Significance
During chemotaxis, cells recognize an extracellular chemical gradient and produce amplified intracellular responses independently of the actin cytoskeleton. This process is called directional sensing and observed as the activation of Ras GTPase and the production of phosphatidylinositol (3,4,5)-triphosphate (PIP3) toward higher concentrations of chemoattractants. How directional sensing is controlled is largely unknown. In our current study, we demonstrate that a Rho GTPase (RacE) and a Rho guanine nucleotide exchange factor (GxcT) are required for the orientation of directional sensing independently of feedback from the actin cytoskeleton and cell polarity in Dictyostelium, and reveal a previously unknown role for Rho GTPases in intracellular signaling upstream of Ras activation and PIP3 production.
Abstract
During chemotaxis, cells sense extracellular chemical gradients and position Ras GTPase activation and phosphatidylinositol (3,4,5)-triphosphate (PIP3) production toward chemoattractants. These two major signaling events are visualized by biosensors in a crescent-like zone at the plasma membrane. Here, we show that a Dictyostelium Rho GTPase, RacE, and a guanine nucleotide exchange factor, GxcT, stabilize the orientation of Ras activation and PIP3 production in response to chemoattractant gradients, and this regulation occurred independently of the actin cytoskeleton and cell polarity. Cells lacking RacE or GxcT fail to persistently direct Ras activation and PIP3 production toward chemoattractants, leading to lateral pseudopod extension and impaired chemotaxis. Constitutively active forms of RacE and human RhoA are located on the portion of the plasma membrane that faces lower concentrations of chemoattractants, opposite of PIP3 production. Mechanisms that control the localization of the constitutively active form of RacE require its effector domain, but not PIP3. Our findings reveal a critical role for Rho GTPases in positioning Ras activation and thereby establishing the accuracy of directional sensing.
Chemotaxis plays an important role in many biological processes, including pattern formation during development, wiring of the neural network, and immune responses (1–4). In addition to its physiological roles, alterations in chemotaxis contribute to the pathophysiology of cancer metastasis, inflammation, and allergies. During chemotaxis, cells sense shallow, extracellular chemical gradients and persistently move toward higher concentrations of chemoattractants through the localized activation of intracellular signaling cascades and the extension of pseudopods at the leading edge (5, 6).
The accuracy of chemotaxis is remarkably high, and cells can migrate with tremendous persistence in shallow chemical gradients, even when the concentration difference is as low as 2% across the length of the cell (7, 8). Such extreme precision requires directional sensing and polarization: Directional sensing is the ability of a cell to detect a chemoattractant gradient and produce amplified intracellular responses, whereas polarization establishes an elongated, polarized cell morphology, which is characterized by distinct posterior and anterior regions that contain different molecular components (9). Directional sensing and polarization are interconnected, but they are separable: Directional sensing can be observed in cells treated with Latrunculin A (LatA), which disrupts the actin cytoskeleton, whereas polarity can be formed in response to global chemoattractant stimulation without concentration gradients. During chemotaxis, the actin cytoskeleton stabilizes cell polarity and the asymmetric distribution of molecules to the front and back of cells, creating positive feedback systems that maintain directional persistence (10). However, whether cells control the spatial and temporal accuracy of chemotactic signaling at the step of directional sensing remains unknown.
The molecular mechanisms underlying chemotaxis are evolutionarily conserved and have been studied extensively using the single-celled amoeba Dictyostelium discoideum as a model system (8, 11). During Dictyostelium development, which is initiated upon starvation, free-moving amoeboid cells chemotax toward aggregation centers that release the chemoattractant cAMP, resulting in the formation of stress-resistant, multicellular structures called fruiting bodies that contain spore cells. cAMP binds to seven-transmembrane domain receptors on the plasma membrane and activates the associated underlying heterotrimeric G proteins. cAMP receptors are uniformly distributed along the plasma membrane, whereas heterotrimeric G protein activation reflects the receptor occupancy by the ligand without any signal amplification (12–14). However, the activation of heterotrimeric G proteins leads to the robust, local activation of Ras GTPases, as shown by the recruitment of a biosensor for activated Ras GTPase to the leading edge of chemotaxing cells (15). Similarly, a biosensor for the short-lived, lipid second messenger phosphatidylinositol (3,4,5)-triphosphate (PIP3) is also highly localized to the leading edge upon heterotrimeric G protein activation (16, 17). Ras activation and PIP3 production appear to act in parallel but are interconnected, as Ras GTPases modulate the accumulation of PIP3 by regulating the activity of PI3-kinase, likely through direct protein interactions (18). Ras activation and PIP3 production lead to remodeling of the actin cytoskeleton by promoting the polymerization of actin at the leading edge (17, 19). Directional sensing converts extracellular chemical gradients into the local activation of signaling events and functions as a central step of chemotaxis (20–22). Because their restriction to the portion of the plasma membrane facing higher concentrations of chemoattractants occurs independently of the actin cytoskeleton, biosensors for Ras activation and PIP3 production have been used to directly measure directional sensing without feedback from cytoskeletal-mediated events (13, 15, 20, 21, 23–25).
In mammals, it has been shown that Rho family GTPases, including Rho, Rac, and Cdc42, act as downstream effectors of Ras GTPases and PIP3 to control distinct types of actin cytoskeleton remodeling (26, 27). Like many other small GTPases, the activation of Rho, Rac, and Cdc42 is meditated by the binding of GTP, whereas their inactivation is mediated by the hydrolysis of GTP to GDP. Therefore, guanine nucleotide exchange factors (GEFs), which facilitate the exchange of GDP for GTP, stimulate these GTPases, whereas GTPase activating proteins (GAPs) turn them off. Many GEFs and GAPs contain pleckstrin homology (PH) domains, some of which bind to phosphatidylinositol lipids, such as PIP3 (28, 29). RhoA is activated at the front, where it promotes membrane protrusion, as well as at the rear of cells, where it facilitates contraction of the actin cytoskeleton to move the cytoplasm forward in migrating fibroblasts (30). In amoeboid cells such as neutrophils, RhoA activity is preferentially observed at the rear of polarized cells (31). In contrast, Rac1 is activated at the leading edge of both fibroblasts and neutrophils and promotes actin polymerization, causing lamellipodia formation (32, 33). Similar to Rac1, Cdc42 is also activated at the leading edge of fibroblasts and forms filopodia (34). These Rho family proteins are essential for directed cell migration as regulators of the actin cytoskeleton. However, it is unknown whether these proteins are required for directional sensing during chemotaxis.
Here, we have systematically deleted the individual genes that encode Rho GTPases and its potential GEFs in Dictyostelium to understand their function in chemotaxis. We found chemotactic defects in cells lacking the Rho GTPase RacE or a GEF, called GxcT. Further analyses of racE− and gxcT− cells showed that RacE and GxcT stabilize the orientation of Ras activation and PIP3 production in response to chemoattractant gradients, and this regulation occurred independently of the actin cytoskeleton. As a consequence of unstable directional sensing, racE− and gxcT− cells abnormally extended pseudopods from the lateral sides of cells, leading to decreased chemotactic efficiency. Constitutively active mutants of RacE and human RhoA were located at the side of cells facing away from the source of chemoattractants in the absence of the actin cytoskeleton. Our findings define a unique mechanism that regulates directional sensing and reveal a molecular function for Rho GTPases in intracellular signaling upstream of Ras activation and PIP3 production.
Results
GxcT, a Putative RhoGEF, Is Required for Growth and Development.
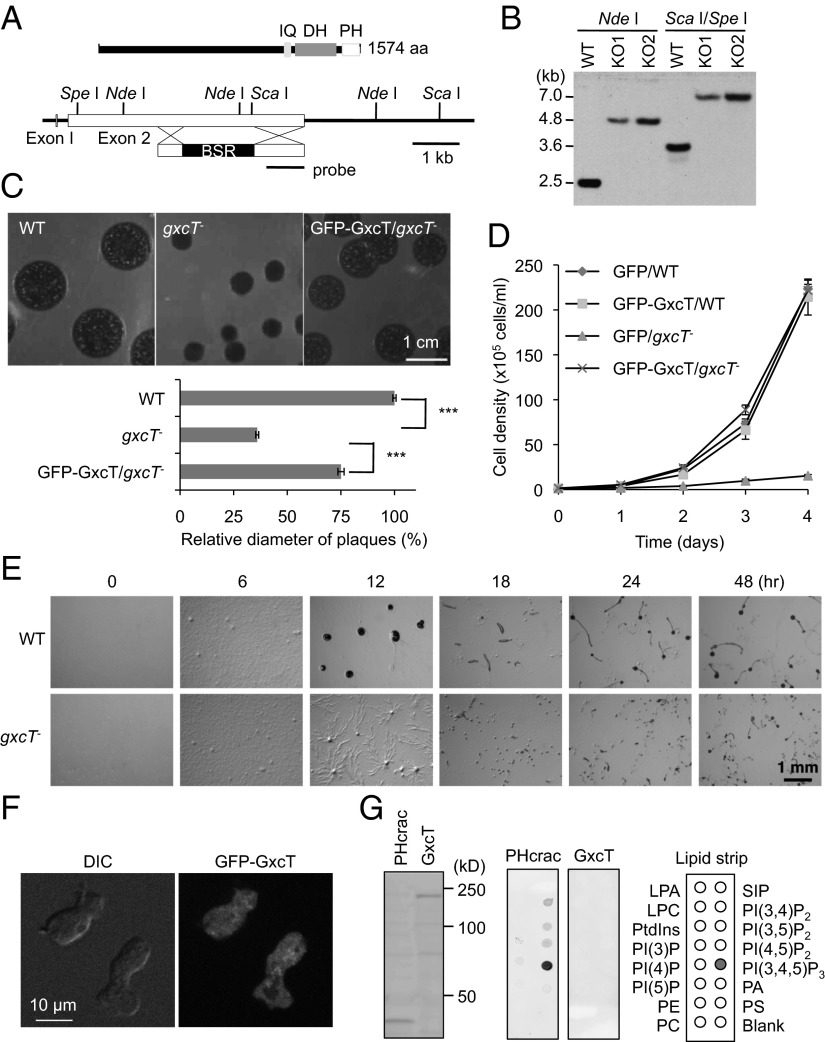
There are 46 potential RhoGEFs that contain a Dbl-homologous domain (RhoGEF domain) in the Dictyostelium genome (35). To study the role of these GEFs in chemotaxis, we systematically deleted the corresponding individual genes by homologous recombination. So far, 20 GEFs have been disrupted (SI Appendix, Table S1). None of the knockout strains showed noticeable impairments in cell growth or development except for gxcT− cells, which exhibited dramatic growth defects both on bacterial lawns and in synthetic media (Fig. 1 A–D). Growth defects in synthetic media appear to be due to impaired cytokinesis as gxcT− cells are multinucleated under the same conditions (SI Appendix, Fig. S1 A and B). GxcT is a previously uncharacterized 177-kDa protein (1,574 amino acids) that contains RhoGEF, IQ calmodulin-binding, and PH domains (Fig. 1A). In addition to their slow-growth phenotype, gxcT− cells also displayed abnormal development upon starvation. When WT cells were placed on nonnutrient agar, they aggregated within 12 h and formed fruiting bodies by 24 h (Fig. 1E). In contrast, gxcT− cells failed to normally aggregate and formed smaller fruiting bodies. Exogenous expression of a GFP-GxcT fusion protein rescued the growth and developmental defects of gxcT− cells, suggesting that GFP-GxcT is functional (Fig. 1 C and D and SI Appendix, Fig. S1C). Fluorescence microscopy showed that GFP-GxcT was present in the cytosol of cells in the presence or absence of cAMP stimulation (Fig. 1F). Similarly, GFP, fused to the PH domain of GxcT, was found in the cytosol (SI Appendix, Fig. S1D). Although some PH domains are known to bind to phosphatidylinositols (36, 37), GFP-GxcT did not interact with any such lipids in dot blot assays (Fig. 1G), consistent with its cytosolic localization in cells. As a positive control, GFP, fused to the PH domain from Crac (PHcrac-GFP), showed strong binding to PIP3 (Fig. 1G) as previously reported (38, 39).
Fig. 1.
GxcT is necessary for normal cell growth and development. (A) GxcT contains calmodulin-binding (IQ), RhoGEF (DH), and PH domains. The majority of the gxcT gene was replaced by a blasticidin resistance cassette (BSR) flanked by DNA sequences homologous to gxcT. (B) Genomic DNA was digested with the indicated restriction enzymes and analyzed by Southern blotting with a DNA fragment corresponding to the region designated as the probe in A. After digestion with NdeI, WT cells showed a 2.5-kb band as expected, whereas gxcT− cells showed a 4.8-kb band. After digestion with ScaI and SpeI, WT and gxcT− cells showed the expected 3.5- and 7-kb bands, respectively. (C) gxcT− cells formed smaller plaques when grown clonally on bacterial lawns. Expressing a plasmid carrying GFP-GxcT in gxcT− cells nearly restored the WT phenotype (GFP-GxcT/gxcT−). Relative diameter of plaques was quantified (n = 22). (D) WT and gxcT− cells expressing the indicated proteins were cultured in HL5 medium and counted daily with a hemocytometer. Values represent the mean ± SEM (n ≧ 3). (E) WT and gxcT− cells were plated on nonnutrient DB agar and examined over time for development. (F) gxcT− cells expressing GFP-GxcT were observed by fluorescence microscopy. (G) Whole cell lysates were prepared from Dictyostelium cells expressing PHcrac-GFP or GFP-GxcT and immunoblotted using anti-GFP antibodies (Left). Similar whole cell lysates were used for lipid dot blot assays, which showed that GFP-GxcT did not bind any of the indicated lipids (Center and Right).
gxcT− Cells Are Defective in Chemotaxis.
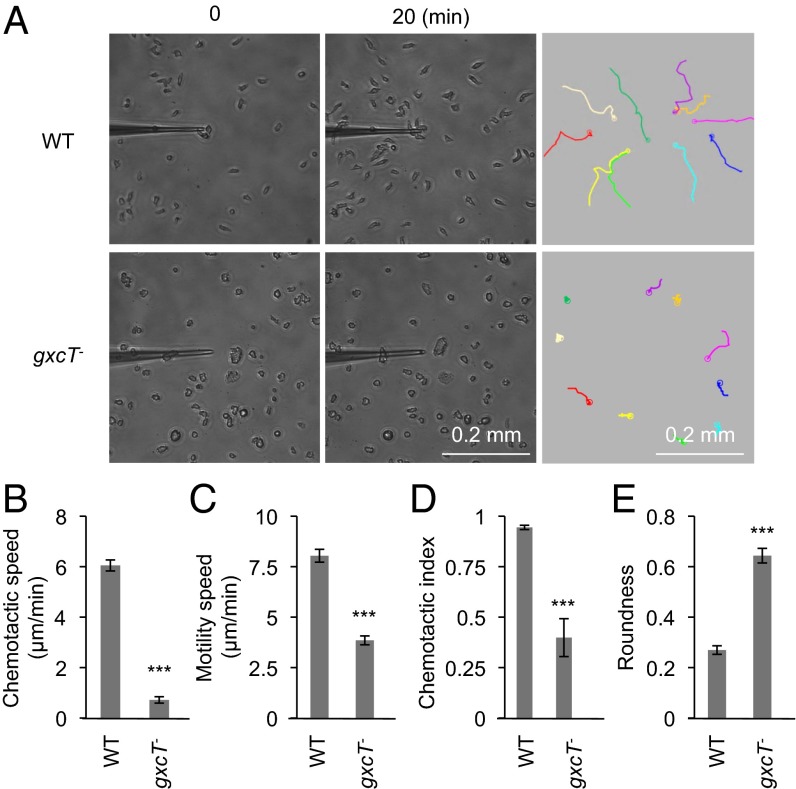
The impaired aggregation during development of gxcT− cells suggested a defect in chemotaxis toward cAMP. To test this possibility, WT and gxcT− cells were developed for 5 h and placed in a gradient of cAMP, which was continuously released from a micropipette. WT cells moved toward the tip of the micropipette, but gxcT− cells failed to do so (Fig. 2A). To exclude the possibility of a developmental delay in gxcT− cells, we developed gxcT− cells for 8 h and observed similar chemotactic defects (SI Appendix, Fig. S2A). Quantitative analyses of the resulting chemotactic behaviors showed that gxcT− cells had significant reductions in both chemotactic speed (the rate of cell movement along the direction of the cAMP gradient) and motility speed (the rate of cell movement regardless of the direction) compared with WT cells (Fig. 2 B and C and SI Appendix, Fig. S2 B and C). The chemotactic index, which indicates the directional accuracy of cell migration, was also decreased in gxcT− cells (Fig. 2D). In addition, gxcT− cells were less polarized and rounder than the WT controls (Fig. 2E and SI Appendix, Fig. S2D). These chemotaxis defects were not simply due to impaired expression of developmentally regulated genes, because cAMP receptor 1 was present at normal abundances in gxcT− cells during development (SI Appendix, Fig. S2E).
Fig. 2.
Chemotaxis requires GxcT. (A) Developed WT and gxcT− cells were placed in a chemoattractant gradient, established by a micropipette releasing cAMP, and observed for 20 min by phase contrast microscopy. The trajectories of cell migration are shown. These chemotaxis assays were then quantified (B–E). (B) Chemotactic speed was calculated as the distance traveled toward the micropipette divided by the elapsed time (20 min). (C) Motility speed, defined as the total distance traveled divided by the elapsed time, was determined by measuring the position of the centroid every 30 s for a period of 20 min. (D) Chemotactic index was defined as the distance traveled in the direction of the gradient divided by the total distance traveled in 20 min. (E) Roundness was determined by calculating the ratio of the short (As) and long (Al) axes of cells (As/Al). For B–E, values represent the mean ± SEM (n = 3). At least 20 cells were analyzed for each experiment. ***P < 0.001.
Normal Ras Activation and PIP3 Production upon Uniform cAMP Stimulation Occur in gxcT− Cells.
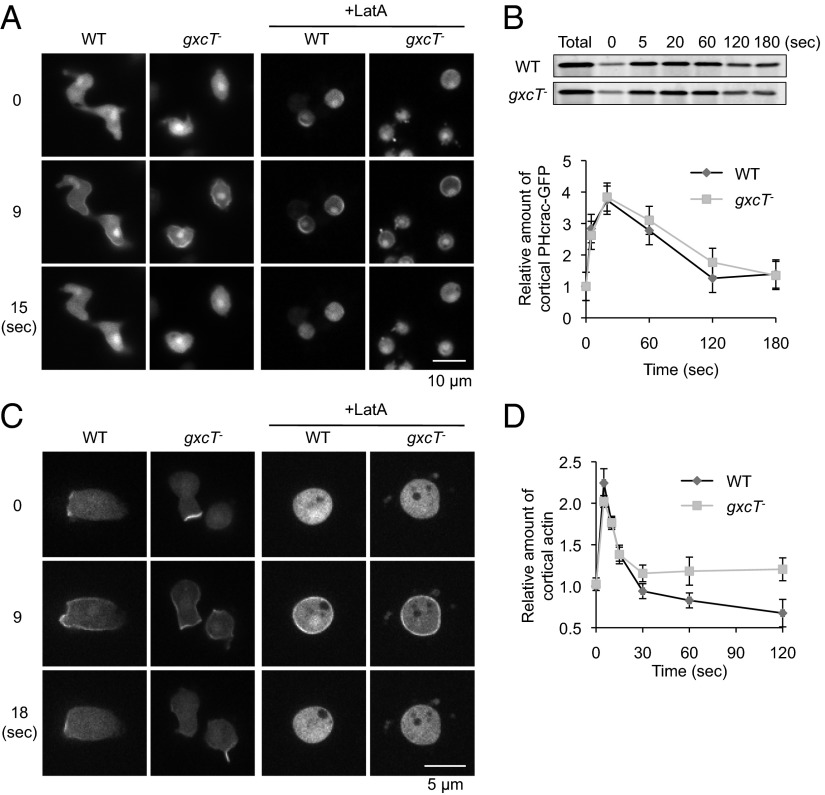
PIP3, one of the signaling molecules that regulate actin polymerization, is transiently generated in the plasma membrane upon cAMP stimulation (16, 17). We examined the production of PIP3 by live cell imaging using the PIP3 biosensor PHcrac-GFP. In response to a uniform cAMP stimulus, PHcrac-GFP was recruited to the plasma membrane with similar kinetics in WT and gxcT− cells, suggesting that PIP3 was produced normally in the mutant strain (Fig. 3A). It has been reported that cAMP-stimulated PIP3 production is independent of the actin cytoskeleton (13). The kinetics of PIP3 production upon stimulation with uniform cAMP were similar for WT and gxcT− cells in the presence of Latrunculin A, which disrupts the actin cytoskeleton (Fig. 3A, +LatA). Furthermore, the normal production of PIP3 in gxcT− cells was confirmed biochemically by quantifying the amount of cAMP-induced PHcrac-GFP that was associated with the membrane fraction (Fig. 3B).
Fig. 3.
GxcT is dispensible for PIP3 production and Ras activation in response to global chemoattractant stimulation. (A) WT and gxcT− cells expressing the PIP3 biosensor PHcrac-GFP were uniformly stimulated with 1 µM cAMP in the presence or absence of 5 µM Latrunculin A (LatA). Cells were observed by fluorescence microscopy at the indicated time points, relative to the addition of cAMP. (B) At the indicated time points after uniform cAMP stimulation, WT and gxcT− cells expressing PHcrac-GFP were collected and filter lysed. The membrane fraction was isolated and immunoblotted using anti-GFP antibodies (Upper). The total amount of PHcrac-GFP in whole cell lysates is also shown. Band intensity was quantified by densitometry and plotted over time (Lower). Values represent the mean ± SEM (n = 3). (C) Cells expressing a biosensor, RBD-GFP, for Ras activation were uniformly simulated with cAMP in the presence or absence of Latrunculin A (LatA) and observed by fluorescence microscopy as in A. Images are shown for the indicated time points after cAMP addition. (D) At the indicated time points after cAMP stimulation, WT and gxcT− cells were lysed, and the cortical actin was isolated and quantified as described in Experimental Procedures. Values represent the mean ± SEM (n = 3). There are no significant differences between WT and gxcT− cells.
Ras GTPases are activated by cAMP downstream of heterotrimeric G protein activation. Activated Ras in turn stimulates both PIP3 production and TORC2 activation (19). To examine the activation of Ras GTPases, the biosensor Ras binding domain (RBD)-GFP, which binds to GTP-bound, active forms of Ras (15), was expressed in WT and gxcT− cells. As previously reported (15, 23), RBD-GFP was located at the leading edge of developed, unstimulated WT cells, and was further recruited to the plasma membrane upon uniform cAMP stimulation (Fig. 3C). Similar to PIP3 production, cAMP-stimulated membrane recruitment of RBD-GFP is independent of the actin cytoskeleton (Fig. 3C, +LatA). RBD-GFP behaved similarly in WT and gxcT− cells, both in the presence or absence of Latrunculin A (Fig. 3C). Consistent with the normal activation of both the Ras and PIP3 pathways, cAMP-stimulated actin polymerization was comparable in WT and gxcT− cells (Fig. 3D), suggesting that it may be the spatial regulation of actin polymerization that caused the observed polarity and migration phenotypes in gxcT− cells.
The Spatial Distribution of PIP3 Production and Ras Activation Is Unstable in gxcT− Cells.
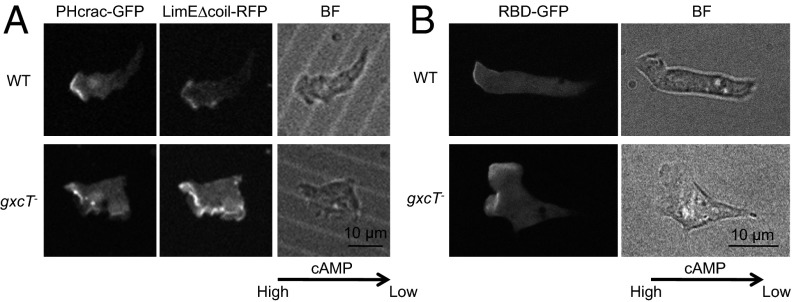
To further examine the chemotactic defects of gxcT− cells, PHcrac-GFP was used to analyze PIP3 production in cells that were chemotaxing toward a micropipette releasing cAMP. Pseudopods were labeled by coexpressing the F-actin biosensor LimE∆coil-RFP (39, 40). In WT cells, PHcrac-GFP was concentrated at pseudopods, marked by LimE∆coil-RFP, which were extended mostly in the direction of the higher concentrations of cAMP coming from the micropipette (Fig. 4A). In contrast, PHcrac-GFP was less oriented toward the cAMP gradient in gxcT− cells, which were less polarized and displayed wider pseudopods than WT cells (Fig. 4A). In addition, these pseudopods were often extended from the lateral sides of the cell, consistent with the increase in roundness described above in Fig. 2E. Similarly, Ras activation, as revealed by RBD-GFP, was not well directed toward the cAMP gradient in gxcT− cells (Fig. 4B).
Fig. 4.
GxcT is required for normal distribution of PIP3 production and Ras activation in chemotactic cells. (A) WT and gxcT− cells coexpressing PHcrac-GFP and the F-actin biosensor LimE∆coil-RFP were observed in a cAMP gradient using fluorescence and bright field (BF) microscopy. The direction of the gradient is indicated by the arrow. (B) Cells expressing RBD-GFP were examined in a cAMP gradient as in A.
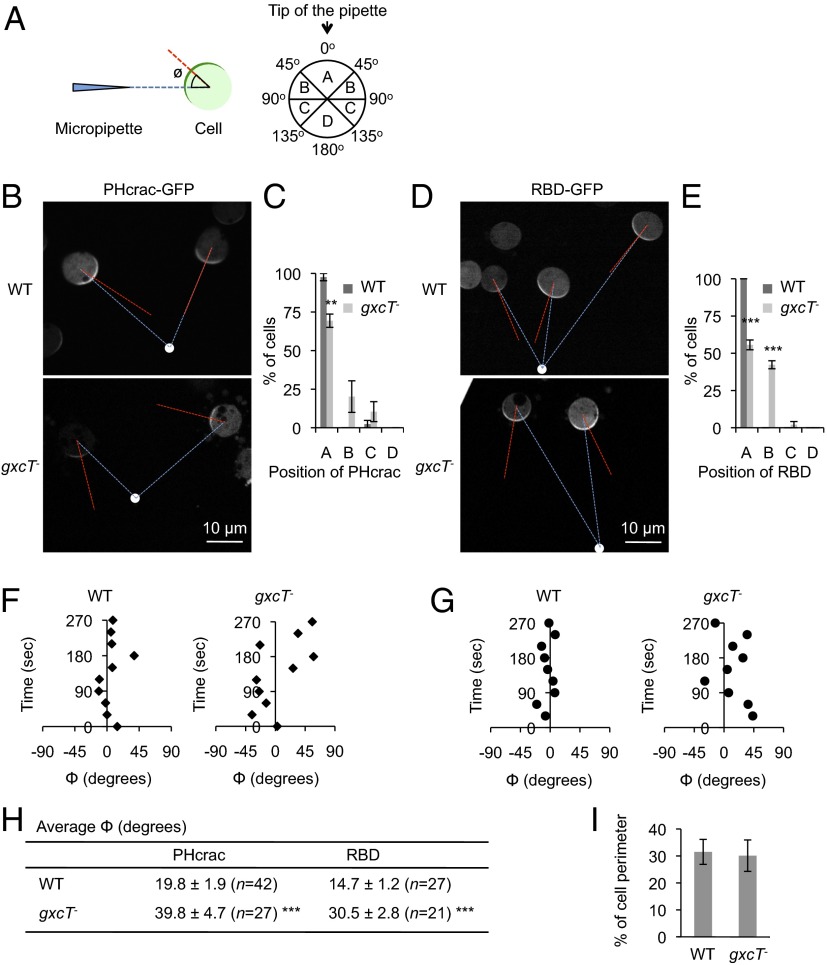
To address why PIP3 production and Ras activation in response to a cAMP gradient were less oriented in gxcT− cells than in WT cells, directional sensing was examined more straightforwardly using an established directional sensing assay (13, 15, 20, 21, 23, 24). In this assay, cells expressing PHcrac-GFP and RBD-GFP were treated with Latrunculin A and then observed by live cell imaging 1 min after being placed in a cAMP gradient. Because Latrunculin A inhibits actin polymerization, this treatment would suppress any effects that feedback from the actin cytoskeleton has on gradient sensing. PHcrac-GFP and RBD-GFP have been shown to localize to the plasma membrane in a crescent-like pattern facing higher concentrations of cAMP in WT cells that have been treated with Latrunculin A (Fig. 5A) (13, 15, 20, 21, 23, 24). Consistent with these previous observations, PIP3 production was localized to the edge of the membrane that faced the tip of a micropipette releasing cAMP, resulting in a crescent-like PHcrac-GFP distribution (Fig. 5B). To quantify the position of PHcrac-GFP crescents, Φ was calculated as the angle formed between two lines: the line drawn between the centroid of the cell and the center of the PHcrac-GFP crescent, and the line drawn between the centroid of the cell and the tip of the micropipette (Fig. 5A). Quantification of the angle (Φ) showed that, in almost all WT cells, the center of the PHcrac-GFP crescent was located within 45° of the micropipette tip position (Fig. 5C). In contrast, only about 70% of gxcT− cells had PHcrac-GFP crescents that were centered within 45° of the micropipette position; the remaining 30% of cells showed angles ranging from 45° to 135°. Similar to the distribution of PHcrac-GFP, the localization of the RBD-GFP crescents was not as spatially targeted toward high cAMP concentrations in gxcT− cells as in WT cells. Almost 100% of WT, Latrunculin A-treated cells positioned the center of the RBD-GFP crescent to within a 45° angle (Φ) from the source of the cAMP gradient (Fig. 5 D and E). However, only ∼50% of gxcT− cells had RBD-GFP crescents centered within this range. The difference between RBD and PHcrac in gxcT− cells was not statistically significant. Without cAMP gradients, PHcrac-GFP remained in the cytosol in WT and gxcT− cells. Consistent with the function of Ras activation upstream of PIP3 production, we found that the localization of RBD-GFP was independent of PIP3 in Latrunculin A-treated WT cells (SI Appendix, Fig. S3).
Fig. 5.
GxcT orients the position of PIP3 production and Ras activation in a chemoattractant gradient. (A) Gradient sensing assay. Cells expressing PHcrac-GFP or RBD-GFP were placed in a cAMP gradient in the presence of Latrunculin A. To quantify the position of PHcrac and RBD crescents, the angle Φ was defined by calculating the angle that is formed by two lines: the line drawn between the centroid of the cell and the center of the crescent (red) and the line drawn between the centroid of the cell and the tip of the micropipette (blue). The edges of each crescent were defined as the points at which the fluorescence intensity on the membrane was 1.5-fold higher than that in the cytoplasm. (B and D) WT and gxcT− cells expressing PHcrac-GFP (B) or RBD-GFP (D) were exposed to a cAMP gradient in the presence of Latrunculin A. Images were taken 1 min after the cAMP gradient was formed. White dots indicate the position of the micropipette tip that was releasing cAMP. (C and E) The position of PHcrac-GFP (C) and RBD-GFP (E) crescents was quantified by calculating the angle (Φ), as described in A. The distribution of the resulting angles for individual cells is shown. Values represent the mean ± SEM (n ≥ 3). More than 25 cells were analyzed in each experiment. ***P < 0.001. (F and G) Time-lapse analyses of PHcrac-GFP (F) and RBD-GFP (G) localization were performed in the presence of a cAMP gradient and Latrunculin A. Images were taken every 30 s for 270 s. The position of the resulting fluorescent crescents was quantified by calculating the angle Φ at the indicated time points. Examples of plots from a single cell are shown. (H) The average of the absolute value of Φ for PHcrac-GFP and RBD-GFP crescents was calculated from the time-lapse analyses in F and G. The average value of Φ was determined from 10 time points for each cell. Values represent the mean ± SEM. N indicates the number of cells analyzed. (I) The length of PHcrac-GFP crescents on the plasma membrane was determined as a percentage of the whole cell perimeter. Values represent the mean ± SEM (n = 10).
When PHcrac-GFP crescents were observed using time-lapse fluorescence microscopy, we found that their position moved around the axis originating from the tip of the micropipette. The average fluctuation in the angle (Φ) over time was ∼20° in WT cells, whereas this fluctuation increased to ∼40° in gxcT− cells, suggesting that the position of the PHcrac-GFP crescent was significantly less stable in these mutants (Fig. 5 F and H). However, the relative length and intensity of the PHcrac-GFP crescent were indistinguishable in WT and gxcT− cells (Fig. 5I). The localization of RBD-GFP in gxcT− cells was also unstable, with an average angle (Φ) fluctuation of ∼30°, compared with a fluctuation of ∼15° in WT cells (Fig. 5 G and H). These results suggest that GxcT is required for the orientation of chemical sensing.
RacE Is Required for Chemotaxis and the Stable Distribution of Ras GTPase Activation.
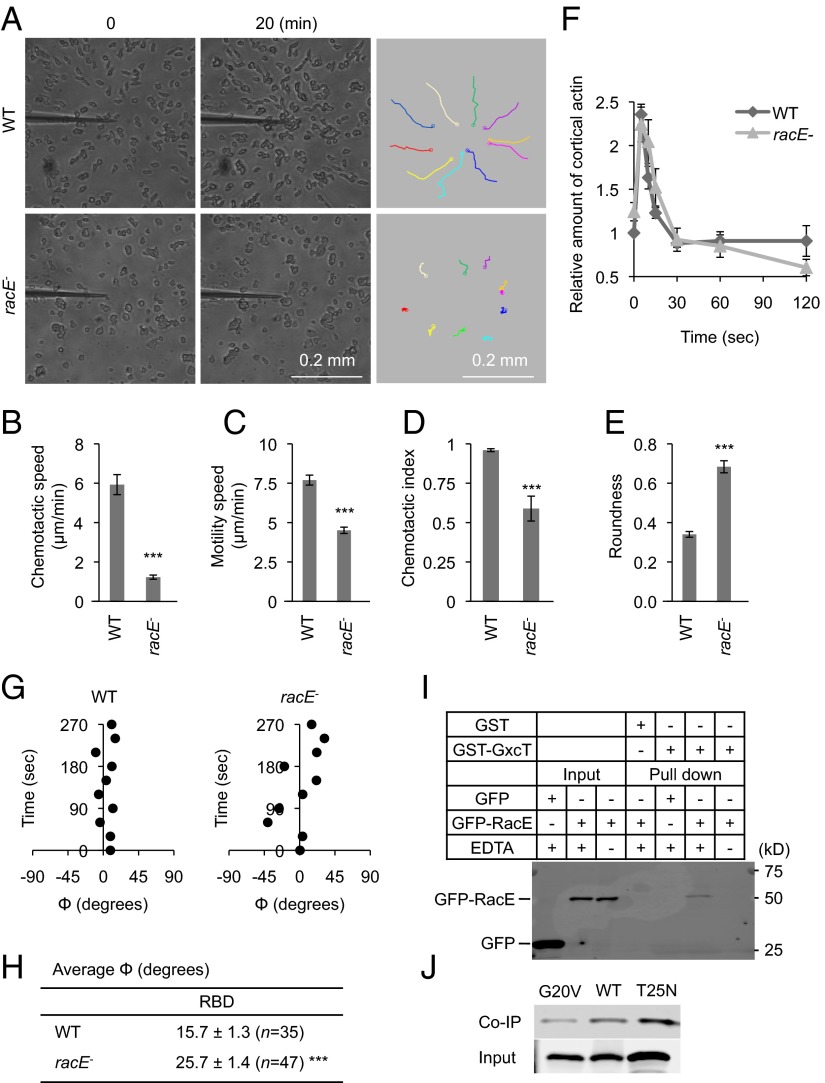
In addition to the GEF genes, we have also individually deleted the Rho GTPase genes in Dictyostelium (13 of 20 genes) (SI Appendix, Table S2 and Fig. S4) (35). Previous studies have shown that most Rho GTPases that have been named Rac in the Dictyostelium genome cannot be grouped into well-defined subfamilies in terms of sequence similarity except that Dictyostelium Rac1A–Rac1C, RacF1, and RacF2 are part of the human Rac subfamily (35, 41). During the characterization of the 13 Rho GTPase knockout strains, only racE− cells showed impaired growth both on bacterial lawns and in culture medium (SI Appendix, Fig. S5 A, D, and E). racE− cells also failed to aggregate normally during development (SI Appendix, Fig. S6). Furthermore, consistent with previous reports that RacE is required for cell division (42, 43), these mutant cells were multinucleated in shaking culture similar to those in gxcT− cells (SI Appendix, Fig. S5 B and C). Exogenous expression of a GFP-RacE fusion protein rescued the growth defects of racE− cells, suggesting that GFP-RacE is functional (SI Appendix, Fig. S7A). Because they were the only Rho GTPase mutants that phenocopied the original GxcT deletion strain, racE− cells were further characterized. When placed in a gradient of cAMP, racE− cells were defective in normal chemotaxis (Fig. 6A), with reduced chemotactic speed (Fig. 6B), motility speed (Fig. 6C), and chemotactic index (Fig. 6D). GFP-RacE rescued the chemotaxis defects of racE− cells (SI Appendix, Fig. S7B). Cell morphology was also abnormal in racE− cells, which had increased roundness compared with WT cells (Fig. 6E). cAMP receptor 1 was normally expressed in racE− cells during development (SI Appendix, Fig. S2E) and their chemotaxis defects were persistent after longer starvation (8 h) (SI Appendix, Fig. S2 A–D). Upon uniform stimulation, cAMP-induced actin polymerization occurred normally in racE− cells, as with gxcT− cells (Fig. 6F). To examine the localization of Ras activation and PIP3 production in racE− cells, WT and mutant cells expressing RBD-GFP or PHcrac-RFP were treated with Latrunculin A and placed in a cAMP gradient. Similar to gxcT− cells, we observed that the position of both RBD-GFP crescents (Fig. 6 G and H) and PHcrac-RFP crescents (SI Appendix, Fig. S8A) was unstable in racE− cells.
Fig. 6.
RacE is important for chemotaxis and binds to GxcT. (A) Developed WT and racE− cells were placed in a cAMP gradient and observed for 20 min. The trajectories of cell migration are shown (Right). (B–E) Chemotactic behaviors including chemotactic speed (B), motility speed (C), chemotactic index (D), and roundness (E) were quantified as described in Fig. 2 B–E. Values represent the mean ± SEM (n = 3). More than 20 cells were analyzed in each experiment. ***P < 0.001. (F) At the indicated time points after uniform cAMP stimulation, WT and racE− cells were lysed, and the cortical actin was isolated and quantified as described in Experimental Procedures. Values represent the mean ± SEM (n = 3). There are no significant differences between WT and racE− cells. (G) Time-lapse analyses of RBD-GFP localization were performed in the presence of a cAMP gradient and Latrunculin A. The position of the RBD-GFP crescents was quantified by calculating the angle Φ over time as described in Fig. 4 D–H. (H) The average value of Φ for RBD-GFP crescents was calculated from the time-lapse analyses in G. Values represent the mean ± SEM. N indicates the number of cells analyzed. ***P < 0.001. (I) Recombinant GST-GxcT was purified from E. coli and incubated, in the presence or absence of EDTA, with whole cell lysates prepared from Dictyostelium cells that were expressing either GFP-RacE or GFP. GST-GxcT was precipitated using glutathione beads, and the bound fraction was analyzed by immunoblotting using anti-GFP antibodies. GST was purified and precipitated as a negative control. (J) GST-GxcT was incubated with cell lysates expressing either GFP-RacE, GFP-RacE(G20V), or GFP-RacE(T25N) and precipitated using glutathione beads. The lysates (input) and bound fractions (coimmunoprecipitation, co-IP) were analyzed by immunoblotting using anti-GFP antibodies.
RacE Binds to GxcT.
We examined interactions between GxcT and RacE using a GST pull-down assay. Purified GST fused to the RhoGEF and PH domains of GxcT (GST-GxcT) was isolated from Escherichia coli and mixed with whole cell lysates that were prepared from Dictyostelium cells expressing either GFP-RacE or GFP alone. After incubation with these lysates, GST-GxcT was pulled down using glutathione beads, and the resulting pellet fraction was analyzed by immunoblotting with anti-GFP antibodies. GFP-RacE, but not GFP, was precipitated along with GST-GxcT in the presence of EDTA (Fig. 6I). Previous studies have shown that the interactions between Rho GTPases and their GEFs are increased by EDTA, which removes magnesium ions from the GTPase, thus causing the release of the bound guanine nucleotide (44). Our data suggest that nucleotide-free RacE preferentially associates with GxcT (Fig. 6I). As another control, GFP-RacE did not precipitate when GST was pulled down. Moreover, we compared interactions of GxcT with the dominant negative mutant of RacE [RacE(T25N)] and the constitutively active mutant [RacE(G20V)] (45). We found that GxcT strongly binds to RacE(T25N) and weakly binds to RacE(G20V), compared with the WT form of RacE (Fig. 6J), suggesting that GxcT is a potential GEF for RacE.
Overexpression of WT or a Constitutively Active Form of RacE, but Not a Dominant Negative Form, Stabilizes PHcrac Crescents.
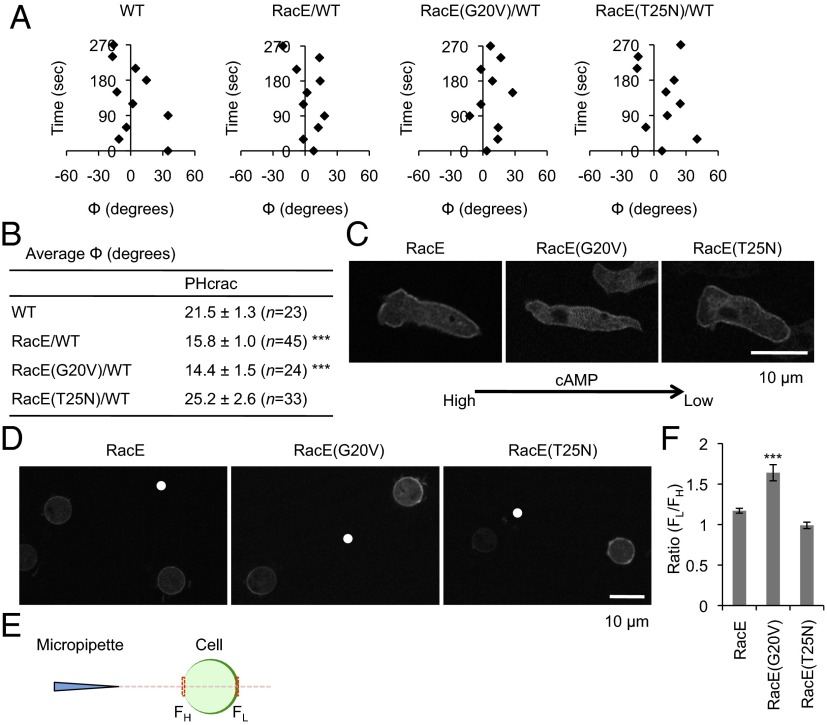
To determine whether overexpression of RacE can stabilize the position of PHcrac crescents, WT cells were cotransfected with PHcrac-RFP and GFP that was fused to different versions of RacE, including WT, RacE(G20V), and RacE(T25N). As shown in Fig. 7A, exogenous expression of either RacE or RacE(G20V) stabilized PHcrac-RFP crescent localization in response to a cAMP gradient by reducing the fluctuation in the angle Φ over time. Furthermore, quantification of these PHcrac-RFP crescents showed that the average value of Φ was significantly reduced from 20° in WT cells to 15° in cells expressing RacE or RacE(G20V) (Fig. 7B). This effect was dependent on RacE activity, as the expression of RacE(T25N) did not enhance the directional accuracy or stability of PHcrac-RFP crescent localization. In addition, the effect of RacE(G20V) required endogenous RacE as RacE(G20V) expression in racE− cells did not stabilize PHcrac-RFP crescents (SI Appendix, Fig. S8A). Finally, we also expressed RacE(G20V) in gxcT− cells and found that its expression does not rescue gradient-sending defects in this mutant (SI Appendix, Fig. S8B), suggesting that GxcT may have other targets in addition to RacE. Consistent with this notion, we found that GxcT binds to Rac1C, RacC, and RacF1, in addition to RacE (SI Appendix, Fig. S9).
Fig. 7.
RacE orients the localization of PHcrac-RFP crescents. (A) The localization of PHcrac-RFP in WT cells coexpressing GFP-RacE, GFP-RacE(G20V), or GFP-RacE(T25N) was analyzed by time-lapse fluorescence microscopy in the presence of a cAMP gradient and Latrunculin A. The position of the PHcrac-RFP crescents was quantified by calculating the angle Φ over time as described in Fig. 4 D–H. (B) The average value of Φ for PHcrac-GFP crescents was calculated from the time-lapse analyses in A, as described in SI Appendix, Fig. S3. Values represent the mean ± SEM. N indicates the number of cells analyzed. ***P < 0.001. (C) WT cells expressing GFP-RacE, GFP-RacE(G20V), or GFP-RacE(T25N) were placed in chemoattractant gradients and assayed by fluorescence microscopy for protein localization. The concentration of cAMP was higher on the left side of each image, as indicated by the arrow. (D) The localizations of GFP-RacE, GFP-RacE(G20V), and GFP-RacE(T25N) were examined by fluorescence microscopy in WT cells that were treated with Latrunculin A and placed in a cAMP gradient. White dots indicate the position of the cAMP releasing micropipette tips. (E and F) The ratio of GFP fluorescence on the plasma membrane facing lower (FL) and higher (FH) concentrations of cAMP was determined (FL/FH) for cells imaged as in D. Values represent the mean ± SEM. More than 25 cells were examined for each. ***P < 0.001.
RacE(G20V) Is Located at the Back of Cells in a cAMP Gradient When the Actin Cytoskeleton Is Disrupted.
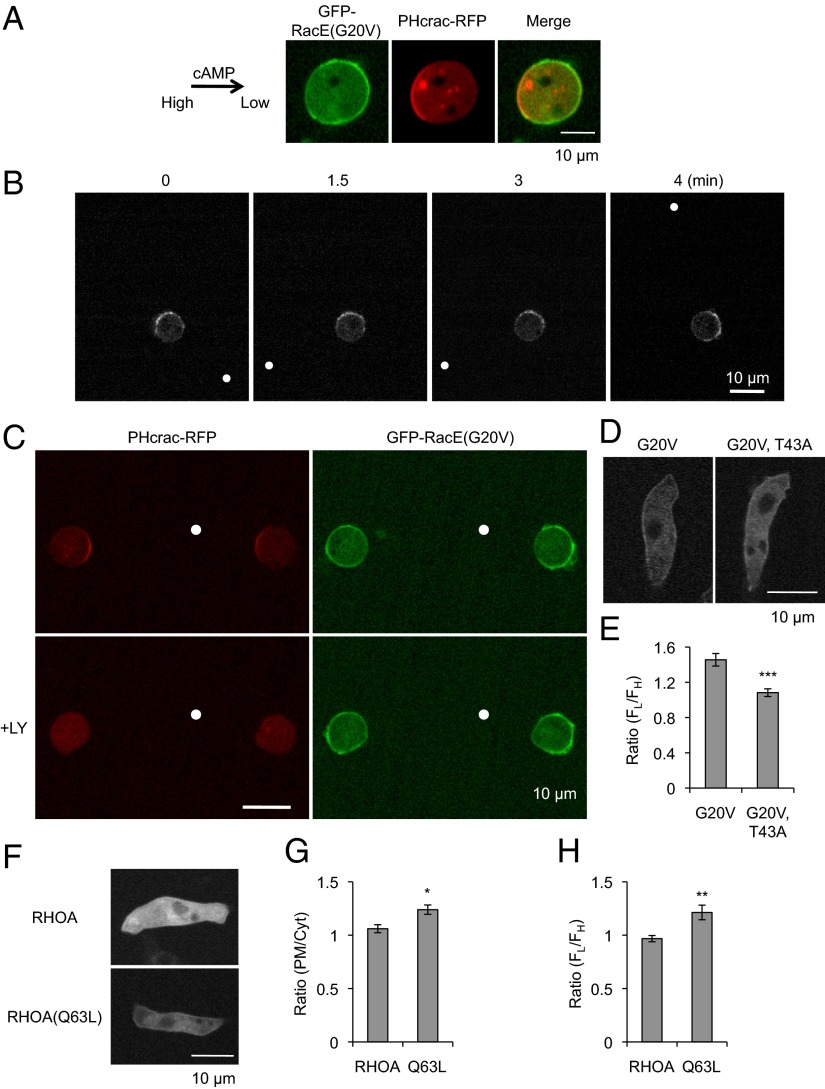
RacE has a CAAX motif and undergoes posttranslational lipid modifications for targeting to the plasma membrane (46). It has been reported that RacE, RacE(G20V), and RacE(T25N) are located uniformly on the plasma membrane in migrating cells (45). Consistent with previous findings, these RacE proteins, fused to GFP, were evenly distributed along the plasma membrane in cells undergoing chemotaxis to cAMP (Fig. 7C and SI Appendix, Fig. S10A). Remarkably, when cells were placed in a cAMP gradient after treatment with Latrunculin A, GFP-RacE(G20V) was located at the side facing away from the tip of the micropipette (Fig. 7D). When cells were treated with different concentrations of Latrunculin A, the localization of GFP-RacE(G20V) became polarized as the cell shape became less polarized (SI Appendix, Fig. S10B). In contrast, GFP-RacE and GFP-RacE(T25N) were uniformly distributed on the plasma membrane under the same conditions (Fig. 7D). Quantification of the fluorescence intensity showed a significant increase in the amount of GFP-RacE(G20V) at the side facing away from the micropipette relative to the side facing toward the micropipette, whereas the amount of GFP-RacE, as well as GFP-RacE(T25N), was equivalent at both ends of the cell (Fig. 7 E and F). Additionally, the localizations of GFP-RacE(G20V) and PHcrac-RFP appeared to be mutually exclusive when these proteins were coexpressed in Latrunculin A-treated cells (Fig. 8A). The localization of PHcrac is known to be highly dynamic and it responds to changes in the direction of a chemical gradient by reorienting toward the new chemoattractant source (13, 20). To determine whether the localization of GFP-RacE(G20V) is also dynamic and responsive to changes in cAMP gradients, a micropipette releasing cAMP was moved to different positions around the perimeter of Latrunculin A-treated cells expressing this protein. Under these conditions, GFP-RacE(G20V) consistently relocalized, keeping its position against the cAMP gradient (Fig. 8B).
Fig. 8.
The localization of RacE(G20V) is regulated by chemoattractant gradients in the absence of the actin cytoskeleton. (A) WT cells coexpressing GFP-RacE(G20V) and PHcrac-RFP were treated with Latrunculin A and placed in a cAMP gradient. The arrow indicates the direction of the gradient. (B) WT cells expressing GFP-RacE(G20V) were treated with Latrunculin A, placed in a cAMP gradient, and observed by fluorescence microscopy at the indicated time points. The direction of the gradient was changed at 1 and 3.5 min by repositioning a micropipette filled with cAMP. White dots indicate the position of the micropipette tip. (C) WT cells coexpressing GFP-RacE(G20V) and PHcrac-RFP were treated with Latrunculin A and placed in a cAMP gradient. Images were taken before (Upper) and ∼2 min after (Lower) the addition of the PI3 kinase inhibitor LY294002 (20 µM). White dots indicate the position of the cAMP source. (D) WT cells expressing GFP-RacE(G20V) or GFP-RacE(G20V, T43A) were observed by fluorescence microscopy in a cAMP gradient in the absence of Latrunculin A. The concentration of cAMP was higher on the upper side of each image. (E) The ratio of GFP fluorescence on the plasma membrane facing lower (FL) and higher (FH) concentrations of cAMP was determined (FL/FH) in WT cells expressing GFP-RacE(G20V) or GFP-RacE(G20V, T43A) in the presence of Latrunculin A. Values represent the mean ± SEM. More than 12 cells were examined for each. ***P < 0.001. (F) WT cells expressing GFP-RhoA or GFP-RhoA(Q63L) were placed in chemoattractant gradients in the absence of Latrunculin A. The concentration of cAMP was higher on the left side of each image. (G) The ratio of GFP fluorescence on the plasma membrane (FPM) and in the cytoplasm (FCYT) was determined (FPM/FCYT). Values represent the mean ± SEM. More than five cells were examined for each. *P < 0.05. (H) WT cells expressing GFP-human RhoA or GFP-human RhoA(Q63L) were placed in a cAMP gradient in the presence of Latrunculin A. FL/FH was determined as described above. Values represent the mean ± SEM. Twelve cells were examined for each. **P < 0.01.
To determine mechanisms that control the localization of GFP-RacE(G20V) in a cAMP gradient in the presence of Latrunculin A, we tested whether PIP3 regulates the dynamics of GFP-RacE(G20V) localization using the PI3 kinase inhibitor LY294002. For Latrunculin A-treated cells in a cAMP gradient, LY294002 addition abolished the localization of PHcrac-RFP, but did not affect the distribution of GFP-RacE(G20V) (Fig. 8C), suggesting that the regulation of the active Rho GTPase occurs upstream of PIP3 signaling. Moreover, we replaced a conserved residue in the effector loop of RacE (threonine at residue 43 was changed to alanine), creating GFP-RacE(G20V, T43A) (47). The amino acid sequence of this domain is highly specific to each type of Rho GTPases and identical between human RhoA and Dictyostelium RacE. An equivalent mutation in RhoA blocks interactions with its effectors (47). Strikingly, this mutation abolished the biased localization of GFP-RacE(G20V), as GFP-RacE(G20V, T43A) was uniformly distributed along the plasma membrane in a cAMP gradient in the presence of Latrunculin A (Fig. 8E and SI Appendix, Fig. S11). In contrast, this mutation did not affect the localization of GFP-RacE(G20V, T43A) in a cAMP gradient in the absence of Latrunculin A (Fig. 8D).
When WT RhoA and its constitutively active mutant, RhoA(Q63L), were expressed as GFP fusions in Dictyostelium cells, GFP-RhoA was mainly located in the cytosol of chemotaxing cells in the absence of Latrunculin A, whereas GFP-RhoA(Q63L) was slightly recruited to the plasma membrane (Fig. 8 F and G). Similar to RacE, GFP-RhoA(Q63L), but not GFP-RhoA, was located away from the cAMP gradient in the presence of Latrunculin A (Fig. 8H). These data suggest that the mechanism underlying the localization of Rho GTPases is evolutionarily conserved in chemical gradient sensing.
Discussion
In the current study, we showed that the Rho GTPase RacE and its potential GEF, GxcT, regulate the directional sensing mechanism independently of morphological polarity and the actin cytoskeleton. In both racE− and gxcT− cells, the accuracy of directional sensing was reduced, and pseudopods were abnormally extended from the lateral sides of migrating cells. RacE and GxcT were necessary for the spatial, but not temporal, accuracy of chemoattractant-induced Ras activation and PIP3 production. When chemoattractant stimuli were applied uniformly, thereby eliminating any spatial information, racE− and gxcT− cells exhibited normal Ras activation and PIP3 production. Furthermore, overexpression of RacE or RacE(G20V) (a constitutively active form), but not RacE(T25N) (a dominant negative form) further stabilized the accuracy of directional sensing in WT cells, suggesting that the activation of RacE is critical for this process. These data indicate the specific requirement for RacE and GxcT in regulating the stability of spatial sensing during chemotaxis. Our data also suggest that RacE and GxcT do not participate in restricting the localization of Ras and PIP3 signals to a crescent because the size and shape of PHcrac or RBD crescents are unaltered in racE− and gxcT− cells. Instead, it is the localization of the entire given crescent toward the gradient, which is stabilized by these proteins. Therefore, RacE and GxcT determine the orientation of the signal crescent.
Previous studies have shown that PTEN and a RasGAP called “NfaA” are involved in directional sensing. The roles of these proteins in directional sensing are distinct from that of RacE. PTEN dephosphorylates PIP3 and turns off PIP3 signaling (15, 17). Thus, pten− cells maintain increased amounts of PIP3 and the length of PHcrac crescents become longer in directional sensing studies. nfaA− cells fail to normally shut off Ras activation, leading to sustained activation of Ras GTPase and PIP3 signaling (25). In mammals, Leukemia-associated RhoGEF (LARG)-related GEFs, which contain the regulator of G protein signaling (RGS) domain, play roles in the activation of RhoA (48). In contrast, the Dictyostelium genome does not have LARG-related GEFs (35); therefore, it would be important to define mechanisms underlying the activation of GEFs for Rho GTPases in Dictyostelium cells in future studies.
The role of RacE has been studied during cytokinesis (49, 50). These studies have shown that RacE regulates the cell cortex via 14-3-3 and myosin II. Although our current study shows that cells lacking RacE are defective in spatial Ras and PIP3 activation independently of the cytoskeleton, we do not rule out the possibility that RacE also plays additional roles for chemotaxis through regulation of the cell cortex and myosin II assembly.
Rho GTPases contain a CAAX motif and are subjected to lipid modifications that anchor these proteins to the membrane; this membrane targeting is essential for GTPase function (46, 51). We showed that, in the absence of the actin cytoskeleton, RacE(G20V) and human RhoA(Q63L) were located on the portion of the plasma membrane that faced lower concentrations of chemoattractant in a gradient. The localization of RacE(G20V) depends on an intact effector domain, which is highly conserved in RacE and human RhoA. It would be important to identify components that bind to RacE(G20V) through the effector domain in future studies. In contrast, in the presence of the actin cytoskeleton, RacE was uniformly distributed along the plasma membrane, regardless of its activation status (Fig. 7C) (45, 52). The actin cytoskeleton either inhibits this recruitment or continuously redistributes the active form of RacE from the rear of the cell to other regions of the membrane, possibly through either active lateral movement or endocytosis- and exocytosis-based membrane trafficking. Such redistribution may be part of a feedback mechanism that amplifies the signaling response induced by chemical gradients. We speculate that active RacE at the front and back of Dictyostelium cells may play distinct roles in cell migration. RacE stabilizes directional sensing at the rear, whereas RacE may control the extension of pseudopods at the front. Supporting this notion, previous studies have shown that RhoA is active in the leading edge and retracting tail of fibroblasts and neurtrophils (30, 53, 54). RhoA also regulates PIP3 signaling by both activating and anchoring PTEN to the back of migrating neutrophils (55). These studies have suggested that RhoA is activated at different subcellular locations, where it exerts distinct functions through its specific interactions with different upstream and downstream components (56).
As GxcT was uniformly distributed in the cytoplasm, RacE signaling might convey spatial information by one of two possible mechanisms: GxcT may be locally activated, or RacE may translocate to the rear of cells once it is activated. As discussed above, our observation that the constitutively active mutant RacE(G20V), which does not rely on GxcT for activation, was enriched at the rear of cells supports the latter possibility. Because RacE(G20V) and PHcrac have opposing intracellular localizations under certain conditions, we propose that RacE inhibits the activation of Ras GTPases, thereby preventing the production of PIP3 at the rear of cells. In summary, our findings describe a unique function of RacE acting upstream of Ras/PIP3 signaling and actin cytoskeleton remodeling to regulate directional sensing. We propose that Rho GTPases play a critical role in persistent chemotactic migration by establishing accurate directional sensing.
Experimental Procedures
Details of all experimental procedures for lipid dot blot, actin polymerization, and GST pull-down assays are described in SI Appendix, SI Methods.
Cell Culture, Gene Knockout, and Plasmids.
All D. discoideum cell lines were cultured in Dictyostelium standard culture medium (HL5) medium at 22 °C. Genes were disrupted by homologous recombination using the blasticidin resistance cassette (39). Gene disruption was confirmed by PCR and a Southern blot. The primers used for the construction of the gene targeting vectors are listed in SI Appendix, Tables S2 and S3. The protein expression plasmids and primes used for plasmid construction are listed in SI Appendix, Tables S3 and S5, respectively.
Cell Growth and Development.
To examine cell growth, 25 mL of cells (4 × 105 cells/mL) were cultured in a 250-mL flask on a rotary shaker at 180 rpm at 22 °C and cells were counted daily with a hemocytometer. To assess developmental phenotypes, cells growing exponentially were washed twice in developmental buffer (DB) (10 mM phosphate buffer, 2 mM MgSO4, 0.2 mM CaCl2) and plated on 1% nonnutrient DB agar (5 × 105 cells/cm2).
Chemotaxis.
A chemotaxis assay was performed as described (17, 38, 39, 57). Cells were cultured in HL5 medium on Petri dishes, washed twice with DB, resuspended to 2 × 107 cells/mL, and shaken for 1 h before being induced to differentiate with 100 nM cAMP pulses at 6-min intervals for 4 h. Differentiated cells were plated on a chambered coverslip (Lab-Tek; Nalgen Nunc). A cAMP gradient was generated by a micropipette (Femtotips; Eppendorf) containing 1 µM cAMP and a microinjector with a compensation pressure of 100 hPa (FemtoJet; Eppendorf). Images of moving cells were recorded at 30-s intervals for 20 min using an Olympus CKX41 inverted microscope equipped with a 10× objective connected to a digital camera (CFW-1308M). ImageJ software was used to collect and process data.
Directional Sensing Assay.
Directional sensing was assessed as described (13, 15, 20, 23, 24, 58). After developed for 5 h, cells expressing PHcrac-GFP, PHcrac-RFP, or RBD-GFP were plated on a chambered coverslip and treated with 5 μM Latrunculin A for 10 min. A cAMP gradient was generated by a micropipette containing 1 µM cAMP. Cells were observed through a microscope consisting of a fully automated DMI6000 (Leica) and a Yokogawa CSU10 spinning disk confocal.
Statistical Analysis.
Results were statistically analyzed using the t test.
Supplementary Material
Acknowledgments
We are grateful to P. N. Devreotes and K. F. Swaney for critically reading the manuscript and members of the M.I. and H. Sesaki laboratories for helpful discussion. This work was supported by National Institutes of Health Grant GM084015 (to M.I.) and Grant GM089853 (to H. Sesaki).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312540110/-/DCSupplemental.
References
- 1.Roussos ET, Condeelis JS, Patsialou A. Chemotaxis in cancer. Nat Rev Cancer. 2011;11(8):573–587. doi: 10.1038/nrc3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang F. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb Perspect Biol. 2009;1(4):a002980. doi: 10.1101/cshperspect.a002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341(1):20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Heng JI, Chariot A, Nguyen L. Molecular layers underlying cytoskeletal remodelling during cortical development. Trends Neurosci. 2010;33(1):38–47. doi: 10.1016/j.tins.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Insall RH, Machesky LM. Actin dynamics at the leading edge: From simple machinery to complex networks. Dev Cell. 2009;17(3):310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10(8):538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: A network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Chen CL, Iijima M. Signaling mechanisms for chemotaxis. Dev Growth Differ. 2011;53(4):495–502. doi: 10.1111/j.1440-169X.2011.01265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J Biol Chem. 2003;278(23):20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 10.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9(11):846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 11.Van Haastert PJ, Devreotes PN. Chemotaxis: Signalling the way forward. Nat Rev Mol Cell Biol. 2004;5(8):626–634. doi: 10.1038/nrm1435. [DOI] [PubMed] [Google Scholar]
- 12.Xiao Z, Zhang N, Murphy DB, Devreotes PN. Dynamic distribution of chemoattractant receptors in living cells during chemotaxis and persistent stimulation. J Cell Biol. 1997;139(2):365–374. doi: 10.1083/jcb.139.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin T, Zhang N, Long Y, Parent CA, Devreotes PN. Localization of the G protein betagamma complex in living cells during chemotaxis. Science. 2000;287(5455):1034–1036. doi: 10.1126/science.287.5455.1034. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Meier-Schellersheim M, Jiao X, Nelson LE, Jin T. Quantitative imaging of single live cells reveals spatiotemporal dynamics of multistep signaling events of chemoattractant gradient sensing in Dictyostelium. Mol Biol Cell. 2005;16(2):676–688. doi: 10.1091/mbc.E04-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167(3):505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95(1):81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 17.Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109(5):599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- 18.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109(5):611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 19.Cai H, et al. Ras-mediated activation of the TORC2-PKB pathway is critical for chemotaxis. J Cell Biol. 2010;190(2):233–245. doi: 10.1083/jcb.201001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janetopoulos C, Ma L, Devreotes PN, Iglesias PA. Chemoattractant-induced phosphatidylinositol 3,4,5-trisphosphate accumulation is spatially amplified and adapts, independent of the actin cytoskeleton. Proc Natl Acad Sci USA. 2004;101(24):8951–8956. doi: 10.1073/pnas.0402152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samadani A, Mettetal J, van Oudenaarden A. Cellular asymmetry and individuality in directional sensing. Proc Natl Acad Sci USA. 2006;103(31):11549–11554. doi: 10.1073/pnas.0601909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Servant G, et al. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287(5455):1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kortholt A, et al. Dictyostelium chemotaxis: Essential Ras activation and accessory signalling pathways for amplification. EMBO Rep. 2011;12(12):1273–1279. doi: 10.1038/embor.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iijima M, Huang YE, Luo HR, Vazquez F, Devreotes PN. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J Biol Chem. 2004;279(16):16606–16613. doi: 10.1074/jbc.M312098200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18(20):1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sit ST, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci. 2011;124(Pt 5):679–683. doi: 10.1242/jcs.064964. [DOI] [PubMed] [Google Scholar]
- 27.Jaffe AB, Hall A. Rho GTPases: Biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 28.Cain RJ, Ridley AJ. Phosphoinositide 3-kinases in cell migration. Biol Cell. 2009;101(1):13–29. doi: 10.1042/BC20080079. [DOI] [PubMed] [Google Scholar]
- 29.Vanhaesebroeck B, Stephens L, Hawkins P. PI3K signalling: The path to discovery and understanding. Nat Rev Mol Cell Biol. 2012;13(3):195–203. doi: 10.1038/nrm3290. [DOI] [PubMed] [Google Scholar]
- 30.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440(7087):1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 31.Wong K, Pertz O, Hahn K, Bourne H. Neutrophil polarization: Spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc Natl Acad Sci USA. 2006;103(10):3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290(5490):333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 33.Van Keymeulen A, et al. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J Cell Biol. 2006;174(3):437–445. doi: 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305(5690):1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 35.Vlahou G, Rivero F. Rho GTPase signaling in Dictyostelium discoideum: Insights from the genome. Eur J Cell Biol. 2006;85(9-10):947–959. doi: 10.1016/j.ejcb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through Class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34(Pt 5):647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 37.Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9(2):99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 38.Zhang P, Wang Y, Sesaki H, Iijima M. Proteomic identification of phosphatidylinositol (3,4,5) triphosphate-binding proteins in Dictyostelium discoideum. Proc Natl Acad Sci USA. 2010;107(26):11829–11834. doi: 10.1073/pnas.1006153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen CL, Wang Y, Sesaki H, Iijima M. Myosin I links PIP3 signaling to remodeling of the actin cytoskeleton in chemotaxis. Sci Signal. 2012;5(209):ra10. doi: 10.1126/scisignal.2002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bretschneider T, et al. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14(1):1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 41.Rivero F, Dislich H, Glöckner G, Noegel AA. The Dictyostelium discoideum family of Rho-related proteins. Nucleic Acids Res. 2001;29(5):1068–1079. doi: 10.1093/nar/29.5.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larochelle DA, Vithalani KK, De Lozanne A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J Cell Biol. 1996;133(6):1321–1329. doi: 10.1083/jcb.133.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson DN, Spudich JA. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J Cell Biol. 2000;150(4):823–838. doi: 10.1083/jcb.150.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiering D, Hodgson L. Dynamics of the Rho-family small GTPases in actin regulation and motility. Cell Adhes Migr. 2011;5(2):170–180. doi: 10.4161/cam.5.2.14403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larochelle DA, Vithalani KK, De Lozanne A. Role of Dictyostelium racE in cytokinesis: Mutational analysis and localization studies by use of green fluorescent protein. Mol Biol Cell. 1997;8(5):935–944. doi: 10.1091/mbc.8.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5(5):405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- 47.Kato K, et al. The inositol 5-phosphatase SHIP2 is an effector of RhoA and is involved in cell polarity and migration. Mol Biol Cell. 2012;23(13):2593–2604. doi: 10.1091/mbc.E11-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi Y, et al. The mDial formin is required for neutrophil polarization, migration, and activation of the LARG/RhoA/ROCK signaling axis during chemotaxis. J Immunol. 2009;182(6):3837–3845. doi: 10.4049/jimmunol.0803838. [DOI] [PubMed] [Google Scholar]
- 49.Zhou Q, et al. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr Biol. 2010;20(21):1881–1889. doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerald N, Dai J, Ting-Beall HP, De Lozanne A. A role for Dictyostelium racE in cortical tension and cleavage furrow progression. J Cell Biol. 1998;141(2):483–492. doi: 10.1083/jcb.141.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: New insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 52.Yonemura S, Hirao-Minakuchi K, Nishimura Y. Rho localization in cells and tissues. Exp Cell Res. 2004;295(2):300–314. doi: 10.1016/j.yexcr.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461(7260):99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu J, et al. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114(2):201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, et al. Regulation of PTEN by Rho small GTPases. Nat Cell Biol. 2005;7(4):399–404. doi: 10.1038/ncb1236. [DOI] [PubMed] [Google Scholar]
- 56.Pertz O. Spatio-temporal Rho GTPase signaling: Where are we now? J Cell Sci. 2010;123(Pt 11):1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 57.Wang Y, et al. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22(13):2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai H, Huang CH, Devreotes PN, Iijima M. Analysis of chemotaxis in Dictyostelium. Methods Mol Biol. 2012;757:451–468. doi: 10.1007/978-1-61779-166-6_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.