Abstract
Understanding antibody function is often enhanced by knowledge of the specific binding epitope. Here, we describe a computer algorithm that permits epitope prediction based on a collection of random peptide epitopes (mimotopes) isolated by antibody affinity purification. We applied this methodology to the prediction of epitopes for five monoclonal antibodies against the West Nile virus (WNV) E protein, two of which exhibit therapeutic activity in vivo. This strategy was validated by comparison of our results with existing F(ab)-E protein crystal structures and mutational analysis by yeast surface display. We demonstrate that by combining the results of the mimotope method with our data from mutational analysis, epitopes could be predicted with greater certainty. The two methods displayed great complementarity as the mutational analysis facilitated epitope prediction when the results with the mimotope method were equivocal and the mimotope method revealed a broader number of residues within the epitope than the mutational analysis. Our results demonstrate that the combination of these two prediction strategies provides a robust platform for epitope characterization.
Keywords: Epitope mapping, Monoclonal antibody, Phage display, Neutralization, Flavivirus
1. Introduction
Structural assessment of target epitopes can provide important insight into antibody (Ab) function. As an example, studies using a panel of mouse monoclonal antibodies (mAbs) to the West Nile virus (WNV) E protein have revealed that Abs which bind to a specific site in domain III (DIII) provide the greatest neutralizing activity in vitro and in vivo (Oliphant et al., 2007). Interestingly, while the E protein is the dominant target for humoral immunity following exposure to WNV, the major response in humans is not targeted to the most potently neutralizing DIII-specific epitope (Throsby et al., 2006; Oliphant et al., 2007). In this regard, knowledge of specific epitopes is useful to understand and manipulate humoral responses. Indeed, epitope mapping studies have uncovered novel neutralizing targets for hepatitis C virus (HCV) and HIV, which possess broad activity across multiple virus genotypes (Johansson et al., 2007; Gustchina et al., 2007).
For targeted vaccine development, structural characterization of Ab recognition sites provides an opportunity to construct immunogens that generate epitope-specific immunity (Meloen et al., 1991; Petrakou et al., 1998). For example, as a result of mapping of the HIV-specific 2F5 monoclonal antibody (mAb) epitope, an immunogen was designed that contained a conformational epitope mimic which successfully elicited a broadly neutralizing anti-HIV-1 antibody response (Ho et al., 2002). The development of epitopespecific vaccines may be particularly important for viruses, such as flaviviruses, where antibody-mediated immune enhancement of infection may contribute to pathogenesis. Development of such vaccines, however, is dependent upon accurate characterization of the mAb epitope.
Several methods have been developed for mapping of Ab epitopes including antigen mutagenesis, screening of random peptide libraries, antibody-antigen co-crystallography and nuclear magnetic resonance spectroscopy (Carter, 1994; Chan et al., 2006; Kwong et al., 1998; Meloen et al., 2003; Oliphant et al., 2006; Scott and Smith, 1990; Shiota et al., 2007; Smith, 1991). Previously, we have mapped large panels of neutralizing and non-neutralizing anti-WNV mAbs using libraries of yeast expressing random E protein mutants on their surface (Oliphant et al., 2005) (Oliphant et al., 2006). However, interpretation of the results from mutational studies can be complicated by conformational effects on distal protein elements (Chien et al., 1989; Huang et al., 2007; Kotik and Zuber, 1993; Nall et al., 1989). To corroborate and extend the findings of our mutational analysis, we employed an alternate mapping strategy that defines epitopes using affinity-selected random peptides (Smith, 1991). Manipulation and identification of random peptides is facilitated by the use of phage display libraries where random 10-mer peptides flanked by cysteine residues are presented as circularized peptides on the phage surface (Scott and Smith, 1990).
Assignment of the random peptides to specific regions on the target protein requires the development of sophisticated computer algorithms that identify unique characteristics within the peptide sequences and the target protein. In this manuscript, we describe a computer algorithm that facilitates epitope mapping using sequence data from affinity-selected random peptides (mimotopes). We have applied this novel algorithm to the prediction of epitopes for five mAbs against the WNV E protein that were previously mapped by yeast surface display and/or X-ray crystallography.
2. Materials and methods
2.1. Monoclonal antibodies
Five monoclonal antibodies were characterized in this manuscript: E16, E24, E60, E113 and E121 (Oliphant et al., 2005) (Oliphant et al., 2006). The antibodies were harvested from hybridoma supernatants, precipitated using NH4SO4, and purified by protein A affinity chromatography, as described previously (Oliphant et al., 2005).
2.2. Screening of phage display epitope library with mAbs
To define mAb epitopes, we screened a library of circularized 10-mer peptides for high-affinity binding to random peptides that were specific for individual mAbs. We used a phage display library presenting random peptides with the structure XC(X)10CX inserted into N-terminus of pVIII major coat protein of filamentous bacteriophage, where X represents any amino acid (kindly provided by Dr. J. Scott, Simon Fraser University) (Bonnycastle et al., 1996). Screening of phage library was performed according to the method described by Smith (Smith, 2006). Candidate peptides were selected by two rounds of iterative affinity selection and specificity of phage for individual mAbs was assessed by dot blot. Briefly, phages were immobilized onto nitrocellulose membrane, the membranes were blocked with 5% non-fat milk in Tris-buffered saline (TBS, pH 7.4) and probed with mAbs (1 μg/ml in TBS/0.5% non-fat milk) followed by incubation with horseradish peroxidase conjugated goat anti-mouse IgG (Jackson Immunoresearch Laboratories Inc. West Grove, Pennsylvania, U.S.A.) and development using ECL (Amersham Biosciences, New Jersey, U.S.A.). Phages that specifically bound individual antibodies were purified and the random inserts were sequenced.
2.3. Molecular modeling
Molecular modeling was performed using Chem3D Ultra10 (CambridgeSoft, Cambridge, Massachusetts U.S.A.) and Swiss-Pdb viewer (GlaxoSmithKline, Brentford, Middlesex, United Kingdom). Three-dimensional models were created by RasMol (http://www.bernstein-plus-sons.com/software/rasmol/doc/ rasmol.html).
2.4. Computer algorithm
Amino acid sequences of affinity-selected phages were used as a database for the computer algorithm to predict mAb epitopes. The original version of our algorithm has been described (Enshell-Seijffers et al., 2003). The rationale of the algorithm is based on the assumption that affinity-selected peptides must share key contact residues with the true epitope in the native protein. In the three-dimensional structure of a protein, amino acids creating the epitope can be discontinuous and become juxtaposed at the surface of the antigen through protein folding. Therefore, we assume that neighboring amino acids within the selected phage-displayed peptides may represent pairs of juxtaposed yet non-linear amino acid residues on the protein surface. Additionally, we assume that the most frequently observed amino acid sequence pairs within a given set of affinity-selected peptides likely represent the contact amino acids within the epitope.
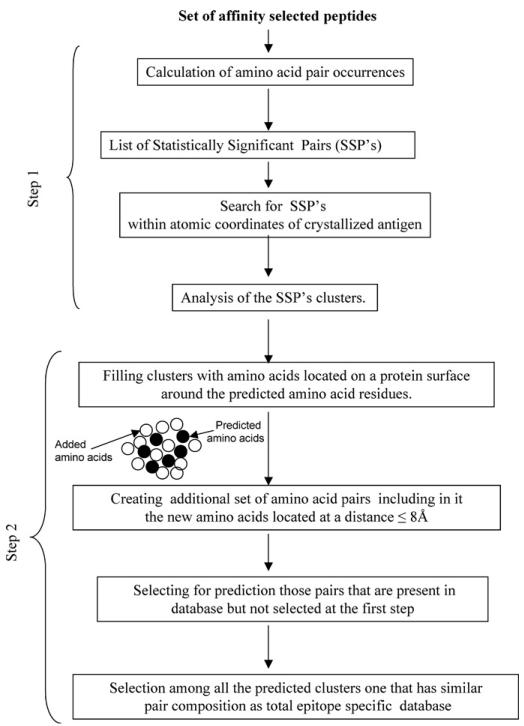
Epitope prediction using the original method is based on statistical considerations and, therefore, a large collection of affinity-selected peptides is required. Furthermore, the predictions from this method were often equivocal. Parameters such as cluster size (number of amino acid pairs predicted to be in the cluster) and diversity (number of discrete amino acid pairings) are very helpful to identify the most probable epitopes. Diversity is a particularly important parameter of the cluster since this parameter reflects epitope complexity and reveals a greater number of discrete residues within the epitope. However, this method typically predicts only a fraction of the epitope. To extend our prediction and encompass a greater fraction of the epitope, we designed a novel algorithm that “fills” the clusters with amino acids located on a protein surface around the amino acid residues predicted by the original algorithm. This second step creates an additional set of amino acid pairs for every predicted cluster based on the mimotope sequences, which includes all amino acids identified in the “filling” step and separated by distance ≤8 Å. This new dataset is then compared to the total amino acid pair composition of the affinity-selected mimotopes. The cluster analysis is then repeated using the new pairs that were present in the original database but were not selected initially because they did not meet the statistical threshold. Finally, the epitope is defined based on the largest predicted clusters that match the pair composition within the complete database of mimotopes. A scheme of this novel algorithm is presented in Fig. 1.
Fig. 1.
Scheme of the computer algorithm.
3. Results and discussion
3.1. Epitope prediction for mAbs E16 and E24
Our previous mapping studies using yeast surface display predicted that E16 and E24 would bind to a similar region within the lateral ridge of domain III (DIII) of WNVE protein. In particular, three amino acids K307, T330 and T332 were predicted as essential for E16 and E24 recognition. These two mAbs are of significant interest because they rapidly neutralize WNV infection in vitro and in vivo. The epitope target of E16 was further defined by X-ray crystallography, validating the results of the yeast display (Nybakken et al., 2005); in addition to the amino acid residues identified by yeast display that accounted for ~80% of hydrogen bond interactions, crystallography defined additional 12 contact residues revealing a discontinuous structural epitope that spans four loops.
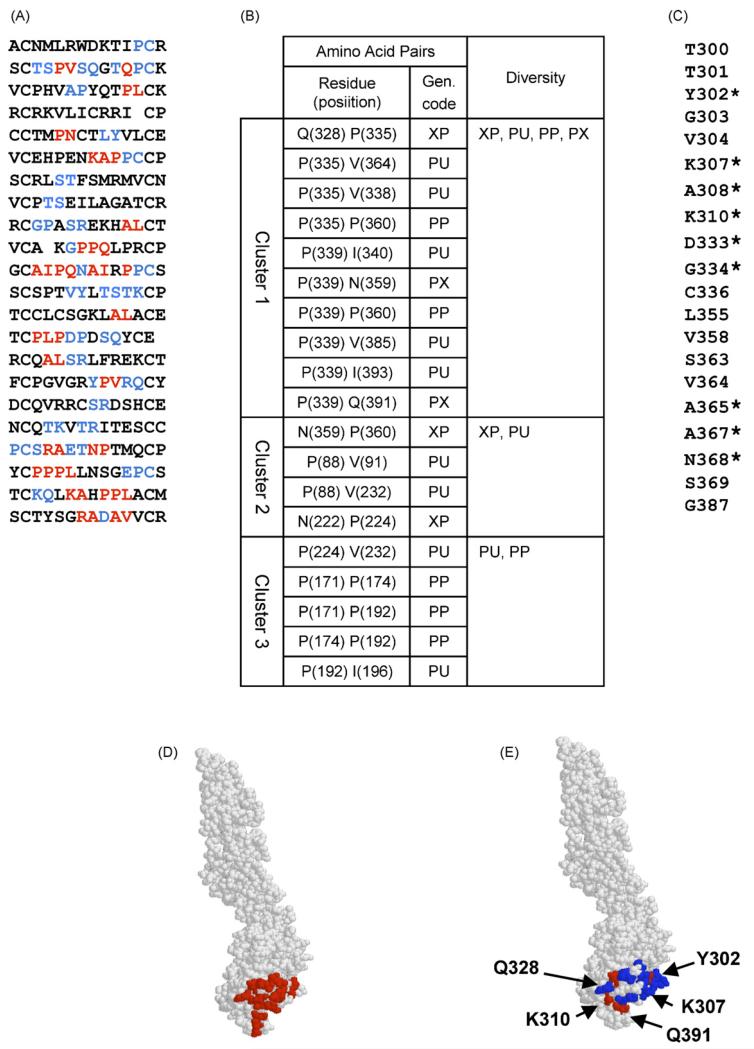
Given the available high-resolution structural data, we chose to initially map the epitope for E16 to confirm the utility of our novel strategy. Immunoaffinity purification of phages using E16 yielded 22 different peptides following exhaustive library screening (Fig. 2A). The epitope prediction based on these sequences was elucidated initially using our original algorithm to identify statistically significant amino acid “pairs” located within close proximity on the surface of the folded protein (an example of the results is presented in Fig. 2B). Cluster analysis revealed that the majority of these pairs fell into the same region of the protein (Fig. 2B, Cluster 1) and this region coincides with the predicted epitope defined by X-ray crystallography. At this stage we identified significant pairs in only 15 of the 22 peptides (Fig. 2A, red letters). Interestingly, all of the significant pairs in this cluster involved one of three proline residues (P335, P339, and P360). Although prolines were not shown to be involved in antibody contact, these residues may have an important indirect role in maintaining the epitope structure.
Fig. 2.
Prediction of the E16 epitope. (A) Peptide sequences found by phage library screening. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (B) Amino acid clusters predicted by the algorithm. Gen. code = generalized code (Enshell-Seijffers et al., 2003)]. (C) List of amino acids added at the second step. Contact residues are indicated by asterisk. (D) E16 epitope estimated by X-ray crystallographic analysis (Nybakken et al., 2005), amino acids of the epitope are shown in red. (E) Epitope predicted by our affinity-selected mimotopes. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
Even though the cluster analysis successfully identified the correct region for the epitope, we only uncovered three of the contact residues defined by crystallography [Q328, Q391, I393]. Many of the residues within the collection of affinity-selected mimotopes are likely to provide important geometric and physico-chemical (charge and polarity) properties to the epitope but are difficult to assign using our original algorithm. Given this limitation, we developed a novel algorithm, which refines the prediction. In this second step, the area around the predicted cluster is “filled” with the neighboring amino acids by calculating the distance between the amino acids predicted by the original algorithm and spatially-proximal neighbors (≤8 Å away). By applying this novel algorithm to the collection of E16 peptides, we identified additional residues (listed in Fig. 2C and shown as blue letters in Fig. 2A; asterisks denote contact residues defined by crystallography), which did not satisfy the strict statistical requirements of the original algorithm. We now find that 21 of the 22 peptides (Fig. 2A) carry relevant amino acids for epitope prediction. Using this additional set, we can more accurately predict the epitope and we confirmed most of the contact residues predicted by the X-ray structure: T300, Y302, S306, K307, A308, K310, T330, T332, D333, G334, A365, A367, N368, Q391, and I393 (Fig. 2D) were also predicted by our second step algorithm (Fig. 2E). Nevertheless, there were a few amino acids (F309, T366 and E390) that were defined by crystal-lography but not predicted using either prediction strategy (i.e. mimotope selection or presentation of random mutants by yeast display).
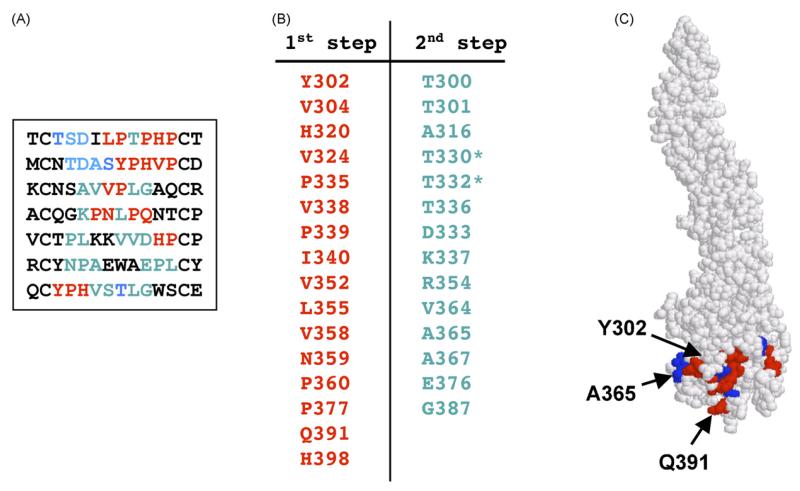
With mAb E24, only 7 different peptides were isolated following exhaustive library screening (Fig. 3A). Interestingly, one of the peptides was overrepresented in our database (MCNTDASYPHVPCD) and appeared more than 30 times during the screening process. The high frequency of this peptide was not due to overrepresentation of a single phage because multiple phages were isolated which expressed this peptide but with distinct nucleotide sequences. In the first step of our prediction method, only two clusters were identified (not shown). Cluster 1 contained greater numbers of significant pairs than Cluster 2 (15 vs. 6) and greater diversity (6 generalized pairs compared to 2 generalized pairs). The first step again successfully predicting the region in which the epitope was found (residues predicted by the first stage are displayed in red in Fig. 3). However, only a limited number of residues within the epitope (Y302, Q391) were identified, similar to the results with E16. By applying the second step of our algorithm, we identified additional amino acids that contributed to the E24 epitope: T332, D333, A365, A367 (displayed in Fig. 3 using blue lettering). Notably, T330 and T332 were previously identified as key constituents of the E24 epitope using mutational analysis (Oliphant et al., 2005). By contrast, the mimotope method did not identify K307 as part of the epitope although mutation of this residue strongly abrogates binding by E24. Consistent with our previous results, our novel algorithm mapped the E24 epitope to the same area as the E16 epitope. Similar to the results with E16, all the significant pairs identified by our first step included proline residues, further emphasizing an important role for proline in maintaining the scaffold of the neutralizing epitope.
Fig. 3.
Prediction of the E24 epitope. (A) Peptide sequences found by phage library screening. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (B) Amino acids predicted by the algorithm (red) and added at the second step (blue). Amino acids found by mutational analysis are indicated by asterisk. (C) Epitope predicted by our affinity-selected mimotopes. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
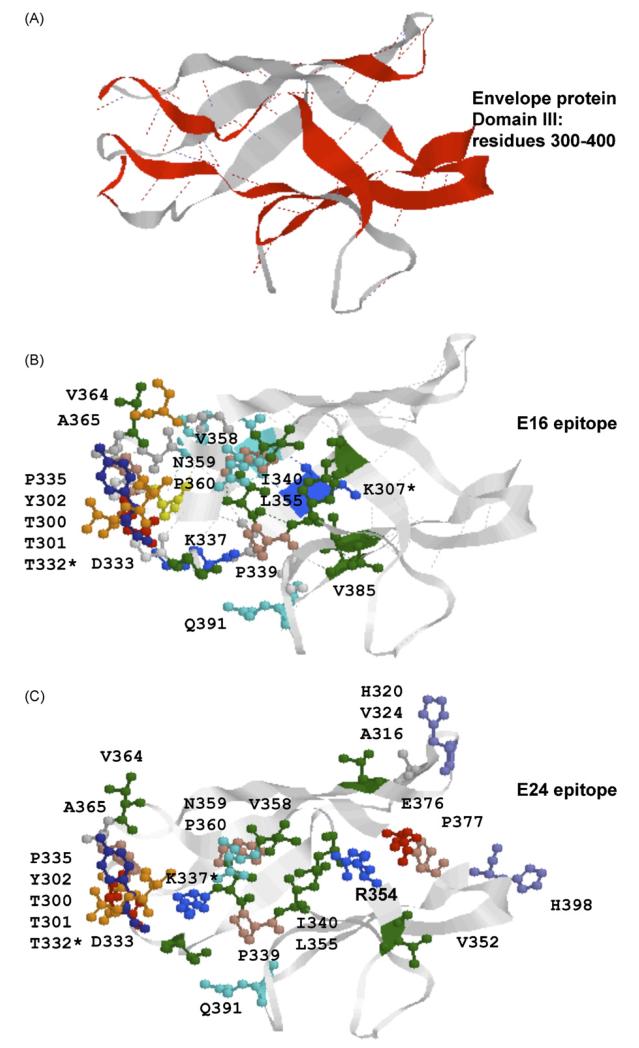
It is noteworthy that E16 and E24 immunoselection did not identify any common phages; this is consistent with our prediction that these two mAbs bind the same region on DIII but not the exact same epitope. The predicted epitope for E24 and the known epitope for E16 are shown side-by-side in Fig. 4 for comparison. The main contact residues are situated on the turns connecting eight β-strands (Fig. 4). Some of the predicted residues are not likely to be involved in antibody contact, such as hydrophobic amino acids (V338, I393) and prolines (P335, P339, P360), but may contribute to maintaining epitope conformation.
Fig. 4.
Structural comparison of the E16 epitope and predicted E24 epitope. (A) 3D model of domain III of envelope protein. β-strands connecting E16 epitope amino acid contact residues are shown in red. (B) 3D model of epitope E16. (C) 3D model of epitope E24. Colours—red: acidic amino acids, green: hydrophobic amino acids, blue: basic amino acids, orange: serine and threonine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.2. Epitope prediction for mAb E121
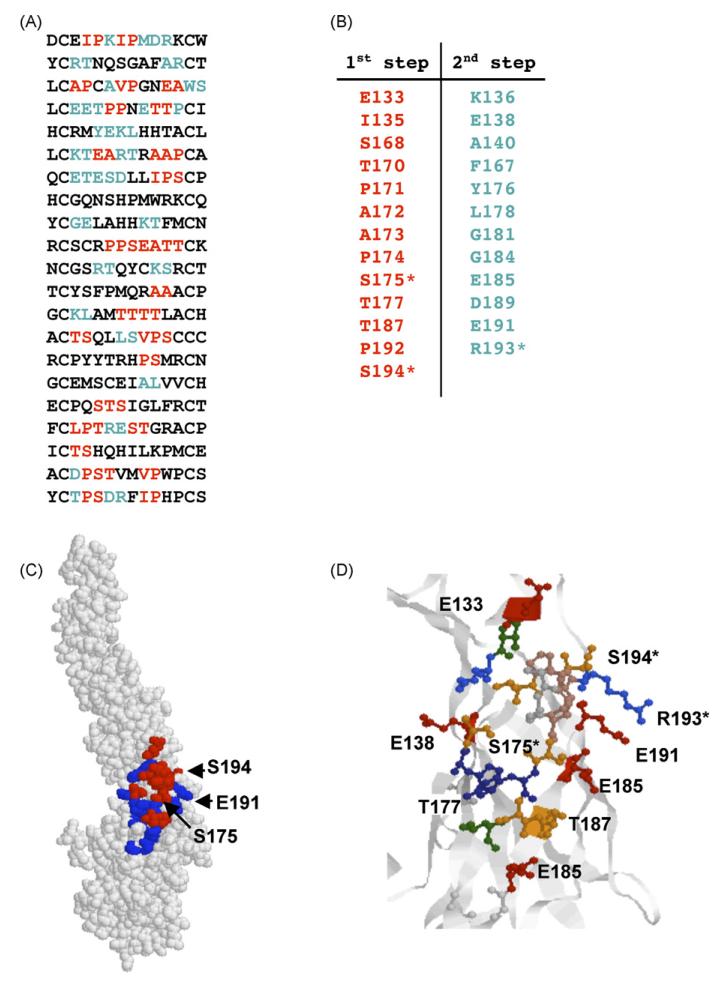
Immunoselection with E121 identified 21 different peptides (Fig. 5A). Based on these sequences, two clusters were predicted that were similar in size. However, Cluster 1 had greater diversity of amino acid residues and, therefore, provided a stronger prediction. Indeed, Cluster 1 matched the region predicted to contain the E121 epitope by yeast display and included residues previously determined to be important for E121 binding (S175, E191, R193, S194) (Oliphant et al., 2006). Again, a greater number of residues within the epitope were defined following the second step (Fig. 5A and B; blue letters) including one a critical residue for E121 binding (R193), reinforcing the value of this second step to the overall prediction. The algorithm suggests that E121 mAb epitope lies within the hinge region between DI and DII and its amino acid residues are located on four β-strands (Fig. 5E); similar structures have been defined for a number of different epitopes (Housset et al., 1997; Kwong et al., 1998). Some of the predicted amino acids within the epitope satisfy conditions for typical β-sheet regularity (n, n + 2): E185, T187, D189, S175, T177, S168, and S170. Amino acids S193 and R194 are situated on a linker with random coil structure between DI and DII.
Fig. 5.
Prediction of the E121 epitope. (A) Peptide sequences found by phage library screening. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (B) Amino acids predicted by the algorithm (red) and added at the second step (blue). Amino acids found by mutational analysis are indicated by asterisks. (C) Epitope predicted by our affinity-selected mimotopes. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (D) Folding of predicted E121 epitope. Colours—red: acidic amino acids, green: hydrophobic amino acids, blue: basic amino acids, orange: serine and threonine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.3. Epitope prediction of mAb E113
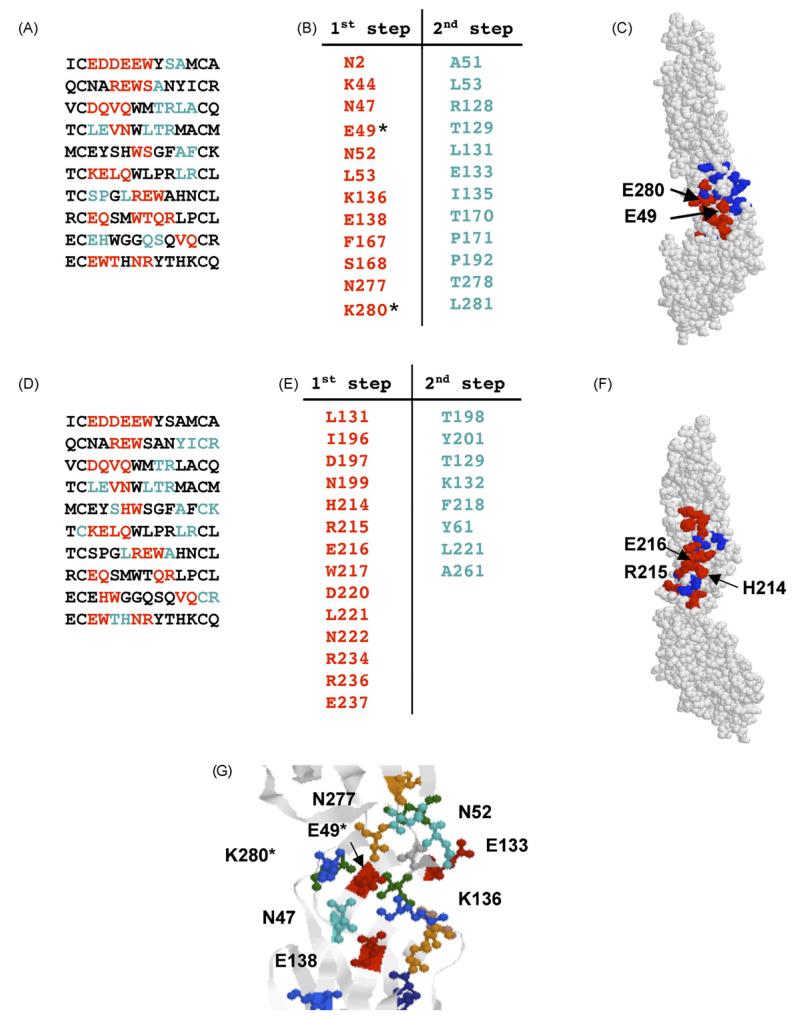
Ten different peptide sequences were discovered following exhaustive screening of the library with mAb E113 (Fig. 6A). While seven unique sequences were sufficient to accurately predict the epitope for E24, the 10 E113-specific sequences failed to yield a conclusive prediction. Rather, two clusters were identified with similar attributes that were spatially adjacent to each other (Fig. 6C and F). In this instance, affinity-selected random peptides failed to produce a single epitope prediction. The first cluster Fig. 6(A–C) spanned a region formed by four β-strands (Fig. 6G) and included E49 and K280 which were shown to be important for E113 binding (Oliphant et al., 2006). Furthermore, a number of sites predicted within the second cluster (W217, N222, and R236) were mutated in our previous report (Oliphant et al., 2006) and did not influence E113 binding; thereby reducing our confidence that this second cluster accurately predicts the epitope. Therefore, in consideration of the data with directed mutagenesis, we believe that the epitope predicted by the first cluster represents the most likely target. This example reveals the complementarity of the methods we have employed for epitope prediction; mutational analysis facilitates the selection of otherwise equivocal predictions and the mimotope method reveals a broader number of discrete residues within the epitope (our previous work did not identify N277, N47, K136, or S168) and provides a better prediction of the epitope structure (four β-strands).
Fig. 6.
Prediction of the E113 epitope. (A) Peptide sequences found by phage library screening and modeled on Cluster 1. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (B) Amino acids associated with Cluster 1 (red) and added at the second step (blue) Amino acids found by mutational analysis are indicated by asterisk. (C) Epitope predicted by Cluster 1. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (D) Peptide sequences found by phage library screening and modeled on Cluster 2. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (E) Amino acids associated with Cluster 2 (red) and added at the second step (blue). (F) Epitope predicted by Cluster 2. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (G) Folding of predicted E113 epitope. Colours—red: acidic amino acids, green: hydrophobic amino acids, blue: basic amino acids, orange: serine and threonine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.4. Epitope prediction for mAbs E60
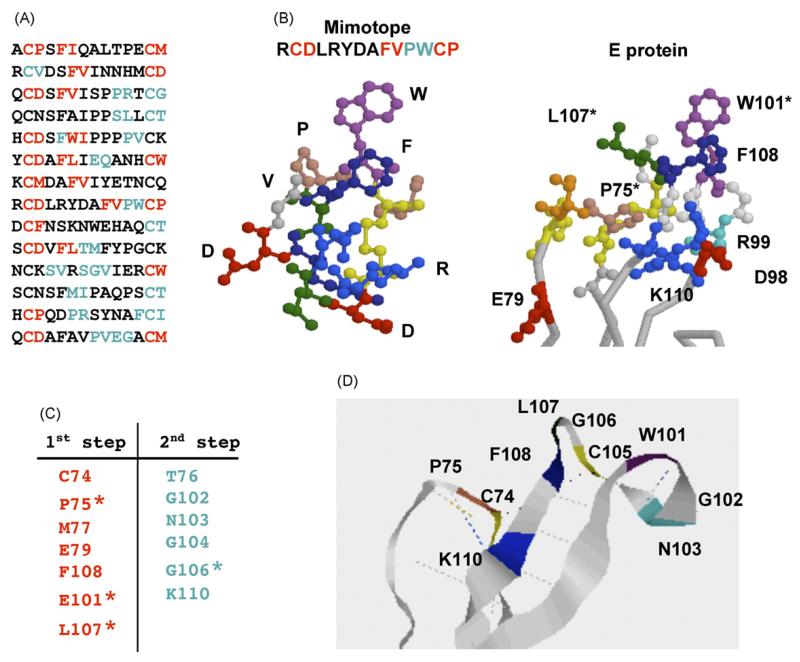
The results with mAb E60 are an additional excellent example of the importance of combining the random mutational screening method described in our previous manuscripts (Oliphant et al., 2006) and the analysis of affinity-selected mimotopes. With E60, 14 unique peptides were isolated (Fig. 7A). The initial epitope predicted by our algorithm did not overlap with the region identified using mutational analysis (data not shown). The epitope for E60 was previously predicted to lie within the fusion loop as mutations of amino acids W101, G106, and L107 abrogated binding of E60 (Oliphant et al., 2006). However, unlike the predictions described above, none of these key residues were predicted in our initial cluster analysis. E60 exhibited strong reactivity for the selected mimotopes, so we were confident that the selected peptides were of sufficient affinity for E60 to be used in our analysis. Interestingly, molecular modeling of the E60 mimotope which was most commonly isolated by affinity selection revealed a structure that it is similar to the fusion loop but with some deviations in atomic coordinates (Fig. 7B). Indeed, similar modeling with 2 other mimotopes from this collection also revealed comparable structures (data not shown). Therefore, it seemed likely that the affinity-selected mimotopes were characteristic of the fusion loop however our algorithm was unable to identify these qualities.
Fig. 7.
Prediction of the E60 epitope. (A) Peptide sequences found by phage library screening. Amino acids predicted at the first stage are shown in red and those predicted at the second step are shown in blue. (B) 3D Molecular models of E60 mimotope and the fusion loop of envelope protein. (C) Amino acids clusters predicted by the algorithm (red) and added at the second step (blue). (D) Ribbon diagram of the E60 epitope. Colours—red: acidic amino acids, green: hydrophobic amino acids, blue: basic amino acids, orange: serine and threonine. Hydrogen and disulfide bonds are shown in dotted lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
We hypothesized that there may be differences between the atomic coordinates defined by X-ray crystallography and the actual protein conformation recognized by E60 in solution. As a result, our computer algorithm, which makes predictions based on the distances between amino acid side chains, would not provide an accurate prediction. To accommodate this possibility, we modified the conditions to accept a maximal distance between amino acid neighbors of 9 Å to reduce the stringency of our analysis. We also included pairing with all possible amino acid neighbors, regardless of their accessibility of the protein surface. To avoid prediction of huge clusters, we only used amino acid pairs with very high statistical probability after the first step of the algorithm. The results of the prediction are presented in Fig. 7C. We identified three small clusters using these modifications to our selection criteria (data not shown). The clusters were small due to the high stringency applied to the selection of significant residues. Nevertheless, a cluster was defined that contained two of the amino acids previously identified by our mutational method (W107, L107) and one additional residue within the fusion loop (F108) (Oliphant et al., 2006). Following the second step of the algorithm, we also predicted that G106, the third key residue defined by mutational analysis, lies within the E60 epitope. Most of the highly statistically significant amino acid pairs identified through the algorithm included residues P75 and C74 (both located outside the fusion loop). C74 makes disulfide bond with C105 thus stabilizing local conformation of the loop. Proline is often important for sustaining protein conformation and P75 was actually determined to be important for the epitope integrity through mutational analysis (Oliphant et al., 2005). It is of interest that E60 is a flavivirus cross-reactive antibody and binds strongly to Dengue virus E protein, which contains conserved residues identified in our prediction (C74, P75, E79, W101, G106, L107). Predicted residues T76, G102, N103, G104 and K110 are also conserved in Dengue virus E protein and any, or all, of these amino acids could contribute to antibody contact. Thus, the case of E60 clearly demonstrates the importance of combining random mutagenesis screening of epitope binding sites with the mimotope methodology. A ribbon diagram of the predicted structure is shown in Fig. 7E. The epitope structure seems to be stabilized by a single disulfide bond and multiple hydrogen bonds (shown in Fig. 7E).
Overall, the results of this study indicate that our novel computer algorithm for epitope prediction is functional and provides useful insight into the nature of the antibody recognition. Screening of the random peptide libraries for epitope analysis yielded information that was not previously defined by crystallographic or mutational studies. For example, an important role for proline residues in maintaining the scaffold of the E16 epitope was suggested. Although further investigations will be required to fully define the role of the non-contact residue identified by our approach, the method provides a useful tool for hypothesis generation regarding epitope structure and the data provide an important blueprint for the development of epitope-specific vaccines. Our results demonstrate that while both yeast display of randomly mutated domains and phage display of random peptides are effective methods for epitope prediction, the combined methodologies provide greater information and confidence in epitope prediction. Whereas random mutagenesis may not reveal all of the residues within a particular epitope, it allows the operator to pinpoint key regions without a priori knowledge of the epitope location. Such a tool is highly complementary to the mimotope method, which can define many residues within an epitope but commonly returns equivocal predictions that require validation through mutagenesis.
Acknowledgments
This work was supported by NIH NIAID Contract# N01-AI-40066 (to J.L.B. and M.B.L.) and the NIH NIAID grant #AI–061373 (to M.S.D.)
References
- Bonnycastle LL, Mehroke JS, Rashed M, Gong X, Scott JK. Probing the basis of antibody reactivity with a panel of constrained peptide libraries displayed by filamentous phage. J. Mol. Biol. 1996;258:747–762. doi: 10.1006/jmbi.1996.0284. [DOI] [PubMed] [Google Scholar]
- Carter JM. Epitope mapping of a protein using the Geysen (PEPSCAN) procedure. Methods Mol. Biol. 1994;36:207–223. doi: 10.1385/0-89603-274-4:207. [DOI] [PubMed] [Google Scholar]
- Chan SL, Ong ST, Ong SY, Chew FT, Mok YK. Nuclear magnetic resonance structure-based epitope mapping and modulation of dust mite group 13 allergen as a hypoallergen. J. Immunol. 2006;176:4852–4860. doi: 10.4049/jimmunol.176.8.4852. [DOI] [PubMed] [Google Scholar]
- Chien NC, Roberts VA, Giusti AM, Scharff MD, Getzoff ED. Significant structural and functional change of an antigen-binding site by a distant amino acid substitution: proposal of a structural mechanism. Proc. Natl. Acad. Sci. U.S.A. 1989;86:5532–5536. doi: 10.1073/pnas.86.14.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enshell-Seijffers D, Denisov D, Groisman B, Smelyanski L, Meyuhas R, Gross G, Denisova G, Gershoni JM. The mapping and reconstitution of a conformational discontinuous B-cell epitope of HIV-1. J. Mol. Biol. 2003;334:87–101. doi: 10.1016/j.jmb.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Gustchina E, Louis JM, Lam SN, Bewley CA, Clore GM. A monoclonal Fab derived from a human nonimmune phage library reveals a new epitope on gp41 and neutralizes diverse human immunodeficiency virus type 1 strains. J. Virol. 2007;81:12946–12953. doi: 10.1128/JVI.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J, MacDonald KS, Barber BH. Construction of recombinant targeting immunogens incorporating an HIV-1 neutralizing epitope into sites of differing conformational constraint. Vaccine. 2002;20:1169–1180. doi: 10.1016/s0264-410x(01)00441-8. [DOI] [PubMed] [Google Scholar]
- Housset D, Mazza G, Gregoire C, Piras C, Malissen B, Fontecilla-Camps JC. The three-dimensional structure of a T-cell antigen receptor V alpha V beta heterodimer reveals a novel arrangement of the V beta domain. EMBO J. 1997;16:4205–4216. doi: 10.1093/emboj/16.14.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LT, Gromiha MM, Ho SY. Sequence analysis and rule development of predicting protein stability change upon mutation using decision tree model. J. Mol. Model. 2007;13:879–890. doi: 10.1007/s00894-007-0197-4. [DOI] [PubMed] [Google Scholar]
- Johansson DX, Voisset C, Tarr AW, Aung M, Ball JK, Dubuisson J, Persson MA. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc. Natl. Acad. Sci. U.S.A. 2007;104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotik M, Zuber H. Mutations that significantly change the stability, flexibility and quaternary structure of the l-lactate dehydrogenase from Bacillus megaterium. Eur. J. Biochem. 1993;211:267–280. doi: 10.1111/j.1432-1033.1993.tb19895.x. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloen RH, Puijk WC, Langeveld JP, Langedijk JP, Timmerman P. Design of synthetic peptides for diagnostics. Curr. Protein Pept. Sci. 2003;4:253–260. doi: 10.2174/1389203033487144. [DOI] [PubMed] [Google Scholar]
- Meloen RH, Amerongen AV, Hage-Van Noort M, Langedijk JP, Posthumus WP, Puyk WC, Plasman H, Lenstra JA, Langeveld JP. The use of peptides to reconstruct conformational determinants; a brief review. Ann. Biol. Clin. (Paris) 1991;49:231–241. [PubMed] [Google Scholar]
- Nall BT, Zuniga EH, White TB, Wood LC, Ramdas L. Replacement of a conserved proline and the alkaline conformational change in iso-2-cytochrome c. Biochemistry. 1989;28:9834–9839. doi: 10.1021/bi00451a043. [DOI] [PubMed] [Google Scholar]
- Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–769. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Austin SK, Xu Q, Bramson J, Loeb M, Throsby M, Fremont DH, Pierson TC, Diamond MS. Induction of epitope-specific neutralizing antibodies against West Nile virus. J. Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J. Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrakou E, Murray A, Rosamund C, Graves L, Price MR. Evaluation of Pepscan analyses for epitope mapping of anti-MUC1 monoclonal antibodies–a comparative study and review of five antibodies. Anticancer Res. 1998;18:4419–4421. [PubMed] [Google Scholar]
- Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Shiota T, Okame M, Takanashi S, Khamrin P, Takagi M, Satou K, Masuoka Y, Yagyu F, Shimizu Y, Kohno H, Mizuguchi M, Okitsu S, Ushijima H. Characterization of a broadly reactive monoclonal antibody against norovirus genogroups I and II: recognition of a novel conformational epitope. J. Virol. 2007;81:12298–12306. doi: 10.1128/JVI.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Surface presentation of protein epitopes using bacteriophage expression systems. Curr. Opin. Biotechnol. 1991;2:668–673. doi: 10.1016/0958-1669(91)90032-z. [DOI] [PubMed] [Google Scholar]
- Smith GP. Phage-Display Vectors and Libraries Based on Filamentous Phage Strain fd-tet. 2006 http://www.biosci.missouri.edu/smithgp/PhageDisplayWebsite/PhageDisplayWebsiteIndex.html.
- Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile Virus. J Virol. 2006;80:6982–6992. doi: 10.1128/JVI.00551-06. [DOI] [PMC free article] [PubMed] [Google Scholar]