Abstract
The rat is the preferred animal model in many areas of biomedical research and drug development. Genetic manipulation in rats has lagged behind that in mice due to the lack of efficient gene targeting tools. Previously, we generated a knockout rat via conventional homologous recombination in rat embryonic stem (ES) cells. Here, we show that efficient gene targeting in rat ES cells can be achieved quickly through transcription activator-like effector nuclease (TALEN)-mediated DNA double-strand breaks. Using the Golden Gate cloning technique, we constructed a pair of TALEN targeting vectors for the gene of interest in 5 days. After gene transfection, the targeted rat ES cell colonies were isolated, screened, and confirmed by PCR without the need of drug selection. Our results suggest that TALEN-mediated gene targeting is a superior means of establishing genetically modified rat ES cell lines with high efficiency and short turnaround time.
Keywords: TAL effector, BMPR2, Rat ES cells, Gene-targeting
1. INTRODUCTION
Embryonic stem (ES) cells were first isolated from mouse embryos three decades ago. ES cells can be maintained in culture indefinitely while retaining the capacity to generate nearly any type of cell in the body. The pluripotency of ES cells and the availability of gene-targeting technology have enabled the creation of mouse animal models with a variety of genetic modifications. These models have become important tools for understanding gene function and modeling human diseases (Austin et al., 2004). Although the rat is the preferred animal model in many human health-related research fields, mainly due to its bigger size, more-developed brain and other humanized features (Abbott, 2004), the unavailability of authentic rat ES cells had, until recently, prevented the application of gene-targeting technology in this species. Instead, several alternative methods had been developed to generate genetically modified rats without using ES cells. Those methods include N-ethyl-N-nitrosourea (ENU) mutagenesis (Zan et al., 2003), transposon-tagged mutagenesis (Kitada et al., 2007), and zinc finger nuclease (ZFN)-based pronuclear injection (Geurts et al., 2009). Unfortunately, all of these non-ES methods are expensive and/or difficult to apply.
By using small-molecule inhibitors to block mitogen activated protein kinase (MAPK) and glycogen synthase kinase 3 (GSK3), we successfully derived ES cells from rat embryos in 2008 (Li et al., 2008). A gene knockout rat was generated 2 years later via conventional homologous recombination in rat ES cells (Tong et al., 2010). The application of conventional gene-targeting methodology in rat ES cells makes it possible for researchers to achieve any type of genetic modification in rats, as has been the case in mice for decades. However, the generation of knockout animals via the traditional method is a time-consuming and laborious process, and therefore, a more-efficient tool is preferred for generating knockout rats.
Recently, two independent groups reported that transcription activator-like (TAL) effector, a protein secreted by Xanthomonas bacteria, can bind to specific DNA sequences via repetitive amino acid residues in the central domain (Boch et al., 2009; Moscou and Bogdanove, 2009). It has been shown that the 12th and 13th amino acid residues in sequential repeats actually determine the DNA binding specificity and thereby the TAL effector’s target site. The simple relationship between amino acids in the TAL effector and the DNA bases in its target provides the possibility of engineering TAL effector proteins with an affinity for a pre-determined DNA sequence. Fusion proteins carrying the DNA binding domain of the TAL effector and the DNA cleavage domain of restriction enzyme Fok I can create a double-strand break at a particular genomic site among a wide range of species, from yeast to humans (Li et al., 2011; Miller et al., 2011). These engineered TAL effector nucleases (TALENs) have been successfully applied to disrupt gene function in the rat through pronuclear injection (Tesson et al., 2011).
The assembly of the repeat variable di-residues (RVD) containing a highly conserved repetitive sequence in TALENs, however, is challenging for researchers using the regular cloning method, and chemical synthesis of the entire RVD region is relatively expensive. In 2009, a type IIs restriction enzyme-based DNA cloning method called Golden Gate Shuffling was reported (Engler et al., 2009). Golden Gate cloning allows a plasmid to be assembled from 10 separate input plasmids without the introduction of any mutation. This feature of the technique makes it possible to assemble more than 20 RVDs in just two rounds of cloning. The first successful assembly of pre-designed TALENs using the Golden Gate cloning method was recently performed to target a promoter sequence driving GFP expression in a transgenic plant (Weber et al., 2011). Here, we modified the Golden Gate cloning system and applied it to construct TALENs that can be used to generate gene-targeted rat ES cells with high efficiency.
2. MATERIALS AND METHODS
2.1. DNA cloning
Escherichia coli (E. coli) strains carrying each plasmid included in the Golden Gate cloning system for TALEN construction were kindly provided by Dr. Daniel F. Voytas. Plasmid DNA was extracted using a Mini-prep kit from Qiagen Inc. (USA). Concentrations of all plasmids were adjusted to 150 ng/mL with ddH2O for Golden Gate cloning. All DNA cloning procedures were performed according to the protocols provided on the Addgene Website (http://www.addgene.org/TALeffector/goldengate/voytas/). To appropriately modify the backbone plasmid pTAL3 in the Golden Gate cloning system, pCAG-EGFP-ires-Pac was digested with BamH I and EcoR I to remove the EGFP-ires-Pac cassette and was then ligated with the bGHpolyA fragment generated by PCR amplification to obtain plasmid pCAG-bGHpolyA. The fragment extending from restriction site Bgl II to Afl II in the pTAL3, including the selection marker gene LacZ and cleavage domain of Fok I, was subcloned into pCAG-bGHpolyA via Afl II and BamH I to form pCAG-TAL3. Golden Gate TALNs assembly was carried out as previously described (Cermak et al., 2011).
2.2. Rat ES cell and hepatocellular carcinoma (HCC) cell culture
Rat ES cells were routinely cultured on γ-irradiated mouse embryonic fibroblasts (MEFs) in N2B27 medium supplemented with 3 μmol/L of CHIR99021 and 1 μmol/L of PD0325901 (2i medium). Rat ES cells were passaged every 2–3 days as described previously (Tong et al., 2011). The rat HCC cell line derived from diethylnitrosamine (DEN)-induced rat liver tumors was maintained in N2B27 medium supplemented with 10 ng/mL of EGF, 10 ng/mL of bFGF and 3 μmol/L of CHIR99021. Rat HCC cells were cultured on Matrigel-coated dishes and passaged every 3 days or until 70% confluent.
2.3. Amaxa nucleofection
Rat ES cells were treated with 0.025% trypsin for 5 min to obtain a single cell suspension. After neutralization of trypsin with 10% fetal bovine serum (FBS), 106 rat ES cells were washed with 10 mL of PBS twice. Nucleofection was performed according to the instructions in the Amaxa Mouse ES Cell Nucleofector Kit (Amaxa, VPH-1001, USA). Briefly, 90 μL of Nucleofector solution was mixed with 20 μL of supplement, and 1.5 μg each of the TALEN-expressing plasmid pair pCAG-TAL-F and pCAG-TAL-R was dissolved into 10 μL of the Nucleofector solution mixture. The cell pellet was resuspended with 100 μL of the mixture and then combined with the plasmid solution. Nucleofection program A-13 was applied to introduce pCAG-TAL-F and pCAG-TAL-R into the rat ES cells. After transfection, rat ES cells were seeded into four 100 mm dishes plated with MEF feeder cells and cultured in 10 mL of 2i medium. Single colonies emerged 5 days after nucleofection and were ready for picking up.
2.4. Genomic PCR screening in 96-well plates
Genomic DNA of the isolated rat ES cell colonies was prepared using ZR-96 quick gDNA kit (Zymo Research, USA, D3012) and eluted in 30 μL of ddH2O. 5 μL of DNA solution was used to provide 50–200 ng of genomic DNA for the PCR template. Paq5000 DNA polymerase was used for amplifying the bone morphogenetic protein receptor 2 (BMPR2) target region with the primer pairs F: 5′-AGATGCCATACCCA GATGAGAC-3′ and R: 5′-AGGCTCTGCTGCATTGATTG-3′. PCR thermocycling was as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 30 s and 72°C for 45 s. The expected size of the PCR product was 506 bp and was visualized on 1.5% TAE agarose gel with SYBR (1:10,000) (Invitrogen).
2.5. Histochemical analysis of stem cell markers
The expression of pluripotency markers Oct4 and Nanog was detected by immuno-cytochemistry staining. Cells were fixed in 4% paraformaldehyde and then treated with blocking solution for 2 h, followed by incubation with Oct4 and Nanog primary antibodies overnight at 4°C. Alexa fluorophore-conjugated secondary antibodies were incubated with cells for 2 h at 1:1000 dilutions in a dark room. The cells were washed with 1× PBS-T three times and then examined using fluorescence microscopy.
3. RESULTS AND DISCUSSION
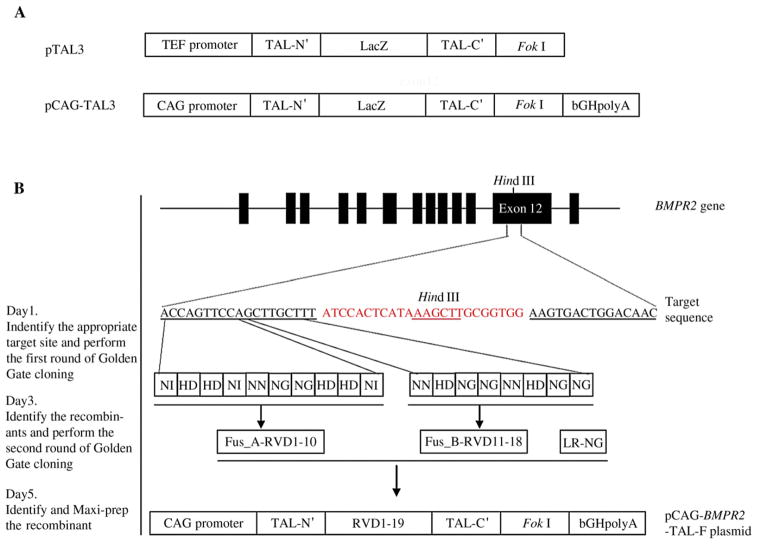
The Golden Gate cloning system allows the construction of TALEN-expressing plasmid pairs in just 5 days (Cermak et al., 2011). However, the system was originally designed for use in yeast and therefore had to be modified for mammalian cell gene targeting applications. To achieve this aim, we first replaced the yeast TEF promoter in the backbone plasmid pTAL3 with a cytomegalovirus early enhancer/chicken β-actin (CAG) promoter. The CAG promoter is frequently used to drive a high level of gene expression in almost all types of mammalian cells, especially ES cells. We then inserted the polyadenylation sequence of the bovine growth hormone (bGHployA) downstream of the TALEN expression cassette (Fig. 1A). bGHpolyA is required for producing mature mRNA in mammalian cells. To ascertain whether this TALEN construct could be successfully applied to rat ES cell gene targeting, we chose to disrupt exon 12 of the rat BMPR2 gene. The DNA sequence of exon 12 was retrieved from the UCSC Genome Browser and input into the website http://boglabx.plp.iastate.edu/TALENT/. A 58 bp target region including a Hind III site was identified by the on-line software, and RVDs were assembled based on the target sequence. The assembled RVDs were cloned into pCAG-TAL3 to generate a TALEN plasmid pair pCAG-BMPR2-TAL-F and pCAG-BMPR2-TAL-R, using two rounds of Golden Gate cloning (Fig. 1B).
Fig. 1. Schematic diagram of the strategy for constructing TALENs via Golden Gate cloning method.
A: the structure of plasmid pTAL3 and pCAG-TAL3. B: the TALEN target is located in exon 12 of BMPR2 with a Hind III site in the spacer region (red). The left and right TALENs binding sequences are underlined. The pCAG-BMPR2-TAL-F was constructed using two rounds of Golden Gate cloning.
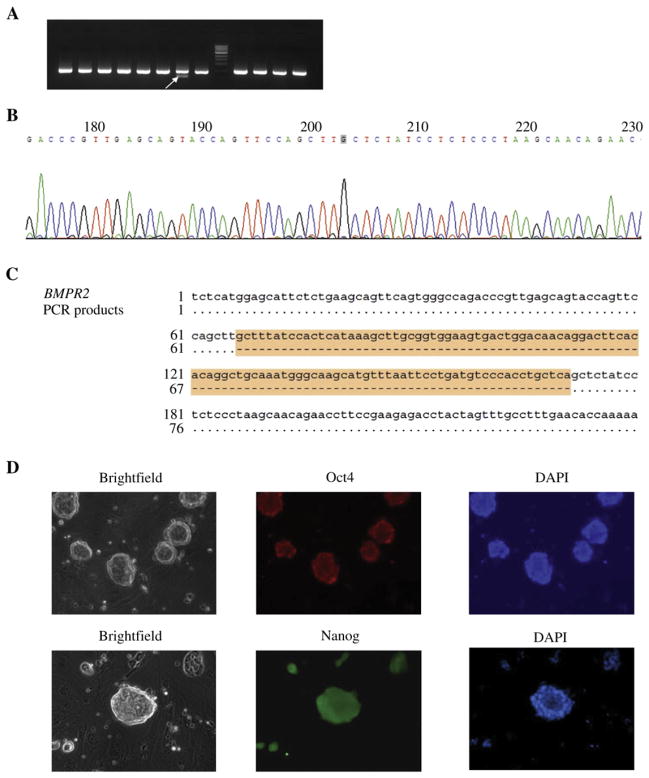
We first tested the ability of the modified TALENs to alter the BMPR2 locus in rat HCC cells. This cell line is characterized by high transfection efficiency. The HCC cells were co-transfected with plasmids expressing the TALEN pair. Genomic DNA was extracted from HCC cells on day 4 after transfection. A 506 bp fragment containing the TALEN target site (Fig. 2A) was amplified by PCR and digested by restriction enzyme Hind III. The PCR products from wild-type rat ES cells were used as a control. Some of the PCR products from TALEN-transfected samples were resistant to Hind III digestion even when a 50-fold excess of the nuclease was used, indicating the targeting event had occurred. PCR products from non-transfected samples were completely digested by Hind III (Fig. 2B). DNA sequencing confirmed that a 9 base-pair TCATAAAGC deletion was created by TALENs (Fig. 2C).
Fig. 2. Confirmation of BMPR2 gene disruption in HCC cells.
A: the PCR strategy for identification of targeting events. As indicated, the primer pair F and R were used to amplify the 506 bp DNA fragment including the TALENs target site. B: the products from samples transfected with TALENs-expressing plasmids were resistant to Hind III digestion (lane 1), whereas non-transfected sample products were completely digested (lane 2). Untreated PCR products were loaded as a control (lane 3 and lane4). M1, 100 bp DNA ladder; M2, 1 kb DNA ladder. C: DNA sequencing results indicated a 9 bp deletion in the spacer region. The two TALEN binding sites are highlighted.
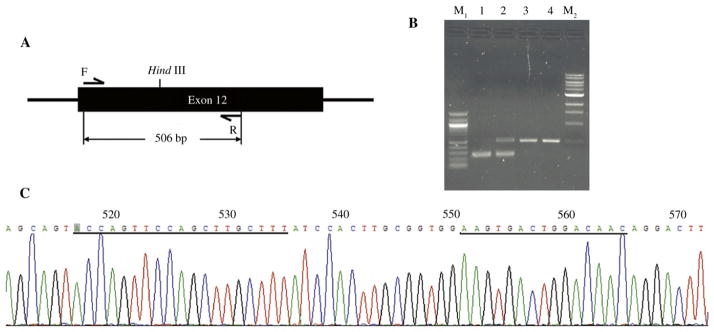
We then turned to carrying out TALEN-mediated gene targeting in rat ES cells. Plasmids expressing TALEN pairs were co-transfected into rat ES cells by nucleofection. The transfected cells were seeded at clonal density. Five days after transfection, we randomly picked up colonies and expanded them individually in 96-well plates. Genomic DNA from each colony was extracted and PCR was performed to amplify the region containing the TALEN target sequence. Among 96 samples on the gel, we found that three of them had an extra band as shown in Fig. 3A, which was gel-retrieved and sequenced. The mutations created by TALENs were confirmed by sequencing the PCR products of extra bands from the DNA samples of the single ES cell colonies. A 104 bp deletion including the whole target site of BMPR2 was found by sequence alignment of the PCR product with rat genome sequence data retrieved from the UCSC website (Fig. 3B and C.). The gene-targeted rat ES cells retained the typical morphology of undifferentiated ES cells and expressed pluripotency markers Oct4 and Nanog (Fig. 3D). Because of the short turnaround time to achieve gene targeting by TALENs, we expect that gene-targeted rat ES cells generated by TAL-ENs will have a greater chance of contributing to the germline than do gene-targeted rat ES cells generated by conventional gene-targeting technologies.
Fig. 3. BMPR2 gene disruption in rat ES cells mediated by TALENs.
A: PCR screening of the rat ES cell colonies. The positive colonies yielded an extra band on the gel, indicated by the arrow. B: DNA sequencing result of the extra band indicated in (A). C: alignment of the sequencing result with the intact BMPR2 sequence indicates a 104 bp deletion (highlighted). D: BMPR2-targeted rat ES cells stained for pluripotency markers Oct4 (top panel) and Nanog (bottom panel). Left panel: phase contrast images; right panel: nuclear counterstain with diamidino-2-phenylindole (DAPI).
In summary, we provide here the first description of a rapid and cost-effective method of gene targeting in rat ES cells by TALEN-mediated DNA double-stand breaks. Unlike the construction of ZFN vectors, construction of TALEN-expressing vectors can be rapidly and conveniently accomplished. Drug selection is unnecessary in TALEN-mediated gene targeting, in contrast with conventional gene targeting by homologous recombination. Moreover, using the modified Golden Gate cloning system, construction of a pair of TALENs targeting any sequence of interest can be completed in just 5 days. The combination of TALEN-mediated gene targeting and ES cell technologies will greatly reduce the time expenditure and cost for generating transgenic and gene knockout rat animal models.
Acknowledgments
We thank Dr. Daniel F. Votas for providing the Golden Gate cloning system and Dr. Colby Starker for technical assistance. This work was supported by a NIH grant to Qi-Long Ying (R01OD010926).
References
- Abbott A. Laboratory animals: the renaissance rat. Nature. 2004;428:464–466. doi: 10.1038/428464a. [DOI] [PubMed] [Google Scholar]
- Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, Grieder FB, Heintz N, Hicks G, Insel TR, Joyner A, Koller BH, Lloyd KC, Magnuson T, Moore MW, Nagy A, Pollock JD, Roses AD, Sands AT, Seed B, Skarnes WC, Snoddy J, Soriano P, Stewart DJ, Stewart F, Stillman B, Varmus H, Varticovski L, Verma IM, Vogt TF, Melchner HV, Witkowski J, Woychik RP, Wurst W, Yancopoulos GD, Young SG, Zambrowicz B. The knockout mouse project. Nat Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009;325:433. doi: 10.1126/science.1172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4:131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- Li P, Tong C, Mehrian-Shai R, Jia L, Wu N, Yan Y, Maxson RE, Schulze EN, Song H, Hsieh CL, Pera MF, Ying Q. Germline competent embryonic stem cells derived from rat blastocysts. Cell. 2008;135:1299–1310. doi: 10.1016/j.cell.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and Fok I DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L, Gregory PD, Rebar EJ. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Tesson L, Usal C, Menoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Tong C, Huang G, Ashton C, Li P, Ying QL. Generating gene knockout rats by homologous recombination in embryonic stem cells. Nat Protoc. 2011;6:827–844. doi: 10.1038/nprot.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Li P, Wu NL, Yan Y, Ying QL. Production of p53 gene knockout rats by homologous recombination in embryonic stem cells. Nature. 2010;467:211–213. doi: 10.1038/nature09368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Gruetzner R, Werner S, Engler C, Marillonnet S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE. 2011;6:e19722. doi: 10.1371/journal.pone.0019722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan Y, Haag JD, Chen KS, Shepel LA, Wigington D, Wang YR, Hu R, Lopez-Guajardo CC, Brose HL, Porter KI, Leonard RA, Hitt AA, Schommer SL, Elegbede AF, Gould MN. Production of knockout rats using ENU mutagenesis and a yeast-based screening assay. Nat Biotechnol. 2003;21:645–651. doi: 10.1038/nbt830. [DOI] [PubMed] [Google Scholar]