Abstract
In the past decade, the study of dispersal of marine organisms has shifted from focusing predominantly on the larval stage to a recent interest in adult movement. Antitropical distributions provide a unique system to assess vagility and dispersal. In this study, we have focused on an antitropical wrasse genus, Semicossyphus, which includes the California sheephead, S. pulcher, and Darwin's sheephead, S. darwini. Using a phylogenetic approach based on mitochondrial and nuclear markers, and a population genetic approach based on mitochondrial control region sequences and 10 microsatellite loci, we compared the phylogenetic relationships of these two species, as well as the population genetic characteristics within S. pulcher. While S. pulcher and S. darwini are found in the temperate eastern Pacific regions of the northern and southern hemispheres, respectively, their genetic divergence was very small (estimated to have occurred between 200 and 600 kya). Within S. pulcher, genetic structuring was generally weak, especially along mainland California, but showed weak differentiation between Sea of Cortez and California, and between mainland California and Channel Islands. We highlight the congruence of weak genetic differentiation both within and between species and discuss possible causes for maintenance of high gene flow. In particular, we argue that deep and cooler water refugia are used as stepping stones to connect distant populations, resulting in low levels of genetic differentiation.
Keywords: Antitropicality, microsatellites, Semicossyphus, sheephead wrasse, speciation, stepping stones
Introduction
In marine fishes, population structuring at large scales is generally weak due to high effective population sizes and/or high migration rates. Similarly, speciation in the sea is thought to be counteracted by high gene flow enabled by dispersive larval forms and a rarity of strong physical barriers to dispersal and intermixing (Rocha and Bowen 2008; Puebla 2009; Bernardi 2013). Attempts at predicting population structure and gene flow among populations of marine fishes based on a number of variables, in particular the pelagic larval duration (PLD) of a given species, mostly resulted in contradictory findings (Waples 1987; Doherty et al. 1995; Shulman and Bermingham 1995; Riginos and Victor 2001; Selkoe and Toonen 2011). While correlations have been tenuous, the methods used to test these predictors have been compromised by the inherent constraints of the metrics and methods used rather than a necessarily weak relationship in nature (Weersing and Toonen 2009; Faurby and Barber 2012).
For marine organisms, dispersal was long thought to be principally achieved via a pelagic larval stage, and although larval dispersal likely plays an important role in shaping genetic patterns, evidence accumulated over the past decade indicates that local retention, in particular for fishes, is more important than once thought (Jones et al. 1999; Swearer et al. 1999; Almany et al. 2007; Saenz-Agudelo et al. 2009a; Beldade et al. 2012; Bernardi et al. 2012; Berumen et al. 2012). With the primacy of larval dispersal diminishing, the roles of ecological characteristics and dispersal of adult stages in shaping genetic population structure has in turn, taken a more important place (Schinske et al. 2010; Luiz et al. 2012). Systems where adult dispersal is likely to play a determining role are therefore important to assess. The case of antitropical distributions, for example, has long been puzzling to biogeographers and marine ecologists. For these taxa, which are present at high latitudes but absent from the intertropical regions, several scenarios of dispersal and vicariance have been proposed (Lindberg 1991). While several cases of antitropicality have been described, it has been argued that the tropical eastern pacific (TEP) is a region where tropical submergence (where deeper cooler water is found below the warm surface water) is likely to have played an important role by allowing fish to traverse the equator via the short, steep continental shelf in the eastern Pacific (Hubbs 1952; Lindberg 1991). Indeed, studies on fish species have shown a genetic link between populations in the Southern and Northern Hemisphere in the TEP (Stepien and Rosenblatt 1996; Bowen and Grant 1997). In this study, we assessed the potential for deep-water stepping stones to genetically connect Semicossyphus populations.
Semicossyphus is an antitropical fish genus in the family Labridae (Wrasses). Wrasses include a large number of predominantly coral reef species (approximately 600 species), with a basal tribe, the Hypsigenyines, that includes the temperate genus Semicossyphus, and its close relatives, the genera Bodianus and Clepticus, which are mostly found on coral reefs (Westneat and Alfaro 2005; Beldade et al. 2009). Semicossyphus includes only three species, the Asian sheephead (S. reticulatus), Darwin's sheephead (S. darwini), and the California sheephead (S. pulcher). The Asian sheephead is found in Japan, Korea, and China (Masuda et al. 1984; Froese and Pauly 2000). Darwin's sheephead, one of the few fish species Charles Darwin collected in the Galapagos Islands (Pauly 2004), is found in deeper cooler waters of the southern and western Galapagos Islands, and coastal areas of Ecuador, Peru, and Chile (Allen and Robertson 1994; Grove and Lavenberg 1997). The California sheephead is found from Monterey Bay, California, to the northern Sea of Cortez, Mexico, including the California Channel islands and the isolated Guadalupe Island, Mexico. Semicossyphus pulcher was originally described as a disjunct species, where individuals are found in the northern Sea of Cortez and the northwestern Pacific coast of the Baja California Peninsula but absent from the southern Sea of Cortez and Baja California (Miller and Lea 1972; Present 1987). However, this species does occur (albeit rarely) as far south as Cabo San Lucas (the southern tip of Baja California) (Bernardi et al. 2003) therefore exhibiting a continuous range from Monterey Bay to the northern Sea of Cortez. This occurs most likely via deeper water where food resources and more homogeneous habitat is conducive to adult S. pulcher dispersal (Bernardi et al. 2003).
Sheephead are reef fish that feed mostly on benthic invertebrates such as sea urchins, gastropods, and octopus, which are resources that are found in both shallow and deeper waters, thus permitting sheephead to roam between shallow and deeper habitats (Hamilton et al. 2011). They are protogynous hermaphrodites (like most wrasses, Kazancioğlu and Alonzo 2010), with juvenile and Initial Phase (IP, female) forms looking very similar in the three species, while the Terminal Phase (TP, male) appears to be similar, except for coloration, in S. pulcher and S. darwini, but looks very different in S. reticulatus (Fig. 1). Semicossyphus are broadcast spawners and consequently produce pelagic larvae that remain in the water column for approximately 30 days, thus allowing, at least theoretically, for long-distance dispersal and high gene flow (Warner 1975; Cowen 1985; Victor 1986; Siegel et al. 2003; Andrews and Anderson 2004; Caselle et al. 2011; Hamilton et al. 2011).
Figure 1.
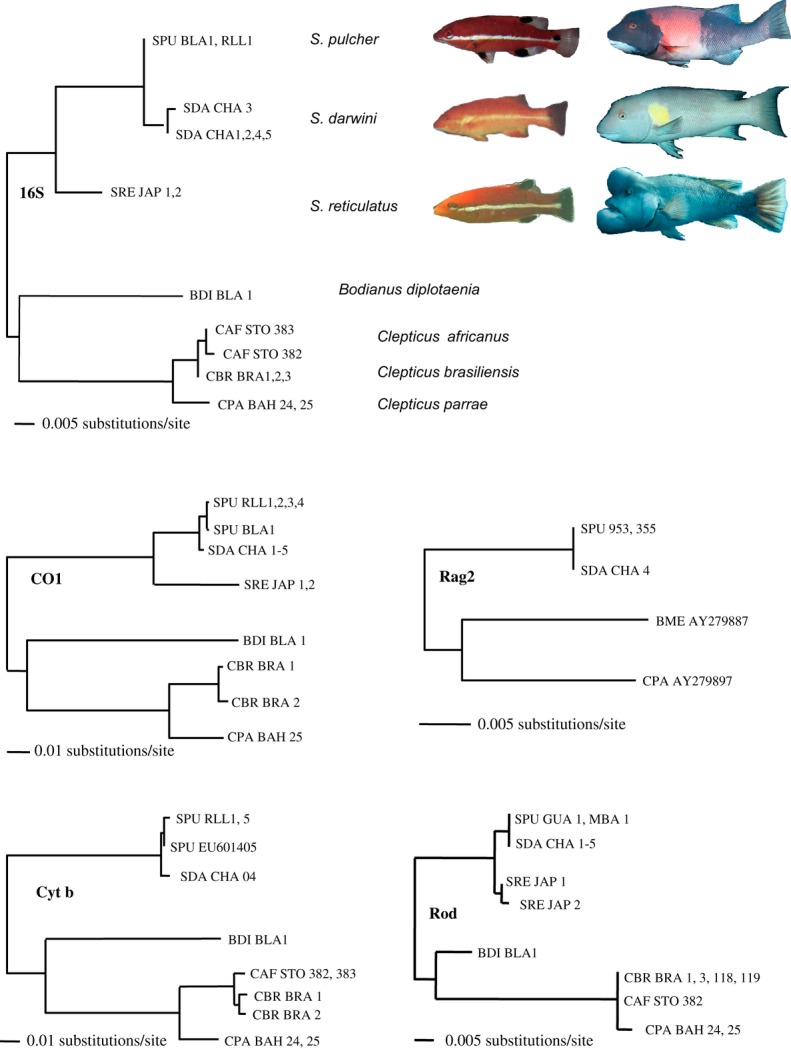
Phylogenetic relationships of the genus Semicossyphus based on three mitochondrial (16SrRNA, 16S; Cytochrome oxydase 1, CO1; Cytochrome b, Cytb) and two nuclear (Recombination activation factor 2, Rag2; Rhodopsin, Rod) markers. All three Semicossyphus species were used (S. pulcher, S. darwini, S. reticulatus). The two closest genera, Bodianus (B. diplotaenia), and Clepticus (C. africanus, C. parrae, C. brasiliensis) were used as outgroups. Pictures of juvenile (left) and terminal phase adult (right) Semicossyphus are shown to emphasise the similarity among juveniles of all three species and adult S. pulcher and S. darwini.
The goal of this study was to assess phylogeographic patterns in the California sheephead, Semicossyphus pulcher and relate these patterns to its antitropical sister taxon, S. darwini. We used a phylogenetic approach using DNA sequences of three mitochondrial and two nuclear markers from all three Semicossyphus species and representatives of the two closest genera, Bodianus and Clepticus as outgroups. We used a population genetic approach using mitochondrial DNA sequences of the hypervariable control region and 10 microsatellite markers on individuals of S. pulcher collected across the entire range of the species, from the Monterey bay to the northern Sea of Cortez, including the California Channel Islands and Guadalupe Island.
Materials and Methods
Collection of samples and DNA extraction
Samples from 499 S. pulcher were collected from 20 locations spanning the entire range of the species, from Monterey Bay, California, to the Sea of Cortez, Mexico, including all major representative offshore islands (Table 1). Samples from five S. darwini were collected from Chile, and samples from two S. reticulatus from Japan (Table 1). Samples of the outgroup species Clepticus africanus, C. braziliensis, C. parrae, and Bodianus diplotaenia were collected from Sao Tomé, Brazil, the Bahamas, and Mexico, respectively (Table 1). DNA was extracted following a standard chloroform protocol (Sambrook et al. 1989).
Table 1.
Sampling of Semicossyphus, Clepticus, and Bodianus. Columns correspond to collection localities, locality codes, and sample numbers for mitochondrial sequences and microsatellite analysis
| Species sampling locality | Code | mtDNA | Microsats |
|---|---|---|---|
| Semicossyphus pulcher (SPU) | California Sheephead | ||
| USA | |||
| California Mainland | |||
| Monterey bay | MOB | 1 | 2 |
| Palos Verdes | PVE | 3 | 54 |
| Point Loma | PTL | 50 | |
| California Channel Islands | |||
| San Miguel | SMI | 19 | |
| Santa Rosa Island | SRI | 33 | |
| Santa Cruz Island | CRU | 43 | |
| Santa Catalina Island | CAT | 20 | 40 |
| San Nicolas Island | SNI | 18 | 46 |
| San Clemente Island | SCL | 20 | 38 |
| Mexico Baja California, Islands | |||
| Isla San Martin | ISM | 38 | |
| Isla Cedros | CED | 20 | |
| Isla Guadalupe | GUA | 13 | 35 |
| Baja California, Pacific Coast | |||
| Bahia Tortugas | BTO | 25 | 48 |
| Bahia Asuncion | ASU | 2 | 2 |
| Punta Canoas | CAN | 18 | |
| Lopez Mateos | LOM | 43 | |
| Sea of Cortez | |||
| Puerto Peñasco | PPE | 4 | 4 |
| Bahia de Los Angeles | BLA | 5 | 10 |
| Bahia San Francisquito | SFR | 5 | 11 |
| Los Frailes | LFR | 2 | 2 |
| Total | SOC | 16 | 27 |
| Semicossyphus darwini (SDA) | Darwin's sheephead | ||
|---|---|---|---|
| Chile | |||
| Caleta Chanaral | CHA | 5 | 5 |
| Semicossyphus reticulatus (SRE) | Asian sheephead | ||
|---|---|---|---|
| Japan | |||
| Iyo, Ehime | SRE | 2 | |
| Outgroups | |||
| Clepticus parrae (CPA) | Creole wrasse | ||
| Bahamas | BAH | 2 | |
| Clepticus braziliensis (CBR) | Brazil's Creole wrasse | ||
| Brazil | BRA | 3 | |
| Clepticus africanus (CAF) | African Creole wrasse | ||
| Sao Tome | STO | 2 | |
| Bodianus diplotaneia (BDI) | Mexican hogfish | ||
| Mexico | |||
| Bahia de Los Angeles | BLA | 1 | |
Phylogenetics
PCR amplification and sequencing
Mitochondrial cytochrome b (CYB), cytochrome oxidase I (CO1), and 16SrRNA (16S) were amplified for a subset of S. pulcher samples (six samples from four locations) and for all samples from the other species via PCR using primers VF2T1 and VR1dT1, 16SAR and 16SBR, and GLUDG-L and CB3H, respectively (Kocher et al. 1989; Palumbi 1996; Ivanova et al. 2007). For the same subset of samples, amplification of the nuclear RAG2 was performed using the primers RAG2F1 and RAG2R3 (Lovejoy 2000). Amplification of the nuclear rhodopsin marker (Rod) followed published nested amplification protocols (Sevilla et al. 2007), with RHO30F and RHO 319R for the first set of primers and Rho F2x and RhoR4n for the second set of primers.
All amplifications were performed in 13 μl reactions containing 0.5 μl of DNA, 0.625 μl of each primer (forward–reverse) and 11.25 ml of Thermo scientific 1.1 × PCR master mix (2.5 mmol/L MgCl2). After an initial denaturation of 1 to 3 min, 30–35 cycles at 94°C for 45 s, followed by 45 s at an annealing temperature of 52–56°C and 60 s at 72°C were conducted, followed by a final extension of 3 min at 72°C. After purification following the manufacturer's protocol (ABI, Perkin-Elmer, Foster City, CA), sequencing was performed with the primers used in the PCR amplification on an ABI 3100 automated sequencer (Applied Biosystems, Foster City, CA) at University of California Berkeley. The putative nature of each sequence was confirmed by BLASTN search. In the case of the nuclear markers, heterozygous individuals were scored using IUPAC ambiguity codes.
Phylogenetic analysis
Sequences were trimmed and aligned using the MAFFT (Katoh et al. 2002) routine implemented in Geneious 5.0 (Biomatters, Auckland, New Zealand). For CYB, CO1, 16S, RAG2, and Rod genes, jModeltest 2 (Guindon and Gascuel 2003; Darriba et al. 2012) was used to determine the substitution model that best fit the data based on the corrected Akaike Information Criterion. Maximum-likelihood analyses of each of these genes were performed in GARLI 2.0 (Zwickl 2006), with priors set to fit the evolutionary model suggested by jModeltest, but allowing the parameters to be recalculated during the run. Each of five independent runs was automatically terminated after 10,000 generations without improvement in topology. The support was evaluated with 100 bootstrap replicates.
Bayesian phylogenetic analyses of these same genes were run in MrBayes 3.1 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003) setting priors to fit the evolutionary model suggested by jModeltest but allowing the parameters to be recalculated during the run.
Population genetics
PCR amplification and genotyping
The hypervariable mitochondrial control region was amplified for a subset of 175 S. pulcher samples from 15 locations (Table 1) and all S. darwini samples using the PCR primers CRA and CRE (Lee et al. 1995). A total of ten microsatellite loci were amplified for all 499 collected S. pulcher samples following published protocols (Poortvliet et al. 2009). Scoring of peaks was performed manually using GENEMAPPER 3.7 (Applied Biosystems). Deviations from Hardy–Weinberg equilibrium and presence of null alleles and linkage disequilibrium were estimated using ARLEQUIN 3.11 (Excoffier et al. 2005) and MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004).
Population genetic analysis
A haplotype network based on mitochondrial control region sequences of S. pulcher samples only (175 samples) was generated in R using HaploNet in the APE 3.0-9 package (Paradis et al. 2004) combined with pie diagrams of haplotype frequencies obtained with APE and ARLEQUIN. Population genetic parameters (Fst and Φst) were calculated with ARLEQUIN, and values of Dst and Jost's D were calculated using GENODIVE (Meirmans and Van Tienderen 2004). Analyses of Molecular Variance (AMOVA) (Excoffier et al. 1992) were computed using the ARLEQUIN package.
To explore and decompose the genetic variability of microsatellite loci into gene pools without providing prior information on the geographic origin of the samples, a Bayesian clustering approach implemented in STRUCTURE 2.2 was used (Pritchard et al. 2000). The program simultaneously defines clusters and assigns individual multilocus genotypes to the defined clusters. Allele frequencies were presumed uncorrelated, and null alleles were coded as recessive to take into account the presence of null alleles in the dataset (Falush et al. 2007). The most likely number of clusters in the dataset was identified based on 10 runs using the Evanno method and visualised in STRUCTURE HARVESTER (Pritchard et al. 2000; Evanno et al. 2005; Earl and VonHoldt 2012). In addition, GENODIVE's K-means clustering was run for number of clusters (K) from 1 to N-2 using AMOVA-based simulated annealing with 50,000 steps and 20 repeats. Cluster membership was examined to determine whether adjacent sampling sites clustered together, illuminating where genetic breaks between regions might exist. Because Fst estimators can be insensitive when gene flow and allelic diversity are high, we also used the program SAShA (Kelly et al. 2010) to detect geographically restricted alleles and test for panmixia. Population structure in control region sequences and microsatellite genotypes was evaluated using an Analysis of Molecular Variance (AMOVA) implemented in ARLEQUIN. Several alternative groupings (California versus Sea of Cortez, California Channel Islands versus all other sampling locations and Southern Mexican islands versus all other sampling locations) were considered.
Results
Phylogenetic reconstructions
All phylogenetic reconstructions showed S. pulcher as a very closely related sister species to S. darwini, with S. reticulatus being distantly related to these two species (Fig. 1), regardless of marker or reconstruction method used. The sequence divergence between S. reticulatus and the other two species varied between 6.1% (CO1) and 4.0% (16S). The sequence divergence between S. pulcher and S. darwini was 0% for the nuclear markers (i.e., no differences at the RAG2 and Rod loci) and less than 1% for the mitochondrial markers (0.57%, 0.61%, and 0.86% for CO1, 16S, and CYB, respectively). Considering a universal substitution rate of 1.5 to 2.5% per million year in fish cytochrome b sequences (Meyer 1994), and a substitution rate of 1.2% per million year based on 19 trans-Isthmian geminate species of fish CO1 sequences (Bermingham et al. 1997; Marko 2002), the divergence time between California sheephead and Darwin sheephead was estimated at approximately 344–573 kya for cytochrome b and 475 kya for CO1.
Mitochondrial control region sequences
Sample numbers, number of haplotypes, haplotype diversity, and nucleotide diversity are given in Table 2. We obtained two sequences for S. reticulatus, five sequences of S. darwini, and 175 sequences of S. pulcher. As for the other molecular markers, S. pulcher and S. darwini were closely related, while S. reticulatus was very distantly related to the two sister species. Sequence divergence between S. reticulatus and S. pulcher + S. darwini was 27.5%. All five S. darwini individuals had different haplotypes from each other and none of these five haplotypes were shared with S. pulcher (Fig. 2). Samples of S. darwini grouped together in a monophyletic assemblage due to five point mutations (3 fixed, 2 nearly fixed) that separated S. darwini from S. pulcher (corresponding to a sequence divergence of 2.1%). Considering a substitution rate of 10% per million years for fish control regions (Domingues et al. 2005, 2006; Drew and Barber 2012), the divergence of S. darwini and S. pulcher based on control region sequences was estimated at approximately 210 kya.
Table 2.
Characteristics of the mitochondrial control region in Semicossyphus pulcher and S. darwini. Locality codes are given in Table 1
| Locality | n | Number of haplotypes | Haplotype diversity (standard deviation) | Nucleotide diversity (standard deviation) |
|---|---|---|---|---|
| MOB | 1 | 1 | 1.0000 (0.0000) | 0.0000 (0.0000) |
| PVE | 3 | 2 | 0.6667 (0.3143) | 0.0017 (0.0021) |
| SMI | 19 | 8 | 0.6725 (0.1190) | 0.0037 (0.0026) |
| CAT | 20 | 7 | 0.5842 (0.1270) | 0.0027 (0.0021) |
| SNI | 18 | 10 | 0.7647 (0.1079) | 0.0034 (0.0025) |
| SCL | 20 | 10 | 0.8316 (0.0751) | 0.0032 (0.0023) |
| CED | 20 | 11 | 0.8053 (0.0903) | 0.0037 (0.0026) |
| GUA | 13 | 5 | 0.6282 (0.1431) | 0.0034 (0.0026) |
| BTO | 25 | 11 | 0.6933 (0.1034) | 0.0139 (0.0024) |
| ASU | 2 | 2 | 1.0000 (0.5000) | 0.0127 (0.0139) |
| CAN | 18 | 4 | 0.3137 (0.1376) | 0.0020 (0.0017) |
| PPE | 4 | 2 | 0.6667 (0.2401) | 0.0017 (0.0019) |
| BLA | 5 | 5 | 1.0000 (0.1265) | 0.0051 (0.0040) |
| SFR | 5 | 5 | 1.0000 (0.1265) | 0.0122 (0.0083) |
| LFR | 2 | 2 | 1.0000 (0.5000) | 0.0101 (0.0113) |
| SDA | 5 | 5 | 1.0000 (0.1265) | 0.0061 (0.0046) |
Figure 2.
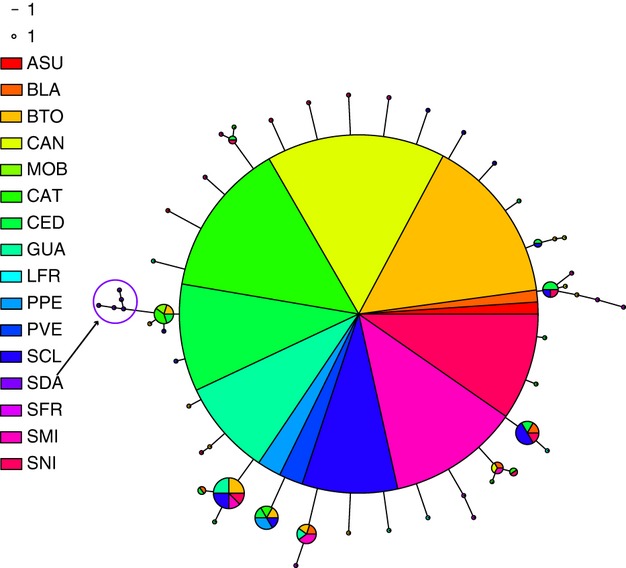
Haplotype network of Semicossyphus pulcher and S. darwini based on the mitochondrial control region (D-loop). Populations are color-coded, the size of the pies are proportional to their corresponding haplotype frequency. Population codes are given in Table 1 and Figure 3.
Microsatellite analyses
We analyzed microsatellites for 504 individuals. We were able to obtain microsatellite scores for all 10 loci for 499 S. pulcher and five S. darwini individuals. Specific characteristics of the microsatellite data used here are provided in Table 3. Loci were neither out of HWE nor in Linkage Disequilibrium.
Table 3.
Microsatellite characteristics for Semicossyphus pulcher and S. darwini. Locality codes are given in Table 1
| Locality (sample #) | A4 | A7 | A109 | C7 | D2 | D101 | D106 | D113 | D118 | D120 | Private alleles | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MOB 2 | Na | 1 | 3 | 4 | 3 | 1 | 1 | 4 | 1 | 3 | 4 | 0 |
| Hobs | N/A | 0.5 | 1 | 1 | N/A | N/A | 1 | N/A | 1 | 1 | ||
| Hexp | N/A | 0.83 | 1 | 0.83 | N/A | N/A | 1 | N/A | 0.83 | 1 | ||
| PVE 54 | Na | 4 | 10 | 15 | 6 | 6 | 7 | 6 | 3 | 7 | 15 | 1 |
| Hobs | 0.61 | 0.91 | 0.74 | 0.72 | 0.41 | 0.59 | 0.61 | 0.59 | 0.65 | 0.89 | ||
| Hexp | 0.54 | 0.85 | 0.77 | 0.64 | 0.41 | 0.64 | 0.72 | 0.51 | 0.69 | 0.91 | ||
| PTL 50 | Na | 3 | 8 | 12 | 6 | 4 | 8 | 6 | 3 | 9 | 14 | 1 |
| Hobs | 0.68 | 0.92 | 0.8 | 0.74 | 0.4 | 0.56 | 0.6 | 0.48 | 0.61 | 0.82 | ||
| Hexp | 0.6 | 0.86 | 0.77 | 0.67 | 0.38 | 0.55 | 0.62 | 0.5 | 0.64 | 0.89 | ||
| SRI 33 | Na | 3 | 10 | 11 | 5 | 4 | 6 | 5 | 3 | 10 | 12 | 2 |
| Hobs | 0.58 | 0.94 | 0.82 | 0.5 | 0.48 | 0.61 | 0.45 | 0.64 | 0.7 | 0.85 | ||
| Hexp | 0.53 | 0.84 | 0.82 | 0.6 | 0.45 | 0.62 | 0.6 | 0.52 | 0.65 | 0.88 | ||
| CRU 43 | Na | 3 | 10 | 13 | 5 | 5 | 7 | 5 | 3 | 11 | 14 | 1 |
| Hobs | 0.58 | 0.88 | 0.72 | 0.58 | 0.51 | 0.61 | 0.56 | 0.49 | 0.72 | 0.98 | ||
| Hexp | 0.6 | 0.84 | 0.83 | 0.6 | 0.42 | 0.55 | 0.65 | 0.5 | 0.64 | 0.9 | ||
| CAT 40 | Na | 3 | 8 | 10 | 5 | 5 | 6 | 6 | 3 | 6 | 14 | 0 |
| Hobs | 0.55 | 0.82 | 0.73 | 0.6 | 0.4 | 0.67 | 0.52 | 0.54 | 0.77 | 0.98 | ||
| Hexp | 0.58 | 0.84 | 0.79 | 0.62 | 0.41 | 0.64 | 0.68 | 0.52 | 0.67 | 0.89 | ||
| SNI 46 | Na | 3 | 10 | 10 | 6 | 5 | 8 | 5 | 3 | 11 | 14 | 1 |
| Hobs | 0.53 | 0.83 | 0.78 | 0.52 | 0.52 | 0.67 | 0.5 | 0.5 | 0.74 | 0.93 | ||
| Hexp | 0.56 | 0.83 | 0.79 | 0.57 | 0.43 | 0.62 | 0.61 | 0.5 | 0.72 | 0.89 | ||
| SCL 38 | Na | 3 | 9 | 8 | 6 | 5 | 7 | 5 | 3 | 8 | 14 | 1 |
| Hobs | 0.7 | 0.84 | 0.71 | 0.55 | 0.57 | 0.53 | 0.79 | 0.74 | 0.78 | 0.87 | ||
| Hexp | 0.59 | 0.83 | 0.73 | 0.5 | 0.53 | 0.59 | 0.72 | 0.54 | 0.67 | 0.91 | ||
| ISM 38 | Na | 3 | 11 | 10 | 5 | 4 | 7 | 6 | 2 | 7 | 14 | 2 |
| Hobs | 0.53 | 0.84 | 0.78 | 0.6 | 0.38 | 0.4 | 0.6 | 0.27 | 0.54 | 0.84 | ||
| Hexp | 0.52 | 0.88 | 0.78 | 0.63 | 0.39 | 0.47 | 0.65 | 0.48 | 0.58 | 0.89 | ||
| GUA 35 | Na | 3 | 9 | 10 | 5 | 4 | 6 | 5 | 3 | 7 | 15 | 1 |
| Hobs | 0.54 | 0.83 | 0.8 | 0.66 | 0.24 | 0.76 | 0.71 | 0.32 | 0.69 | 0.89 | ||
| Hexp | 0.57 | 0.86 | 0.78 | 0.65 | 0.32 | 0.65 | 0.67 | 0.46 | 0.7 | 0.9 | ||
| BTO 48 | Na | 3 | 10 | 10 | 6 | 5 | 6 | 6 | 3 | 10 | 14 | 0 |
| Hobs | 0.42 | 0.81 | 0.9 | 0.64 | 0.44 | 0.51 | 0.53 | 0.5 | 0.64 | 0.94 | ||
| Hexp | 0.57 | 0.86 | 0.81 | 0.68 | 0.38 | 0.53 | 0.68 | 0.49 | 0.64 | 0.91 | ||
| ASU 2 | Na | 2 | 3 | 3 | 3 | 1 | 2 | 2 | 1 | 3 | 4 | 0 |
| Hobs | 1 | 0.5 | 1 | 1 | N/A | 0.5 | 0.5 | N/A | 0.5 | 1 | ||
| Hexp | 0.67 | 0.83 | 0.83 | 0.83 | N/A | 0.5 | 0.5 | N/A | 0.83 | 1 | ||
| LOM 43 | Na | 4 | 9 | 10 | 6 | 4 | 7 | 5 | 3 | 9 | 15 | 1 |
| Hobs | 0.6 | 0.98 | 0.74 | 0.5 | 0.35 | 0.62 | 0.51 | 0.5 | 0.61 | 0.83 | ||
| Hexp | 0.54 | 0.83 | 0.74 | 0.57 | 0.33 | 0.64 | 0.68 | 0.53 | 0.66 | 0.9 | ||
| SOC 27 | Na | 3 | 9 | 10 | 4 | 5 | 7 | 5 | 3 | 5 | 13 | 0 |
| Hobs | 0.59 | 0.7 | 0.78 | 0.38 | 0.44 | 0.58 | 0.44 | 0.48 | 0.59 | 0.74* | ||
| Hexp | 0.61 | 0.84 | 0.74 | 0.61 | 0.4 | 0.6 | 0.59 | 0.58 | 0.58 | 0.88 | ||
| SDA 5 | Na | 1 | 3 | 5 | 3 | 2 | 6 | 4 | 2 | 3 | 8 | 12 |
| Hobs | N/A | 0.2* | 0.5 | 0.2* | 0.2 | 0.6 | 0.6 | 0.2 | 0.6 | 1 | ||
| Hexp | N/A | 0.69 | 0.86 | 0.73 | 0.2 | 0.87 | 0.73 | 0.2 | 0.64 | 0.96 | ||
Na= number of alleles; Hobs= observed heterozygosities; Hexp= expected heterozygosities of microsatellite loci per population.
Indicates significant deviation from Hardy Weinberg Equilibrium after Bonferroni-type corrections.
N/A indicates monomorphic, did not test.
Microsattlite loci names are given in the first row, number of private alleles are given in the right column.
As expected, all approaches separated the two species, S. pulcher and S. darwini, in two genetic clusters based on microsatellite data. The S. darwini samples showed 12 private alleles that are absent in S. pulcher, while the two most differentiated S. pulcher populations, SRI and SMI (northern Channel Islands), had only 2 private alleles (Table 3).
Population genetics
Mitochondrial control region sequences
Sample numbers, number of haplotypes, haplotype diversity, and nucleotide diversity are given in Table 2. One predominant S. pulcher haplotype was found in 93 individuals, from which other haplotypes or groups of haplotypes stemmed (Fig. 2). The haplotype network did not present any obvious geographic pattern; however, haplotype frequencies did show geographic patterns that could be discerned visually when placed on a map (Fig. 3). Indeed, while most locales did include the most common (grey) haplotype, ten of 15 sites had unique (black) haplotypes.
Figure 3.
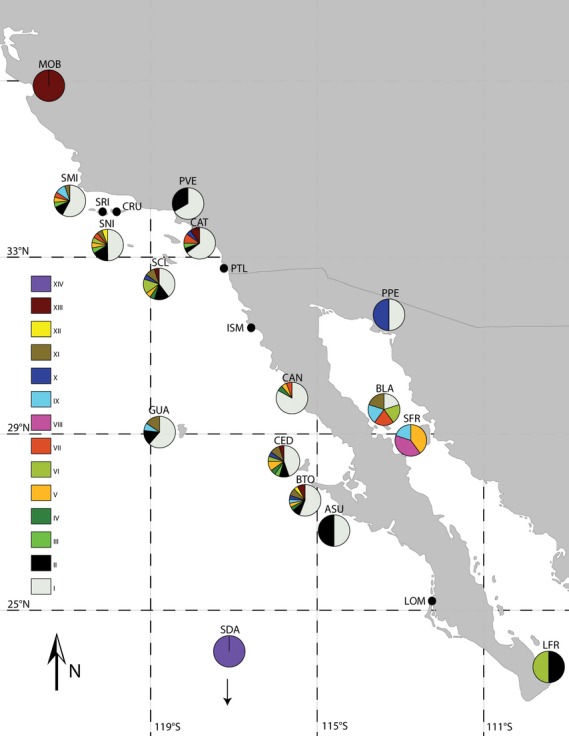
Sampling locations of California sheephead, Semicossyphus pulcher, and mitochondrial control region haplotypes. The most common haplotype is represented in grey, private haplotypes (only found in a given population) are represented in black. The remaining 13 haplotypes are color-coded and shown on Figure S1 as an overlay of the haplotype network of Figure 2. Solid black dots indicate additional sampling locations for microsatellites. Monterey Bay, MOB; San Miguel Island, SMI; Santa Rosa Island, SRI; Santa Cruz Island, CRU; San Nicolas Island, SNI; Santa Catalina Island, CAT; San Clemente Island, SCL; Palos Verdes, PVE; Point Loma, PTL; Isla San Martin, ISM, Punta Canoas, CAN; Isla Guadalupe, GUA; Isla Cedros, CED; Bahia Tortuga, BTO; Bahia Asuncion, ASU; Lopez Mateos, LOM; Los Frailes, LFR; San Francisquito, SFR; Bahia de Los Angeles, BLA; Puerto Peñasco, PPE.
Because S. pulcher has traditionally been considered a Sea of Cortez disjunct species (Present 1987; Bernardi et al. 2003), we first performed an Analysis of Molecular Variance (AMOVA) by separating California and Sea of Cortez populations into two separate groups. In this case, 9.1% of the total variance could be attributed to this partition, which was statistically significant (Φct = 0.091, P = 0.018). We then clustered the California Channel Islands as a separate group, because these islands have also been considered a place where genetic differences may arise (e.g., Bernardi 2000, 2005). There, only 1.1% of the total variance could be assigned to this grouping; yet, this value was still statistically significant ((Φct = 0.011, P = 0.024). Finally, we considered the southern Mexican islands of Cedros and Guadalupe as another group, as recruitment dynamics at Guadalupe Island have been suggested as potentially being linked with major oceanographic shifts (Cowen 1985). In this case, however, the variance attributed to this grouping was zero (Φct = −0.87) and was not statistically significant (P = 0.11). Within California samples, no pairwise comparisons of Φst or Fst performed in ARLEQUIN were found to be statistically significant and neither were measures of Jost's D using GENODIVE.
Microsatellite analyses
Because only 5 Semicossyphus darwini were sampled, these were not included in the population analyses based on microsatellites. AMOVA analyses of S. pulcher microsatellite data did not result in any significant population structure. When we separated California and Sea of Cortez populations into two groups, we found that only 0.02% of the total variance could be attributed to this partition, a result that was also not statistically significant. Clustering the California Channel Islands or the Mexican offshore islands into separate groups also yielded nonsignificant differences, with 0.12% and 0.06% of the variance assignable to these partitions, respectively. Results from STRUCTURE were consistent with the AMOVA (Fig. 4), in showing a lack of population structure of S. pulcher within California and so was the GENODIVE's K-means clustering analysis. The lack of concordance between mitochondrial and microsatellite results, however, is not entirely unusual and has repeatedly been observed before (DiBattista et al. 2012).
Figure 4.
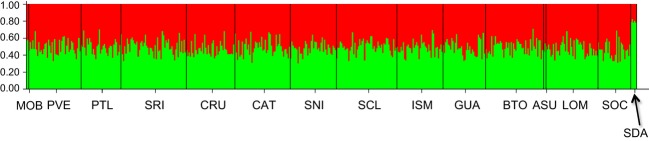
Bayesian population assignment test based on 10 microsatellites loci. Highest likelihood was found when data were partitioned in two clusters (K = 2) represented in green and red. Each vertical line represents one individual and its assignment likelihood to belong to one of the cluster (Y scale) is shown by the color. Black vertical lines represent the limit between predefined groups (populations). Population codes are given in Table 1 and Figure 1.
Discussion
In marine systems, assessing population structure and diversity is complicated by the dispersive (i.e., pelagic) larval stage of most benthic organisms, a life stage that is difficult to fully assess (Paulay and Meyer 2006). Dispersal along the eastern Pacific Coast of South and North America for the sand crab Emerita analoga was ascribed to its long larval phase (Dawson et al. 2011). Similarly, two of three pelagic fishes tested (Stepien and Rosenblatt 1996), as well as sardines (Bowen and Grant 1997), show strong genetic connectivity between Southern and Northern Hemispheres via the coastal Tropical Eastern Pacific. In the case of the sardines, where antitropical populations diverged very recently, some haplotypes were even shared between Chile and California (Bowen and Grant 1997). Here, a very similar pattern of antitropical dispersal was shown for Semicossyphus darwini and S. pulcher, two species that may have diverged as recently as 200–600 kya. Currently, there is no known suitable habitat that intervenes their respective distributions to provide a means for stepping stone gene flow via adult migrations. The species' 1-month pelagic larval duration is unlikely to enable regular larval exchange across the tropics. Nevertheless, the phylogenetic similarity of the species suggests the potential for occasional high dispersal across the tropics, perhaps during extreme storm events. Our analysis of the population genetic structure of S. pulcher also suggests high dispersal potential, providing one example consistent with the idea that gene flow levels within a species may correlate with levels between species.
Population structure and speciation in marine organisms
In general, marine organisms have large effective population sizes and have traditionally been considered mobile or at least with large dispersal potential. This is particularly true at the larval stage, where larvae are carried away from reefs by oceanographic currents. This view, that predicts shallow population structure, was challenged in the late 1990s when fish larvae were found to be retained (self-recruitment) at much higher rates than expected (Jones et al. 1999; Swearer et al. 1999). Indeed, later empirical studies have shown that a significant portion of the larval pool recruits close to the parental habitat and display behaviors that challenge the notion that they are passive dispersers (Jones et al. 2005; Gerlach et al. 2007; Planes et al. 2009; Saenz-Agudelo et al. 2009b; Beldade et al. 2012; Bernardi et al. 2012; Berumen et al. 2012). Thus, combining oceanographic factors together with larval behavior is an approach that is likely to better reflect the mechanisms of dispersal in marine species (Selkoe et al. 2010; White et al. 2010; Alberto et al. 2011; Selkoe and Toonen 2011). In addition, the ecological characteristics of adults, a factor traditionally neglected in attempts to predict population structure in marine organisms, have regained importance (Schinske et al. 2010; Luiz et al. 2012). In turn, such ecological factor may play an important population genetic role over long-time scales that would influence speciation mechanisms. Within this context, the goal of our study was to determine whether speciation-level patterns (macroevolution) matched population-level patterns (microevolution). As discussed above, dispersal across the tropics in sister species of sheephead indicates that occasional long-distance dispersal is likely in these species. Population structure analysis of the California sheephead, S. pulcher, is consistent with this result, with a near absence of any population structure across a geographic area that covers approximately 2,500 km of coastline from Monterey, California, to the northern Sea of Cortez.
Dispersal capabilities and antitropical connections
Several attempts have been made at identifying the factors that influence population structure in marine organisms. While it is clear that in extreme cases, major oceanographic or physical barriers play an overriding role in structuring populations of entire biotas (Bernardi et al. 2003; Lessios and Robertson 2009; Toonen et al. 2011; Von der Heyden et al. 2011), other, more moderate situations may not be so simple. Indeed, predicting population structure has been a challenge. Here, we argue that the ecological and evolutionary mechanisms responsible for the shallow population structure observed in Semicossyphus pulcher are similar to the factors responsible for the low genetic separation between S. pulcher and its sister species S. darwini. We assume here that some key life history, evolutionary, and ecological factors combined with abiotic factors such as historical and oceanographic features, must play an important role.
As noted above, S. pulcher has been considered a disjunct species, with continuous populations in California and the west coast of Baja California, and an isolated disjunct population in the northern Sea of Cortez (Present 1987; Thomson et al. 2000), yet careful field and archival examination showed that this species is found continuously along the Baja California peninsula, albeit in deeper water, where it probably feeds on invertebrates, escapes ecological competition from shallow tropical species, and does not experience warm surface temperatures (Bernardi et al. 2003). In fact, S. pulcher is only seen in the shallow waters of the northern Sea of Cortez during the cold winter months and is absent during the warm summer months, when it presumably migrates to deeper, colder waters (G. Bernardi. pers. obs.). The physical connection of continuous populations via deeper waters is therefore a likely conduit to genetic connectivity in this species, resulting in shallow population structure.
The lack of large genetic separation between S. pulcher and S. darwini, two species that display antitropical distributions, may be due to several potential factors. Stepping stones as a mode of distant connectivity have been evoked for a long time (Kimura 1953; Kimura and Weiss 1964; Hellberg et al. 2002; Purcell et al. 2006; Rocha and Bowen 2008; White et al. 2010) and recently were explicitly tested using oceanographic models in a marine system (Crandall et al. 2012). Similarly to the situation described for S. pulcher, deep-water refugia between the distribution ranges of S. pulcher and S. darwini may have acted as stepping stones in the past. Indeed, oceanographic models of such habitats were analyzed and locations of deep-water refugia in the Tropical Eastern Pacific were predicted (Graham et al. 2007; Santelices 2007). When predictive models were tested in the field, deep-water kelp beds were found by scuba divers, and more relevant to our study, S. darwini individuals were observed there (Graham et al. 2007). It is therefore conceivable that gene flow between antitropical populations remained active until recently, most likely via deep-water refugia, resulting in small genetic divergence between these species in mitochondrial markers and no observed differences in the nuclear markers. In addition, the genetic divergence between S. darwini and S. pulcher, estimated to have occurred approximately 344–573 kya, encompasses the time of glaciating periods (300–455 kya). During these times, the water temperature in the tropical region was lowered, further increasing the potential for temperate fishes to breach that boundary (Chiang and Bitz 2005). The presence of fixed differences in the mitochondrial marker and several private alleles in the microsatellite markers suggests that gene flow between these species has currently stopped.
Adaptation, Fisheries and Conservation
Sheephead are large fishes that are targeted by both commercial and sport fishermen in Chile, California, and Asia (Hamilton et al. 2007; Selkoe et al. 2007; Godoy et al. 2010; Caselle et al. 2011), thus identifying structured populations would inform managers to help conserve different stocks. Our data show a lack of genetic structure for S. pulcher populations in California, suggesting either a genuine panmictic scenario, or structure that is difficult to identify. While we do not know whether California sheephead are panmictic, some significant metrics (e.g., between the Sea of Cortez and the California coast, AMOVA, φct= 0.091, P = 0.018) suggest that structure may be present. In other systems, such as hake or cod, population structure that was not uncovered in early surveys was shown to be present when more powerful molecular markers, such as SNPs, were used, and discrete stocks and fundamental population patterns were revealed (Moen et al. 2008; Milano et al. 2011). Those results, often associated with genes under selection, are indicative of local adaptation, a factor that may ultimately be responsible for the evolution of Sea of Cortez populations and the separation between S. darwini and S. pulcher.
While our study only examines a single system, it provides a rare example where macroevolution mirrors microevolution. The ability of S. pulcher to achieve high gene flow, which reduces population structuring within the species, is also the most likely explanation for the very low divergence between sister species S. darwini and S. pulcher. Lack of large physical distances between suitable habitats likely enables stepping stone dispersal that counteracts divergence due to local adaptation. Despite the appearance of great physical distance between the ranges of S. darwini and S. pulcher, the presence of deep-water habitat may facilitate occasional exchange. The marine environments of Chile and California have often been compared due to their very similar characteristics, with cold currents (Humboldt and California, respectively), high productivity, similar fish assemblages, and kelp forests (Stepien 1990; Boyle and Horn 2006; Perez-Matus et al. 2012). In that respect, the Semicossyphus system offers a unique opportunity to understand which factors are most influential in shaping the structure of marine populations.
Acknowledgments
We would like to thank Stuart Banks and Paul Tompkins for discussions on the ecology of Darwin's sheephead in the Galàpagos Archipelago and Reiji Masuda for discussion on the ecology of the Asian sheephead in Japan. We would like to thank Ziyusei Kanamoto and Keiichi Sakai for providing samples of Semicossyphus reticulatus. We would like to thank Andrea Marchesi, Javier Portocarrero, Scott Hamilton, Milton Love, Donna Schroeder and a large number of PISCO students and technicians for collections in California and Baja California, Niora Fabian and Jessica Holsman for help in the laboratory. We would like to thank Helmo Perez, Universidad Catolica del Norte, Chile for use of Darwin's sheephead picture. Funding for this project was provided by a University of California Marine Council Coastal Environmental Quality Initiative (CEQI) grant, the Biological Resource Division USGS, California Sea Grant, National Marine Sanctuaries MOA 2005-008 / 66832 (KAS), and PISCO, the Partnership for Interdisciplinary Studies of Coastal Oceans, which is supported primarily by the Gordon and Betty Moore Foundation and the David and Lucile Packard Foundation. This is contribution number 442 from PISCO and contribution number 1568 from the Hawaii Institute of Marine Biology.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Haplotype network of Semicossyphus pulcher and S. darwini based on the mitochondrial control region (D-loop), as shown in Figure 2, but where each haplotype is described with a different color. The most common haplotype is in grey. Haplotypes that are unique to a population are in black. This haplotype network was used to generate the colors presented in Figure 3.
References
- Alberto F, Raimondi PT, Reed DC, Watson JR. 2011. pp. 2543–2554. Isolation by oceanographic distance explains genetic structure for Macrocystis pyrifera in the Santa Barbara Channel. [DOI] [PubMed]
- Allen GR,, Robertson DR. Fishes of the tropical eastern Pacific. Honolulu, Hawaii: University of Hawaii Press; 1994. [Google Scholar]
- Almany GR, Berumen ML, Thorrold SR, Planes S, Jones GP. Local replenishment of coral reef fish populations in a marine reserve. Science. 2007;316:742–744. doi: 10.1126/science.1140597. (New York, N.Y.) [DOI] [PubMed] [Google Scholar]
- Andrews KS,, Anderson TW. Habitat-dependent recruitment of two temperate reef fishes at multiple spatial scales. Mar. Ecol. Prog. Ser. 2004;277:231–244. [Google Scholar]
- Beldade R, Heiser JB, Robertson DR, Gasparini L, Floeter S, Bernardi G. Historical biogeography and speciation in the Creole wrasses (Labridae, Clepticus) Mar. Biol. 2009;156:679–687. doi: 10.1007/s00227-008-1118-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beldade R, Holbrook SJ, Schmitt RJ, Planes S, Malone D, Bernardi G. Larger female fish contribute disproportionately more to self-replenishment. Proc. Biol. Sci. 2012;279:2116–2121. doi: 10.1098/rspb.2011.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham E, McCafferty S, Martin AP. Fish biogeography and molecular clocks: perspectives from the Panamanian Isthmus. In: Kocher TD, Stepien CA, editors. Molecular systematics of fishes. New York: Academic Press; 1997. pp. 113–128. [Google Scholar]
- Bernardi G. Barriers to gene flow in Embiotoca jacksoni, a marine fish lacking a pelagic larval stage. Evolution. 2000;54:226–237. doi: 10.1554/0014-3820(2000)054[0226:BTGFIE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Phylogeography and demography of sympatric sister surfperch species, Embiotoca jacksoni and E. lateralis along the California coast: historical versus ecological factors. Evolution. 2005;59:386–394. [PubMed] [Google Scholar]
- Bernardi G. Speciation in fishes. Mol. Ecol. 2013 doi: 10.1111/mec.12494. (in press) [DOI] [PubMed] [Google Scholar]
- Bernardi G, Findley L, Rocha-Olivares A. Vicariance and dispersal across Baja California in disjunct marine fish populations. Evolution. 2003;57:1599–1609. doi: 10.1111/j.0014-3820.2003.tb00367.x. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Beldade R, Holbrook SJ, Schmitt RJ. Full-sibs in cohorts of newly settled coral reef fishes. PLoS ONE. 2012;7:e44953. doi: 10.1371/journal.pone.0044953. (SCA Ferse, Ed,) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berumen ML, Almany GR, Planes S, Jones GP, Saenz-Agudelo P, Thorrold SR. Persistence of self-recruitment and patterns of larval connectivity in a marine protected area network. Ecol. Evol. 2012;2:444–452. doi: 10.1002/ece3.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen BW,, Grant WS. Phylogeography of the sardines (Sardinops SPP.): assessing biogeographic models and population histories in temperate upwelling zones. Evolution. 1997;51:1601–1610. doi: 10.1111/j.1558-5646.1997.tb01483.x. [DOI] [PubMed] [Google Scholar]
- Boyle K,, Horn M. Comparison of feeding guild structure and ecomorphology of intertidal fish assemblages from central California and central Chile. Mar. Ecol. Prog. Ser. 2006;319:65–84. [Google Scholar]
- Caselle JE, Hamilton SL, Schroeder DM, Love MS, Standish JD, Rosales-Casian JA, et al. Geographic variation in density, demography, and life history traits of a harvested, sex-changing, temperate reef fish. Can. J. Fish. Aquat. Sci. 2011;68:288–303. [Google Scholar]
- Chiang JCH,, Bitz CM. Influence of high latitude ice cover on the marine Intertropical Convergence Zone. Clim. Dyn. 2005;25:477–496. [Google Scholar]
- Cowen RK. Large-scale pattern of recruitment by the labrid, Semicossyphus pulcher - Causes and implications. J. Mar. Res. 1985;43:719–742. [Google Scholar]
- Crandall ED, Treml EA, Barber PH. Coalescent and biophysical models of stepping-stone gene flow in neritid snails. Mol. Ecol. 2012;21:5579–5598. doi: 10.1111/mec.12031. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MN, Barber PH, González-Guzmán LI, Toonen RJ, Dugan JE, Grosberg RK. Phylogeography of Emerita analoga (Crustacea, Decapoda, Hippidae), an eastern Pacific Ocean sand crab with long-lived pelagic larvae. J. Biogeogr. 2011;38:1600–1612. [Google Scholar]
- DiBattista JD, Rocha LA, Craig MT, Feldheim KA, Bowen BW. Phylogeography of two closely related Indo-Pacific butterflyfishes reveals divergent evolutionary histories and discordant results from mtDNA and microsatellites. J. Hered. 2012;103:617–629. doi: 10.1093/jhered/ess056. [DOI] [PubMed] [Google Scholar]
- Doherty PJ, Planes S, Mather P. Gene flow and larval duration in seven species of fish from the Great Barrier Reef. Ecology. 1995;76:2373–2391. [Google Scholar]
- Domingues VS, Bucciarelli G, Almada VC, Bernardi G. Historical colonization and demography of the Mediterranean damselfish, Chromis chromis. Mol. Ecol. 2005;14:4051–4063. doi: 10.1111/j.1365-294X.2005.02723.x. [DOI] [PubMed] [Google Scholar]
- Domingues VS, Santos RS, Brito A, Almada VC. Historical population dynamics and demography of the Eastern Atlantic pomacentrid Chromis limbata (Valenciennes, 1833) Mol. Phylogenet. Evol. 2006;40:139–147. doi: 10.1016/j.ympev.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Drew JA, Barber PH. Comparative phylogeography in Fijian coral reef fishes: a multi-taxa approach towards marine reserve design. PLoS ONE. 2012;7:e47710. doi: 10.1371/journal.pone.0047710. (M V. Matz, Ed,) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA,, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012;4:359–361. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol. Ecol. Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurby S,, Barber PH. Theoretical limits to the correlation between pelagic larval duration and population genetic structure. Mol. Ecol. 2012;21:3419–3432. doi: 10.1111/j.1365-294X.2012.05609.x. [DOI] [PubMed] [Google Scholar]
- Froese R,, Pauly D. FishBase 2000: concepts, design and data sources. Philippines, Los Baños, Laguna: ICLARM; 2000. [Google Scholar]
- Gerlach G, Atema J, Kingsford MJ, Black KP, Miller-Sims V. Smelling home can prevent dispersal of reef fish larvae. Proc. Natl Acad. Sci. USA. 2007;104:858–863. doi: 10.1073/pnas.0606777104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy N, Gelcich S, Vasquez JA, Castilla JC. Spearfishing to depletion : evidence from temperate reef fishes in Chile. Ecol. Appl. 2010;20:1504–1511. doi: 10.1890/09-1806.1. [DOI] [PubMed] [Google Scholar]
- Graham MH, Kinlan BP, Druehl LD, Garske LE, Banks S. Deep-water kelp refugia as potential hotspots of tropical marine diversity and productivity. Proc. Natl Acad. Sci. USA. 2007;104:16576–16580. doi: 10.1073/pnas.0704778104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove JS, Lavenberg RJ. The fishes of the galápagos Islands. Stanford, CA: Stanford Univ. Press; 1997. [Google Scholar]
- Guindon S,, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hamilton SL, Caselle JE, Standish JD, Schroeder DM, Love MS, Rosales-Casian JA, et al. Size-selective harvesting alters life histories of a temperate sex-changing fish. Ecol. Appl. 2007;17:2268–2280. doi: 10.1890/06-1930.1. [DOI] [PubMed] [Google Scholar]
- Hamilton S, Caselle J, Lantz C, Egloff TL, Kondo E, Newsome SD, et al. Extensive geographic and ontogenetic variation characterizes the trophic ecology of a temperate reef fish on southern California (USA) rocky reefs. Mar. Ecol. Prog. Ser. 2011;429:227–244. doi: 10.3354/meps09086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg ME, Burton RS, Neigel JE, Palumbi SR. Genetic assessment of connnectivity among marine populations. Bull. Mar. Sci. 2002;70:273–290. [Google Scholar]
- Hubbs CL. 1952;3:324–330. Antitropical distribution of fishes and other organisms. Proceeedings of the 7th Pacific Science Congress. [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. (Oxford, England) [DOI] [PubMed] [Google Scholar]
- Ivanova NV, Zemlak TS, Hanner RH, Hebert PDN. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes. 2007;7:544–548. [Google Scholar]
- Jones GP, Milicich MJ, Emslie MJ, Lunow C. Self-recruitment in a coral reef fish population. Nature. 1999;402:802–804. [Google Scholar]
- Jones GP, Planes S, Thorrold SR. Coral reef fish larvae settle close to home. Curr. Biol. 2005;15:1314–1318. doi: 10.1016/j.cub.2005.06.061. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment basedon fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazancioğlu E,, Alonzo SH. A comparative analysis of sex change in Labridae supports the size advantage hypothesis. Evolution. 2010;64:2254–2264. doi: 10.1111/j.1558-5646.2010.01016.x. [DOI] [PubMed] [Google Scholar]
- Kelly RP, Oliver TA, Sivasundar A, Palumbi SR. A method for detecting population genetic structure in diverse, high gene-flow species. J. Hered. 2010;101:423–436. doi: 10.1093/jhered/esq022. [DOI] [PubMed] [Google Scholar]
- Kimura M. 1953;3:62–63. Stepping-stone model of population. Annual Report of the National Institute of Genetics. [Google Scholar]
- Kimura M,, Weiss GH. The stepping stone model of population structure and the decrease of genetic correlation with distance. Genetics. 1964;49:561–576. doi: 10.1093/genetics/49.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, et al. Dynamics of mitochondrial DNA evolution in animals : amplification and sequencing with conserved primers. Proc. Natl Acad. Sci. USA. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Conroy J, Howell WH, Kocher TD. Structure and evolution of teleost mitochondrial control regions. J. Mol. Evol. 1995;41:54–66. doi: 10.1007/BF00174041. [DOI] [PubMed] [Google Scholar]
- Lessios HA,, Robertson DR. Crossing the impassable : genetic connections in 20 reef fishes across the eastern Pacific barrier Crossing the impassable : genetic connections in 20 reef fishes across the eastern Pacific barrier. Proc. Biol. Sci. 2009;273:2201–2208. doi: 10.1098/rspb.2006.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg DR. Marine biotic interchange between the northern and southern hemispheres. Paleobiology. 1991;17:308–324. [Google Scholar]
- Lovejoy NR. Reinterpreting recapitulation : systematics of needlefishes and their allies (Teleostei: Beloniformes) Evolution. 2000;54:1349–1362. doi: 10.1111/j.0014-3820.2000.tb00567.x. [DOI] [PubMed] [Google Scholar]
- Luiz OJ, Madin JS, Robertson DR, Rocha LA, Wirtz P, Floeter SR. Ecological traits influencing range expansion across large oceanic dispersal barriers: insights from tropical Atlantic reef fishes. Proc. Biol. Soc. B: Biol. Sci. 2012;279:1033–1040. doi: 10.1098/rspb.2011.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko PB. Fossil calibration of molecular clocks and the divergence times of geminate species pairs separated by the isthmus of panama. Mol. Biol. Evol. 2002;19:2005–2021. doi: 10.1093/oxfordjournals.molbev.a004024. [DOI] [PubMed] [Google Scholar]
- Masuda H, Amaoka K, Araga C, Uyeno T, Yoshino T. The fishes of the Japanese Archipelago. Tokyo, Japan: Tokai Univ. Press; 1984. [Google Scholar]
- Meirmans PG, Van Tienderen PH. Genotype and Genodive: two programs for the analysis of genetic diversity of asexual organisms. Mol. Ecol. Notes. 2004;4:792–794. [Google Scholar]
- Meyer A. Shortcomings of the cytochrome b gene as a molecular marker. Trends Ecol. Evol. 1994;9:278–280. doi: 10.1016/0169-5347(94)90028-0. [DOI] [PubMed] [Google Scholar]
- Milano I, Babbucci M, Panitz F, Ogden R, Nielsen RO, Taylor MI, et al. Novel tools for conservation genomics: comparing two high-throughput approaches for SNP discovery in the transcriptome of the European hake. PLoS ONE. 2011;6:e28008. doi: 10.1371/journal.pone.0028008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DJ,, Lea RN. Guide to coastal marine fishes of California. Fish bulletin. Berkeley, CA: Department of Fish and Game; 1972. p. 157. [Google Scholar]
- Moen T, Hayes B, Nilsen F, Delghandi M, Fjalestad KT, Fevolden SE, et al. Identification and characterisation of novel SNP markers in Atlantic cod: evidence for directional selection. BMC Genet. 2008;9:18. doi: 10.1186/1471-2156-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR. PCR and molecular systematics. In: Hillis D, Moritz C, Mable B, editors. Molecular systematics. Sunderland, MA: Sinauer Press; 1996. pp. 205–247. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Paulay G,, Meyer C. Dispersal and divergence across the greatest ocean region: do larvae matter? Integr. Comp. Biol. 2006;46:269–281. doi: 10.1093/icb/icj027. [DOI] [PubMed] [Google Scholar]
- Pauly D. Darwin's fishes: an encyclopedia of ichthyology, ecology and evolution. Cambridge: Cambridge Univ. Press; 2004. [Google Scholar]
- Perez-Matus A, Pledger S, Diaz FJ, Ferry LA, Vasquez JA. Plasticity in feeding selectivity and trophic structure of kelp forest associated fishes from northern Chile. Rev. Chil. Hist. Nat. 2012;85:29–48. [Google Scholar]
- Planes S, Jones GP, Thorrold SR. Larval dispersal connects fish populations in a network of marine protected areas. Proc. Natl Acad. Sci. USA. 2009;106:5693–5697. doi: 10.1073/pnas.0808007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortvliet M, Olsen JL, Selkoe KA, Coyer JA, Bernardi G. Isolation and characterization of 11 microsatellite primers for a temperate reef fish, the California sheephead (Semicossyphus pulcher. Mol. Ecol. Resour. 2009;9:429–430. doi: 10.1111/j.1755-0998.2008.02467.x. [DOI] [PubMed] [Google Scholar]
- Present TMC. Genetic differentiation of disjunct Gulf of California and Pacific outer coast populations of Hypsoblennius jenkinsi. Copeia. 1987;1987:1010–1024. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puebla O. Ecological speciation in marine v. freshwater fishes. J. Fish Biol. 2009;75:960–996. doi: 10.1111/j.1095-8649.2009.02358.x. [DOI] [PubMed] [Google Scholar]
- Purcell JFH, Cowen RK, Hughes CR, Williams DA. Weak genetic structure indicates strong dispersal limits: a tale of two coral reef fish. Proc. Biol. Sci. 2006;273:1483–1490. doi: 10.1098/rspb.2006.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riginos C,, Victor BC. Larval spatial distributions and other early life-history characteristics predict genetic differentiation in eastern Pacific blennioid fishes. Proc. Biol. Sci. 2001;268:1931–1936. doi: 10.1098/rspb.2001.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha LA,, Bowen BW. Speciation in coral-reef fishes. J. Fish Biol. 2008;72:1101–1121. [Google Scholar]
- Ronquist F,, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Saenz-Agudelo P, Jones GP, Thorrold SR, Planes S. Estimating connectivity in marine populations: an empirical evaluation of assignment tests and parentage analysis under different gene flow scenarios. Mol. Ecol. 2009;18:1765–1776. doi: 10.1111/j.1365-294X.2009.04109.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santelices B. The discovery of kelp forests in deep-water habitats of tropical regions. Proc. Natl Acad. Sci. USA. 2007;104:19163–19164. doi: 10.1073/pnas.0708963104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinske JN, Bernardi G, Jacobs DK, EJ R. Phylogeography of the diamond turbot (Hypsopsetta guttulata) across the Baja California Peninsula. Mar. Biol. 2010;157:123–134. doi: 10.1007/s00227-009-1302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe K,, Toonen R. Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 2011;436:291–305. [Google Scholar]
- Selkoe K, Vogel A, Gaines S. Effects of ephemeral circulation on recruitment and connectivity of nearshore fish populations spanning Southern and Baja California. Mar. Ecol. Prog. Ser. 2007;351:209–220. [Google Scholar]
- Selkoe KA, Watson JR, White C, Horin TB, Toonen RJ. Taking the chaos out of genetic patchiness : seascape genetics reveals ecological and oceanographic drivers of genetic patterns in three temperate reef species. Mol. Ecol. 2010;19:3708–3726. doi: 10.1111/j.1365-294X.2010.04658.x. [DOI] [PubMed] [Google Scholar]
- Sevilla RG, Diez A, Norén M, Mouchel O, Jerome M, Verrez-Bagnis V, et al. Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol. Ecol. Notes. 2007;7:730–734. [Google Scholar]
- Shulman MJ,, Bermingham E. Early life histories, ocean currents, and the population genetics of Caribbean reef fishes. Evolution. 1995;49:897–910. doi: 10.1111/j.1558-5646.1995.tb02325.x. [DOI] [PubMed] [Google Scholar]
- Siegel D, Kinlan B, Gaylord B, Gaines S. Lagrangian descriptions of marine larval dispersion. Mar. Ecol. Prog. Ser. 2003;260:83–96. [Google Scholar]
- Stepien CA, Population structure, diets, and biogeographic relationships of the rocky intertidal fish assemblage in Central Chile: high levels of herbivory in a temperate system. Bull. Mar. Sci. 1990;47:598–612. [Google Scholar]
- Stepien CA, Rosenblatt RH. Genetic divergence in antitropical pelagic marine fishes (Trachurus, Merluccius, and Scomber) between North and South America. Copeia. 1996;1996:586–598. [Google Scholar]
- Swearer SE, Caselle JE, Lea DW, Warner RR. Larval retention and recruitment in an island population of a coral-reef fish. Nature. 1999;402:799–802. [Google Scholar]
- Thomson DA, Findley LT, Kerstitch AN. Reef fishes of the Sea of Cortez. The rocky-shore fishes of the Gulf of California. Tucson, Arizona, USA: The University of Arizona Press; 2000. [Google Scholar]
- Toonen RJ, Andrews KR, Baums IB, Bird CE, Concepcion GT, Daly-Engel TS, et al. Defining boundaries for ecosystem-based management: a multispecies case study of marine connectivity across the Hawaiian Archipelago. J. Mar. Biol. 2011;2011:13. doi: 10.1155/2011/460173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 2004;4:535–538. [Google Scholar]
- Victor BC. Duration of the planktonic larval stage of one hundred species of Pacific and Atlantic wrasses (family Labridae) Mar. Biol. 1986;90:317–326. [Google Scholar]
- Von der Heyden S, Bowie RCK, Prochazka K, Bloomer P, Crane NL, Bernardi G. Phylogeographic patterns and cryptic speciation across oceanographic barriers in South African intertidal fishes. J. Evol. Biol. 2011;24:2505–2519. doi: 10.1111/j.1420-9101.2011.02382.x. [DOI] [PubMed] [Google Scholar]
- Waples RS. A multispecies approach to the analysis of gene flow in marine shore fishes. Evolution. 1987;41:385–400. doi: 10.1111/j.1558-5646.1987.tb05805.x. [DOI] [PubMed] [Google Scholar]
- Warner RR. The reproductive biology of the protogynous hermaphrodite Pimelometopon pulchrum (Pisces: Labridae) Fish. Bull. 1975;73:262–283. [Google Scholar]
- Weersing K, Toonen R. Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 2009;393:1–12. [Google Scholar]
- Westneat MW,, Alfaro ME. Phylogenetic relationships and evolutionary history of the reef fish family Labridae. Mol. Phylogenet. Evol. 2005;36:370–390. doi: 10.1016/j.ympev.2005.02.001. [DOI] [PubMed] [Google Scholar]
- White C, Selkoe KA, Watson J, Siegel DA, Zacherl DC, Toonen RJ. Ocean currents help explain population genetic structure. Proc. Biol. Sci. 2010;277:1685–1694. doi: 10.1098/rspb.2009.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin, TX: The University of Texas at Austin; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


