol. 2008;23:38–44. doi: 10.1016/j.tree.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Barrett SCH, Emerson B, Mallet J. The reproductive biology and genetics of island plants [and discussion] Philos. Trans. Royal Soc. B: Biol. Sci. 1996;351:725–733. [Google Scholar]
- Berthouly-Salazar C, Hui BJ, van Rensburg JJ, Le Roux BJ, van Vuuren C. Spatial sorting drives morphological variation in the invasive bird, Acridotheris tristis. PLoS ONE. 2012;7:e38145. doi: 10.1371/journal.pone.0038145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blossey B, Nötzold R. Evolution of increased competitive ability in invasive nonindigenous plants: a hypothesis. J. Ecol. 1995;83 [Google Scholar]
- Bossard C. Invasive plants of California's wildlands. Berkeley: Univ. of California Press; 2000. [Google Scholar]
- Bossdorf O, Prati D, Auge H, Schmid B. Reduced competitive ability in an invasive plant. Ecol. Lett. 2004a;7:346–353. [Google Scholar]
- Bossdorf O, Schröder S, Prati D, Auge H. Palatability and tolerance to simulated herbivory in native and introduced populations of Alliaria petiolata (Brassicaceae) Am. J. Bot. 2004b;91:856–862. doi: 10.3732/ajb.91.6.856. [DOI] [PubMed] [Google Scholar]
- Bossdorf O, Auge H, Lafuma L, Rogers W, Siemann E, Prati D. Phenotypic and genetic differentiation between native and introduced plant populations. Oecologia. 2005;144:1–11. doi: 10.1007/s00442-005-0070-z. [DOI] [PubMed] [Google Scholar]
- Buswell JM, Moles AT, Hartley S. Is rapid evolution common in introduced plant species? J. Ecol. 2011;99:214–224. [Google Scholar]
- Chakraborty R, Nei M. Genetic differentiation of quantitative characters between populations or species: I. Mutation and random genetic drift. Genet. Res. 1982;39:303–314. [Google Scholar]
- Chang C, Smith M. Invasion of an intact plant community: the role of population versus community level diversity. Oecologia. 2012;168:1091–1102. doi: 10.1007/s00442-011-2157-z. [DOI] [PubMed] [Google Scholar]
- Chen Y, Opp S, Berlocher S, Roderick G. Are bottlenecks associated with colonization? Genetic diversity and diapause variation of native and introduced Rhagoletis completa populations. Oecologia. 2006;149:656–667. doi: 10.1007/s00442-006-0482-4. [DOI] [PubMed] [Google Scholar]
- Cheptou P-O, Carrue O, Rouifed S, Cantarel A. Rapid evolution of seed dispersal in an urban environment in the weed Crepis sancta. Proc. Natl Acad. Sci. USA. 2008;105:3796–3799. doi: 10.1073/pnas.0708446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody ML, Overton JM. Short-term evolution of reduced dispersal in island plant populations. J. Ecol. 1996;84:53–61. [Google Scholar]
- Crawford KM, Whitney KD. Population genetic diversity influences colonization success. Mol. Ecol. 2010;19:1253–1263. doi: 10.1111/j.1365-294X.2010.04550.x. [DOI] [PubMed] [Google Scholar]
- DeWalt SJ, Hamrick JL. Genetic variation of introduced Hawaiian and native Costa Rican populations of an invasive tropical shrub, Clidemia hirta (Melastomataceae) Am. J. Bot. 2004;91:1155–1162. doi: 10.3732/ajb.91.8.1155. [DOI] [PubMed] [Google Scholar]
- DeWalt S, Denslow J, Hamrick JL. Biomass allocation, growth, and photosynthesis of genotypes from native and introduced ranges of the tropical shrub Clidemia hirta. Oecologia. 2004;138:521–531. doi: 10.1007/s00442-003-1462-6. [DOI] [PubMed] [Google Scholar]
- Dewar RC, Sherwin WB, Thomas E, Holleley CE, Nichols RA. Predictions of single-nucleotide polymorphism differentiation between two populations in terms of mutual information. Mol. Ecol. 2011;20:3156–3166. doi: 10.1111/j.1365-294X.2011.05171.x. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008a;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- Dlugosch KM, Parker IM. Invading populations of an ornamental shrub show rapid life history evolution despite genetic bottlenecks. Ecol. Lett. 2008b;11:701–709. doi: 10.1111/j.1461-0248.2008.01181.x. [DOI] [PubMed] [Google Scholar]
- Durka W, Bossdorf O, Prati D, Auge H. Molecular evidence for multiple introductions of garlic mustard (Alliaria petiolata, Brassicaceae) to North America. Mol. Ecol. 2005;14:1697–1706. doi: 10.1111/j.1365-294X.2005.02521.x. [DOI] [PubMed] [Google Scholar]
- Egbert JR, Belthoff JR. Wing shape in house finches differs relative to migratory habit in eastern and western North America. The Condor. 2003;105:825–829. [Google Scholar]
- Estoup A, Wilson IJ, Sullivan C, Cornuet J-M, Moritz C. Inferring population history from microsatellite and enzyme data in serially introduced cane toads, Bufo marinus. Genetics. 2001;159:1671–1687. doi: 10.1093/genetics/159.4.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolut. Bioinf. Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankham R. Robertson A. Selection Experiments in Laboratory and Domestic Animals. Slough: Commonwealth Agricultural Bureaux; 1980a. The founder effect and response to artificial selection in Drosophila; pp. 87–90. [Google Scholar]
- Frankham R. Origin of genetic variation in selection lines. In: Robertson A, editor. Selection experiments in laboratory and domestic animals. Slough: Commonwealth Agricultural Bureaux; 1980b. pp. 56–68. [Google Scholar]
- Frankham R. Origin of genetic variation in selection lines. In: Towner R, editor. Proceeding of the thirty-second annual breeders' roundtable. MO: St. Louis; 1983. pp. 1–18. [Google Scholar]
- Frankham R, Lees K, Montgomery ME, England PR, Lowe EH, Briscoe DA. Do population size bottlenecks reduce evolutionary potential? Anim. Conserv. 1999;2:255–260. [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge, U.K: Cambridge Univ. Press; 2010. [Google Scholar]
- GCW database. 2012. Global Compendium of Weeds. Available at: http://www.hear.org/gcw/species. (accessed on 17 August 2012)
- Genton BJ, Shykoff JA, Giraud T. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 2005;14:4275–4285. doi: 10.1111/j.1365-294X.2005.02750.x. [DOI] [PubMed] [Google Scholar]
- Geoghegan JL, Spencer HG. Population-epigenetic models of selection. Theor. Popul. Biol. 2012;81:232–242. doi: 10.1016/j.tpb.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): A computer program to calculate F-Statistics. J. Hered. 1995;86:485–486. [Google Scholar]
- Goudet J. 2002. FSTAT, a program to estimate and test gene diversities and fixation indices version 2.9.3. Available from http://www.unil.ch/izea/softwares/fstat.html. Updated from Goudet (1995)
- GRIN database. 2012. USDA, ARS, National Genetic Resources Program. Germplasm Resources Information Network (GRIN), [Online Database]. National Germplasm Resources Laboratory, Beltsville, Maryland, USA. Available at: http://www.ars-grin.gov/cgi-bin/npgs/html/taxon.pl?415245. (accessed 17 August 2012)
- Harden GJ, editor. Flora of New South Wales. Vol. 3. Kensington, NSW: New South Wales Univ. Press; 1992. [Google Scholar]
- Hardesty BD, Rocha JJ, Le Roux OJ, Meyer J-Y, Westcott D, Wieczorek AM. Getting here from there: testing the genetic paradigm underpinning introduction histories and invasion success. Divers. Distrib. 2012;18:147–157. [Google Scholar]
- Harris C, Dormontt E, Roux J, Lowe A, Leishman M. No consistent association between changes in genetic diversity and adaptive responses of Australian acacias in novel ranges. Evol. Ecol. 2012;26:1–16. [Google Scholar]
- Hawley DM, Hanley D, Dhondt AA, Lovette IJ. Molecular evidence for a founder effect in invasive house finch (Carpodacus mexicanus) populations experiencing an emergent disease epidemic. Mol. Ecol. 2006;15:263–275. doi: 10.1111/j.1365-294X.2005.02767.x. [DOI] [PubMed] [Google Scholar]
- van Heerwaarden B, Willi Y, Kristensen TN, Hoffmann AA. Population bottlenecks increase additive genetic variance but do not break a selection limit in rain forest Drosophila. Genetics. 2008;179:2135–2146. doi: 10.1534/genetics.107.082768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyligers P. Some New South Wales coastal plant distributions: a comparison of herbarium records with transect survey data. Cunninghamia. 1998;5:645–664. [Google Scholar]
- Heyligers P. The role of currents in the dispersal of introduced seashore plants around Australia. Cunninghamia. 2007;10:167–188. [Google Scholar]
- Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54:427–432. [Google Scholar]
- Hill WG. Predictions of response to artificial selection from new mutations. Genet. Res. 1982a;40:255–278. doi: 10.1017/s0016672300019145. [DOI] [PubMed] [Google Scholar]
- Hill WG. Rates of change in quantitative traits from fixation of new mutations. Proc. Natl Acad. Sci. USA. 1982b;79:142–145. doi: 10.1073/pnas.79.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins KA, Rieseberg L. Genetic differentiation in life-history traits of introduced and native common ragweed (Ambrosia artemisiifolia) populations. J. Evol. Biol. 2011;24:2731–2749. doi: 10.1111/j.1420-9101.2011.02404.x. [DOI] [PubMed] [Google Scholar]
- Hovick S, Gümüşer E, Whitney K. Community dominance patterns, not colonizer genetic diversity, drive colonization success in a test using grassland species. Plant Ecol. 2012;213:1365–1380. [Google Scholar]
- Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra Ls. Rapid evolution of a geographic cline in size in an introduced fly. Science. 2000;287:308–309. doi: 10.1126/science.287.5451.308. [DOI] [PubMed] [Google Scholar]
- Hufbauer RA. Biological invasions: paradox lost and paradise gained. Curr. Biol. 2008;18:R246–R247. doi: 10.1016/j.cub.2008.01.038. [DOI] [PubMed] [Google Scholar]
- Jones EI, Gomulkiewicz R. Biotic interactions, rapid evolution, and the establishment of introduced species. Am. Nat. 2012;179:E28–E36. doi: 10.1086/663678. [DOI] [PubMed] [Google Scholar]
- Jost L, DeVries P, Walla T, Greeney H, Chao A, Ricotta C. Partitioning diversity for conservation analyses. Divers. Distrib. 2010;16:65–76. [Google Scholar]
- Kashtan N, Noor E, Alon U. Varying environments can speed up evolution. Proc. Natl Acad. Sci. USA. 2007;104:13711–13716. doi: 10.1073/pnas.0611630104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenring K, Mock K. Genetic diversity, reproductive mode, and dispersal differ between the cryptic invader, Phragmites australis, and its native conspecific. Biol. Invasions. 2012;14:2489–2504. [Google Scholar]
- van Kleunen M, Johnson SD. Effects of self-compatibility on the distribution range of invasive European plants in North America. Conserv. Biol. 2007;21:1537–1544. doi: 10.1111/j.1523-1739.2007.00765.x. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Manning JC, Pasqualetto V, Johnson SD. Phylogenetically independent associations between autonomous self-fertilization and plant invasiveness. Am. Nat. 2008;171:195–201. doi: 10.1086/525057. [DOI] [PubMed] [Google Scholar]
- Kolbe JJ, Glor RE, Rodriguez Schettino L, Lara AC, Larson A, Losos JB. Genetic variation increases during biological invasion by a Cuban lizard. Nature. 2004;431:177–181. doi: 10.1038/nature02807. [DOI] [PubMed] [Google Scholar]
- Koskinen MT, Haugen TO, Primmer CR. Contemporary fisherian life-history evolution in small salmonid populations. Nature. 2002;419:826–830. doi: 10.1038/nature01029. [DOI] [PubMed] [Google Scholar]
- Lavergne S, Molofsky J. Increased genetic variation and evolutionary potential drive the success of an invasive grass. Proc. Natl Acad. Sci. U.S.A. 2007;104:3883–3888. doi: 10.1073/pnas.0607324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre F, Fady B, Fallour-Rubio D, Ghosn D, Bariteau M. Impact of founder population, drift and selection on the genetic diversity of a recently translocated tree population. Heredity. 2004;93:542–550. doi: 10.1038/sj.hdy.6800549. [DOI] [PubMed] [Google Scholar]
- Leinonen T, O'Hara RB, Cano JM, MerilÄ J. Comparative studies of quantitative trait and neutral marker divergence: a meta-analysis. J. Evol. Biol. 2008;21:1–17. doi: 10.1111/j.1420-9101.2007.01445.x. [DOI] [PubMed] [Google Scholar]
- Leinster T, Cobbold CA. Measuring diversity: the importance of species similarity. Ecology. 2011;93:477–489. doi: 10.1890/10-2402.1. [DOI] [PubMed] [Google Scholar]
- Liebl AL, Schrey AW, Richards CL, Martin LB. Patterns of DNA methylation throughout a range expansion of an introduced songbird. Integr. Comp. Biol. 2013;53:351–358. doi: 10.1093/icb/ict007. [DOI] [PubMed] [Google Scholar]
- Mayr E, editor. Summary. The Genetics of Colonizing Species. London: Academic Press; 1965. [Google Scholar]
- Meglécz E, Costedoat C, Dubut V, Gilles A, Malausa T, Pech N, et al. QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics. 2010;26:403–404. doi: 10.1093/bioinformatics/btp670. [DOI] [PubMed] [Google Scholar]
- Mergeay J, Verschuren D, Meester LD. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proceed. Royal Soc. B: Biol. Sci. 2006;273:2839–2844. doi: 10.1098/rspb.2006.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merilä J, Crnokrak P. Comparison of genetic differentiation at marker loci and quantitative traits. J. Evol. Biol. 2001;14:892–903. [Google Scholar]
- Mucina L, Rutherford MC. The vegetation of South Africa, Lesotho and Swaziland. Strelitzia 19. Pretoria: South African Biodiversity Institute; 2006. [Google Scholar]
- Nei M. Genetic distance between populations. Am. Nat. 1972;106:283–292. [Google Scholar]
- Neilan BA, Wilton AN, Jacobs D. A universal procedure for primer labelling of amplicons. Nucleic Acids Res. 1997;25:2938–2939. doi: 10.1093/nar/25.14.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norlindh T. Arctotheca populifolia (Berg.) T. Norl. comb. nova. A South African dune plant. Aquilo Ser. Botanica. 1967;6:84–93. [Google Scholar]
- Novy A, Flory SL, Hartman JM. Evidence for rapid evolution of phenology in an invasive grass. J. Evolut. Biol. 2012a;26:443–450. doi: 10.1111/jeb.12047. [DOI] [PubMed] [Google Scholar]
- Novy A, Flory SL, Honig JA, Bonos S, Hartman JM. Characterization of polymorphic microsatellites for the invasive grass Microstegium vimineum (Poaceae) Am. J. Bot. 2012b;99:e56–e58. doi: 10.3732/ajb.1100337. [DOI] [PubMed] [Google Scholar]
- Noyes RD. Apomixis in the Asteraceae: Diamonds in the rough. Funct. Plant Sci. Biotechnol. 2007;1:207–222. [Google Scholar]
- de Oliveira AK, Cordeiro AR. Adaptation of Drosophila willistoni experimental populations to extreme pH medium. Heredity. 1980;44:123–130. [Google Scholar]
- Pascual M, Aquadro CF, Soto V, Serra L. Microsatellite variation in colonizing and palearctic populations of Drosophila subobscura. Mol. Biol. Evol. 2001;18:731–740. doi: 10.1093/oxfordjournals.molbev.a003855. [DOI] [PubMed] [Google Scholar]
- Peakall R, Smouse PE. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips BL, Brown GP, Webb JK, Shine R. Invasion and the evolution of speed in toads. Nature. 2006;439:803. doi: 10.1038/439803a. [DOI] [PubMed] [Google Scholar]
- Pielou EC. Species-diversity and pattern-diversity in the study of ecological succession. J. Theor. Biol. 1966;10:370–383. doi: 10.1016/0022-5193(66)90133-0. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DH, Frankham R. How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution. 2001;55:1095–1103. doi: 10.1111/j.0014-3820.2001.tb00629.x. [DOI] [PubMed] [Google Scholar]
- Ren MX, Zhang QG, Zhang DY. Random amplified polymorphic DNA markers reveal low genetic variation and a single dominant genotype in Eichhornia crassipes populations throughout China. Weed Res. 2005;45:236–244. [Google Scholar]
- Richards CL, Bossdorf O, Muth NZ, Gurevitch J, Pigliucci M. Jack of all trades, master of some? On the role of phenotypic plasticity in plant invasions. Ecol. Lett. 2006;9:981–993. doi: 10.1111/j.1461-0248.2006.00950.x. [DOI] [PubMed] [Google Scholar]
- Richards CL, Walls RL, Bailey JP, Parameswaran R, George T, Pigliucci M. Plasticity in salt tolerance traits allows for invasion of novel habitat by Japanese knotweed s. l. (Fallopia japonica and F. bohemica, Polygonaceae) Am. J. Bot. 2008;95:931–942. doi: 10.3732/ajb.2007364. [DOI] [PubMed] [Google Scholar]
- Richards CL, Schrey AW, Pigliucci M. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol. Lett. 2012;15:1016–1025. doi: 10.1111/j.1461-0248.2012.01824.x. [DOI] [PubMed] [Google Scholar]
- Ridley CE, Ellstrand NC. Evolution of enhanced reproduction in the hybrid-derived invasive, California wild radish (Raphanus sativus) Biol. Invasions. 2009;11:2251–2264. [Google Scholar]
- Robertson A. A theory of limits in artificial selection. Proceedings of the Royal Society of London. Series B. Biol. Sci. 1960;153:234–249. [Google Scholar]
- Roman J, Darling JA. Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol. Evol. 2007;22:454–464. doi: 10.1016/j.tree.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001;32:305–332. [Google Scholar]
- Saltonstall K, Stevenson JC. The effect of nutrients on seedling growth of native and introduced Phragmites australis. Aquat. Bot. 2007;86:331–336. [Google Scholar]
- Schweingruber F, Borner A, Schulze E-D. Caryophyllaceae. Anatomy of stems in herbs, shrubs and trees: an ecological and systematic approach, Volume 1. Berlin: Springer; 2011. [Google Scholar]
- Scribner KT, Wooten MC, Smith MH, Kennedy PK, Rhodes OE. Variation in life history and genetic traits of Hawaiian mosquitofish populations. J. Evol. Biol. 1992;5:267–288. [Google Scholar]
- Sherwin WB, Jabot F, Rush R, Rossetto M. Measurement of biological information with applications from genes to landscapes. Mol. Ecol. 2006;15:2857–2869. doi: 10.1111/j.1365-294X.2006.02992.x. [DOI] [PubMed] [Google Scholar]
- Siemann E, Rogers WE. Genetic differences in growth of an invasive tree species. Ecol. Lett. 2001;4:514–518. [Google Scholar]
- St. Louis VL, Barlow JC. Genetic differentiation among ancestral and introduced populations of the Eurasian tree sparrow (Passer montanus. Evolution. 1988;42:266–276. doi: 10.1111/j.1558-5646.1988.tb04131.x. [DOI] [PubMed] [Google Scholar]
- Stearns SC. A natural experiment in life-history evolution: field data on the introduction of mosquitofish (Gambusia affinis) to Hawaii. Evolution. 1983;37:601–617. doi: 10.1111/j.1558-5646.1983.tb05577.x. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Self fertilization and population variability in the higher plants. Am. Nat. 1957;91:337–354. [Google Scholar]
- Thomas SM, Murray BG. Breeding systems and hybridization in Petrorhagia sect. KohlrauschiaCaryophyllaceae. Plant Syst. Evol. 1981;139:77–94. [Google Scholar]
- Thompson JA. An improved non-cryogenic transport and storage preservative facilitating DNA extraction from ‘difficult’ plants collected at remote sites. Telopea. 2002;9:755–760. [Google Scholar]
- Tsutsui ND, Suarez AV, Holway DA, Case TJ. Reduced genetic variation and the success of an invasive species. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5948–5953. doi: 10.1073/pnas.100110397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss N, Eckstein RL, Durka W. Range expansion of a selfing polyploid plant despite widespread genetic uniformity. Ann. Bot. 2012;110:585–593. doi: 10.1093/aob/mcs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Caballero A, Keightley PD, Hill WG. Bottleneck effect on genetic variance: a theoretical investigation of the role of dominance. Genetics. 1998;150:435–447. doi: 10.1093/genetics/150.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. Population size and long-term selection. Plant Breed. Rev. 2004;24:249–268. [Google Scholar]
- Willi Y, Hoffmann J, Van Buskirk AA. Limits to the adaptive potential of small populations. Annu. Rev. Ecol. Evol. Syst. 2006;37:433–458. [Google Scholar]
- Williams CK, Moore RJ. Phenotypic adaptation and natural selection in the wild rabbit, Oryctolagus cuniculus, in Australia. J. Anim. Ecol. 1989;58:495–507. [Google Scholar]
- Willis JH, Orr HA. Increased heritable variation following population bottlenecks: the role of dominance. Evolution. 1993;47:949–957. doi: 10.1111/j.1558-5646.1993.tb01249.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The genetical structure of natural populations. Annal. Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Yonekura R, Kawamura K, Uchii K. A peculiar relationship between genetic diversity and adaptability in invasive exotic species: bluegill sunfish as a model species. Ecol. Res. 2007;22:911–919. [Google Scholar]
- Zenger KR, Richardson BJ, Vachot-Griffin AM. A rapid population expansion retains genetic diversity within European rabbits in Australia. Mol. Ecol. 2003;12:789–794. doi: 10.1046/j.1365-294x.2003.01759.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann H, Ritz CM, Hirsch H, Renison D, Wesche K, Hensen I. Highly reduced genetic diversity of Rosa rubiginosa L. populations in the invasive range. Int. J. Plant Sci. 2010;171:435–446. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


