Significance
We describe interaction between the cell division cycle 45 (Cdc45)/minichromosome maintenance 2-7 (Mcm2-7)/Go, Ichi, Nii, and San (GINS) (the CMG complex) and chromosome transmission fidelity 4 (Ctf4). The CMG complex catalyzes parental duplex DNA separation during eukaryotic replication. Ctf4 was reported previously to bind yeast DNA polymerase α (Pol α) and be essential for human DNA replication. Here, we show that human Ctf4 (hCtf4) interacts with human CMG (hCMG) complex to form the dimeric hCtf4–monomeric hCMG complex, which, like hCMG, contains DNA helicase activity. Complex formation involves interactions of hCtf4 with multiple hCMG components. We propose that Ctf4 links the replicative Pol α–primase complex to the replicative helicase, coordinating DNA unwinding and polymerase activities.
Keywords: replisome core structure, dimerization, replication initiation
Abstract
Chromosome transmission fidelity 4 (Ctf4) is a conserved protein required for DNA replication. In this report, interactions between human Ctf4 (hCtf4) and the replicative helicase containing the cell division cycle 45 (Cdc45)/minichromosome maintenance 2-7 (Mcm2-7)/Go, Ichi, Nii, and San (GINS) (CMG) proteins [human CMG (hCMG) complex] were examined. The hCtf4–CMG complex was isolated following in vitro interaction of purified proteins (hCtf4 plus the hCMG complex), coinfection of Spodoptera frugiperda (Sf9) insect cells with viruses expressing the hCMG complex and hCtf4, and from HeLa cell chromatin after benzonase and immunoprecipitation steps. The stability of the hCtf4–CMG complex depends upon interactions between hCtf4 and multiple components of the hCMG complex. The hCtf4–CMG complex, like the hCMG complex, contains DNA helicase activity that is more salt-resistant than the helicase activity of the hCMG complex. We demonstrate that the hCtf4–CMG complex contains a homodimeric hCtf4 and a monomeric hCMG complex and suggest that the homodimeric hCtf4 acts as a platform linking polymerase α to the hCMG complex. The role of the hCMG complex as the core of the replisome is also discussed.
Eukaryotic DNA replication is a stepwise process by which protein complexes are assembled on chromatin. During the G1 phase of the cell cycle, the origin recognition complex (ORC) binds to replication origins and recruits cell division cycle 6 (Cdc6) (1). This complex leads to the association of cdc10 dependent transcript 1 (Cdt1)/Mcm2-7 with chromatin and the loading of the Mcm2-7 complex as a head-to-head dimer (2, 3). At the G1/S transition, this prereplication complex is altered further by a number of replication initiation factors whose actions are facilitated by the cyclin-dependent and cell division cycle 7 (Cdc7)-dumbbell former 4 (Dbf4) kinases (4). These replication initiation factors [which include synthetically lethal with dpb11-1 (Sld)2, Sld3, Sld7, DNA polymerase B possible subunit 11 (Dpb11), and DNA polymerase ε (Pol ε) in budding yeast] play critical roles resulting in the interaction of Cdc45 and GINS with the Mcm2-7 complex and the formation of the replicative DNA helicase complex containing Cdc45/Mcm2-7/GINS (CMG) (5). Other replication factors (including Mcm10 and Ctf4) contribute to the activation of the CMG helicase and association of proteins that recruit the replicative Pols to effect DNA replication (6, 7). Interactions between TopBP1-interacting, replication-stimulating protein (Treslin) (homolog of Sld3) and MDM two binding protein (MTBP) (homolog of Sld7) with topoisomerase-IIbeta-binding protein 1 (TopBP1) (homolog of Dpb11) have been detected in higher eukaryotes, suggesting that the mechanism used by yeast for the chromatin loading of Cdc45 and GINS may be conserved (8). However, in vivo studies in HeLa cells using bimolecular fluorescence complementation assays reported that Mcm10 as well as RecQ protein-like 4 (RecQL4) (homolog of Sld2) and Ctf4 are required for human CMG (hCMG) complex formation (9).
In budding yeast, a large replisome progression complex (RPC) directly involved in DNA replication was identified and found to contain the CMG complex (10). It was shown that the CMG complex acts as the core engine responsible for the movement and activities of the replication fork (11). The CMG complex, composed of stoichiometric levels of Cdc45, Mcm2-7, and GINS, was isolated from Drosophila embryos, as well as insect cells, using the baculovirus-expression system and found to contain 3′→5′ DNA helicase activity (12, 13). Subsequently, the hCMG complex was purified and shown to support the coupled helicase-Pol ε–dependent synthesis of leading strand DNA in vitro (14).
Ctf4 was isolated in budding yeast as a chromosome transmission fidelity factor (15). Although Ctf4 is not essential for budding yeast growth (16), a null mutant of the Ctf4 homolog in fission yeast minichromosome loss 1 (Mcl1) shows severe growth retardation or is lethal (17, 18). Immunodepletion of Ctf4 in Xenopus egg extracts, as well as siRNA targeted destruction of human Ctf4 (hCtf4) in HeLa cells, markedly decreased DNA replication (7, 19). Ctf4 was reported to interact with various replication proteins, including Mcm10, GINS, Pol α, Pol δ, and Pol ε, and was reported to contribute to the stability of the p180 catalytic subunit of Pol α (7, 19–22). Recently, Ctf4 was found to connect Pol α to the budding yeast RPC (21, 22). The association of Ctf4 with the RPC was shown to be dependent on the interaction of Ctf4 with GINS, a component of the CMG complex present in the RPC, suggesting a possible scaffolding function that coordinates the action of the replicative helicase and Pol α during DNA replication.
In the study reported here, we isolated a 12-subunit complex composed of hCtf4 and the hCMG complex. We characterized this complex and showed that it contained DNA helicase activity that is less salt-sensitive than the helicase activity of the hCMG complex. Ctf4 was reported previously to be a homodimer, and we show that the hCtf4–CMG complex is an hCtf4 dimer and hCMG monomer (19, 23). We also show that the interactions between hCtf4 and the hCMG complex lead to a more stable complex than that formed by interactions of hCtf4 with components of the hCMG complex (Cdc45, Mcm2-7, or GINS). Interactions between hCtf4 and the hCMG complex were demonstrated following baculovirus infection of Sf9 cells in vitro with purified components and in HeLa cells. In the latter case, a stable hCtf4–CMG complex was detected following chromatin isolation, digestion of DNA with benzonase, and immunoprecipitation. Collectively, these findings indicate that the Ctf4–CMG complex is formed both in vivo and in vitro in higher eukaryotes.
Results
hCtf4 Forms a Stable Complex with the hCMG Replicative Helicase.
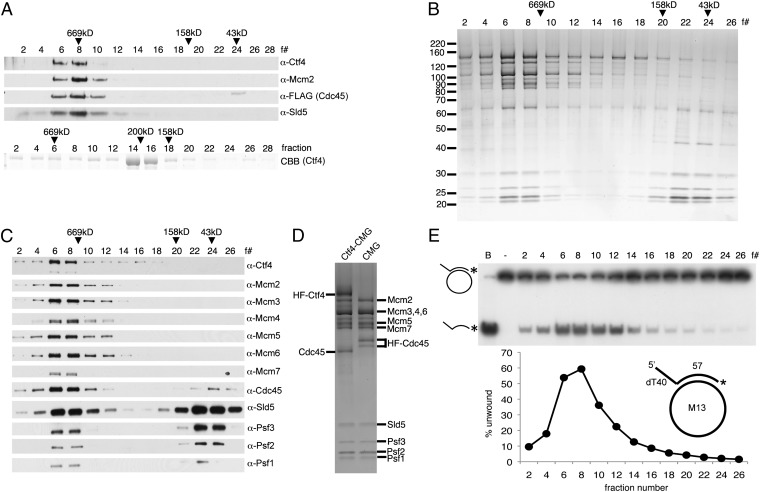
Based on the findings that Ctf4 interacts with components of the RPC (10), we examined whether direct in vitro interaction of purified hCtf4 and the isolated hCMG complex could be detected. For this purpose, untagged hCtf4 and the hCMG complex [His6-FLAG2 (HF)–tagged Cdc45] were mixed; following incubation, the mixture was adsorbed to FLAG M2 agarose beads and FLAG-peptide–eluted proteins subjected to glycerol gradient centrifugation. As shown (Fig. 1A, Upper), hCtf4 sedimented coincidently with the hCMG complex, whereas purified hCtf4 alone sedimented close to the β-amylase marker (200 kDa) (Fig. 1A, Lower), as previously reported (19). The level of hCtf4 and hCMG proteins present in the peak fractions varied, most likely reflecting the higher molecular mass of the hCtf4–CMG complex compared with the hCMG complex and the presence of the hCMG complex free of Ctf4.
Fig. 1.
hCtf4 forms a stable complex with the hCMG complex. (A) Purified hCMG (5 pmol) (HF tag on Cdc45) and untagged hCtf4 (15 pmol, as a dimer) were mixed with anti-FLAG M2 affinity gel. (Upper) Bound proteins were eluted and subjected to glycerol gradient sedimentation. (Lower) As a control, untagged hCtf4 was sedimented separately. CBB, Coomassie brilliant blue. The hCtf4–CMG complex was purified as described in SI Materials and Methods, and each fraction (5 μL) was separated electrophoretically through a 4–20% (wt/vol) polyacrylamide gradient gel (Invitrogen), followed by silver staining (B) or Western blot analysis (C) against all 12 subunits of the hCtf4–CMG complex. The peak positions of protein markers (GE Healthcare Life Sciences) after sedimentation are indicated above fractions (669 kDa, thyroglobulin; 158 kDa, aldolase; 43 kDa, ovalbumin). (D) hCtf4–CMG and hCMG were loaded side by side onto a 4–20% (wt/vol) gel and silver-stained. (E) Helicase activity was measured across the hCtf4–CMG glycerol gradient (0.5-μL fractions), and the substrate unwound (%) was calculated and is shown in the graph. The structure of the helicase substrate containing a 57-mer duplex region and a 5′-dT40 tail (M13 annealed to labeled oligonucleotide no. 2) is shown in the graph. See Table S1 for description of oligonucleotides in this study. *, Location of 32P in substrate. B, boiled substrate.
We next investigated the helicase activity associated with the hCtf4–CMG complex. This complex, devoid of the free hCMG complex, was isolated as described in SI Materials and Methods following infection of Sf9 cells with viruses expressing an HF tag fused to the N terminus of hCtf4 and the hCMG complex that included GST-Sld5. SDS/PAGE analyses of glycerol gradient fractions of the hCtf4–CMG complex (Fig. 1B) revealed the presence of the expected 12 proteins, which were further verified by Western blot analysis (Fig. 1C). Side-by-side SDS/PAGE analysis of hCtf4–CMG (HF-Ctf4) and hCMG (HF-Cdc45) glycerol gradient peak fractions revealed that the isolated complexes differed solely by the presence of hCtf4 (Fig. 1D). The DNA unwinding activity associated with the hCtf4–CMG complex was examined in glycerol gradient fractions using an M13-oligonucleotide substrate. As shown in Fig. 1E, maximal helicase activity and the hCtf4–CMG complex comigrated. These findings indicate that hCtf4 forms a complex with the hCMG complex and that the product contains DNA helicase activity.
hCtf4 Binding Conditionally Affects the hCMG Helicase Activity.
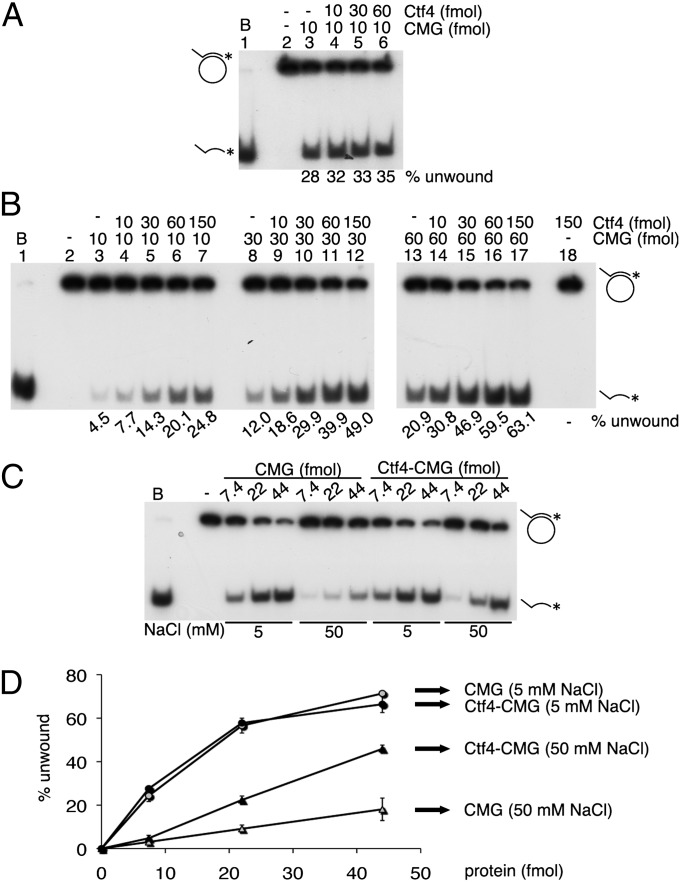
Because purified hCtf4 interacted directly in vitro with the purified hCMG complex, we examined its influence on the DNA helicase activity of the hCMG complex using standard DNA helicase conditions (5 mM NaCl, M13-oligonucleotide substrate). As shown (Fig. 2A), the addition of hCtf4 had little effect on the unwinding activity of the hCMG complex. We noted that at higher NaCl levels (50 mM), the helicase activity of the hCMG complex was reduced ∼sixfold compared with that observed at 5 mM NaCl (compare lanes 3 of Fig. 2 A and B). However, in the presence of 50 mM NaCl, increasing levels of hCtf4 stimulated the hCMG helicase activity maximally 5.5-fold (Fig. 2B, compare lanes 3 and 7). At higher levels of hCMG, however, the stimulation by hCtf4 decreased (Fig. 2B, lanes 8–17). Although the helicase activities of purified hCtf4–CMG and hCMG complexes were the same in the presence of 5 mM NaCl, the hCtf4–CMG complex displaced two- to threefold more DNA than the hCMG complex in the presence of 50 mM NaCl (Fig. 2 C and D), which is somewhat lower than the stimulation described in Fig. 2B. In contrast to observations with M13-oligonucleotide helicase substrates, hCtf4 failed to stimulate the hCMG helicase activity using oligonucleotide substrates (Fig. S1A). We also noted that hCtf4 had no effect on the DNA-independent ATPase activity of the hCMG complex (Fig. S1B). These findings suggest that hCtf4, which possesses ssDNA binding activity (19), might increase the DNA binding properties of the hCMG complex.
Fig. 2.
hCtf4 stimulates the hCMG helicase activity. Indicated levels of hCMG and untagged hCtf4 were incubated using helicase assay conditions containing 5 mM NaCl (A) or 50 mM NaCl (B). (C) Helicase assays were performed with the hCMG and hCtf4–CMG complexes in the presence of 5 or 50 mM NaCl. (D) Substrate unwound (%) from the results presented in C was plotted against the protein added (femtomoles). It should be noted that the amount of hCtf4 indicated in the figures was calculated as a dimer.
Together, these results indicate that the hCtf4–CMG complex contains DNA helicase activity. The association of hCtf4 with hCMG renders the hCMG helicase activity on M13 substrates more salt-resistant than that detected with the hCMG complex alone. The reasons for this difference, however, require further studies.
hCtf4 Associated with the hCMG Complex Is a Dimer.
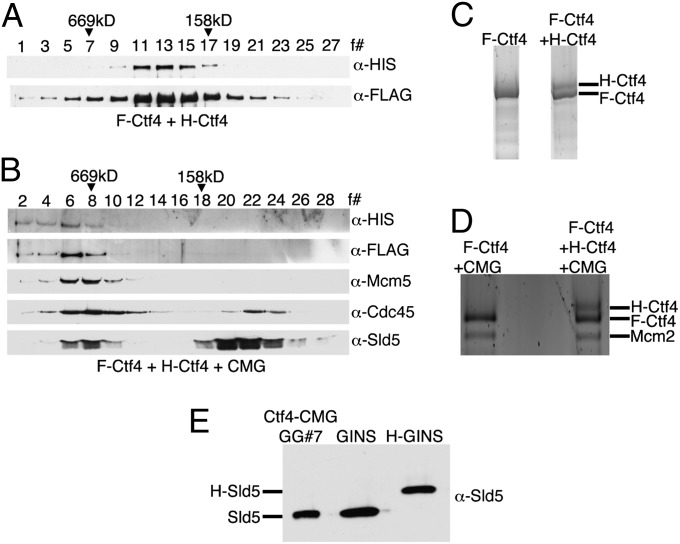
The cloned Drosophila CMG complex was shown to be a stoichiometric complex containing equivalent levels of Cdc45, Mcm2-7, and GINS (13), and we demonstrated that this is also true for the hCMG complex [based on quantitative Western blot analyses (Fig. S2 A–C)]. Ctf4, isolated from both Xenopus and humans, was previously reported to be dimeric, and the region in Ctf4 responsible for this property was mapped to the SepB domain (19, 23). Quantitative Western blot analyses of proteins present in the hCtf4–CMG peak fraction described in Fig. 1B suggested that hCtf4 was dimeric (hCMG/hCtf4 ratio of 1:1.87; Fig. S3 D and E). To validate the structure of hCtf4 in the hCtf4–CMG complex further, two different hCtf4 viruses, each containing a distinct tag [His6 (H) or FLAG (F)], were constructed. Sf9 cells were coinfected with these viruses and those expressing hCMG components. The hCtf4–CMG complex was isolated as described in SI Materials and Methods, in which the initial step, α-FLAG immunoprecipitation, results in the selective pull-down of F-hCtf4 and associated proteins. If hCtf4 were dimeric, both F-hCtf4 and H-hCtf4 would be expected to copurify. Both the hCtf4–CMG and free Ctf4 were isolated and subjected to glycerol gradient separation (steps used in this purification are shown in Fig. S3A). The cosedimentation of F-hCtf4 and H-hCtf4 was detected by Western blot analyses in gradients containing either hCtf4 alone or the hCtf4–CMG complex (Fig. 3 A and B). These findings indicate that both free Ctf4 and hCtf4 complexed to hCMG are dimeric and substantiate this previous conclusion based on hydrodynamic properties (19). We noted that the level of the hCtf4 and hCMG proteins present in the peak fractions varied (Fig. 3B), unlike in previous preparations (Fig. 1 B and C), suggesting that a portion of hCtf4 separated from hCMG during purification for unknown reasons. We also found that F-hCtf4 and H-hCtf4 separated slightly following SDS/PAGE (Fig. S3B), and this difference was also used to verify that hCtf4 is a dimer. For this purpose, F-hCtf4 and the F-hCtf4–CMG complex were isolated (Fig. S3 C and D) and each preparation was subjected to SDS/PAGE analysis side by side with (F-H) hCtf4 and (F-H) hCtf4–CMG isolated from infected Sf9 cells expressing F-Ctf4 and H-Ctf4 (Fig. 3 C and D and Fig. S3 E and F). Silver staining of these gels showed that H-hCtf4 migrated slightly slower than F-hCtf4. These findings further verified that hCtf4 alone and hCtf4 complexed to hCMG are dimeric.
Fig. 3.
hCtf4 is present as a dimer in the hCtf4–CMG complex. Sf9 cells were infected with two differently tagged hCtf4 viruses (H-hCtf4 and F-hCtf4) and viruses expressing hCMG components, and the free hCtf4 and hCtf4–CMG complex formed were purified. Western blot analyses of fractions from the glycerol gradient sedimentation of hCtf4 (A) and hCtf4–CMG (B) are shown. The peak glycerol gradient fractions of the hCtf4 and hCtf4–CMG preparations, shown in A and B, were loaded onto a 4–20% polyacrylamide gel side by side with hCtf4 (C) or hCtf4–CMG (D) purified from cells expressing F-hCtf4 and the hCMG complex. Regions containing hCtf4 bands were cropped in these figures (images from the original gels are shown in Fig. S4 E and F). (E) Sf9 cells were coinfected with F-hCtf4 and hCMG viruses that included two differently tagged Sld5 subunits (GST-Sld5 and H-Sld5), and the hCtf4–CMG complex formed was purified. The peak glycerol gradient fraction was loaded onto gels side by side with untagged and H-hGINS (His6 tag on Sld5) and immunoblotted for Sld5.
To evaluate the stoichiometry of hCMG present in the hCtf4–CMG complex, the hCtf4–CMG complex was isolated after coinfection with two viruses expressing differently tagged Sld5 (GST-Sld5 and H-Sld5; the isolation procedure is described in Fig. S4A). As shown in Fig. S4B, both GST-Sld5 and H-Sld5 were detected through the Q Sepharose column step. However, after GST pull-down and protein elution (by cleavage of Sld5 from GST), only untagged Sld5 was detected (Fig. S4B). The absence of H-Sld5 in the hCtf4–CMG glycerol gradient peak fraction was confirmed by Western blot and silver stain analyses (Fig. 3E and Fig. S4C). Thus, the hCtf4–CMG complex contains two hCtf4 molecules and one hCMG molecule. The implications of these findings are discussed below.
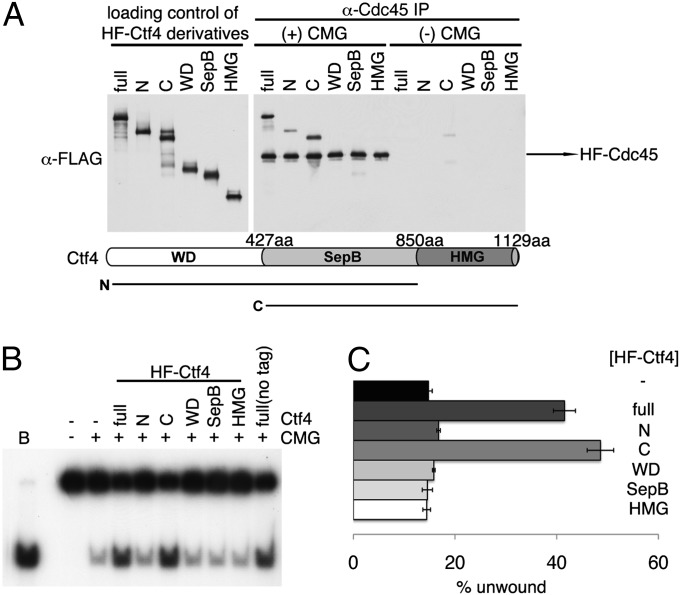
Truncated SepB + HMG Derivative of hCtf4 Binds Efficiently to hCMG and Stimulates Its Helicase Activity.
hCtf4 is composed of three motifs: the N-terminal WD, middle SepB, and C-terminal HMG domains (17). Ctf4 isolated from yeasts, and Drosophila contains the WD and SepB domains but lacks the HMG domain (24). The domain(s) of hCtf4 required for its interaction with hCMG was mapped by in vitro protein binding experiments, followed by α-Cdc45 immunoprecipitation (Fig. 4A). Robust binding of full-length hCtf4 and the C (SepB + HMG) domains to the hCMG complex was observed. Weaker hCMG binding was detected with the N (WD + SepB) domains, whereas the WD, SepB, or HMG domain alone failed to bind the hCMG complex. Previous reports indicated that the budding yeast SepB domain alone mediated an interaction between Ctf4 and GINS (22), suggesting that the HMG domain of higher eukaryotic Ctf4 may have additional binding sites in the CMG complex important for the stability of the hCtf4–CMG complex.
Fig. 4.
Physical and biochemical interactions between different domains of hCtf4 and the hCMG complex. (A) Various derivatives of HF-hCtf4 [300 fmol of full-length, N (WD + SepB), C (SepB + HMG), and SepB domains, which are likely to be dimers, and 600 fmol of WD and HMG domain, which are likely to be monomers] were incubated in the presence (+) or absence (−) of the hCMG complex (100 fmol). Mixtures were then immunoprecipitated with 1 μg of α-Cdc45 antibodies. Loading controls and immunoprecipitated materials were gel-separated and then analyzed by Western blotting against the FLAG tag to detect HF-hCtf4 derivatives and HF-hCdc45. The domains of hCtf4 are shown below the gel. (B) HF-hCtf4 derivatives (50 fmol as dimers and 100 fmol of monomers) used in A and the hCMG complex (100 fmol) were mixed and incubated in standard helicase reaction mixtures containing 50 mM NaCl. Untagged, full-length hCtf4 was also included as a control (last lane). (C) Unwound substrate (%), presented in B, was calculated and is shown in the graph.
We examined the influence of the different hCtf4 domains on the hCMG helicase activity measured in the presence of 50 mM NaCl. The C domain (SepB + HMG) stimulated the helicase activity to the same extent as observed with full-length hCtf4 (Fig. 4 B and C), whereas the other fragments had no effect. The relationship between protein interactions and helicase stimulation suggests that the latter effect occurs through direct binding of hCtf4 to the hCMG complex.
hCtf4 Interacts More Stably with the hCMG Complex than with Components of the hCMG Complex.
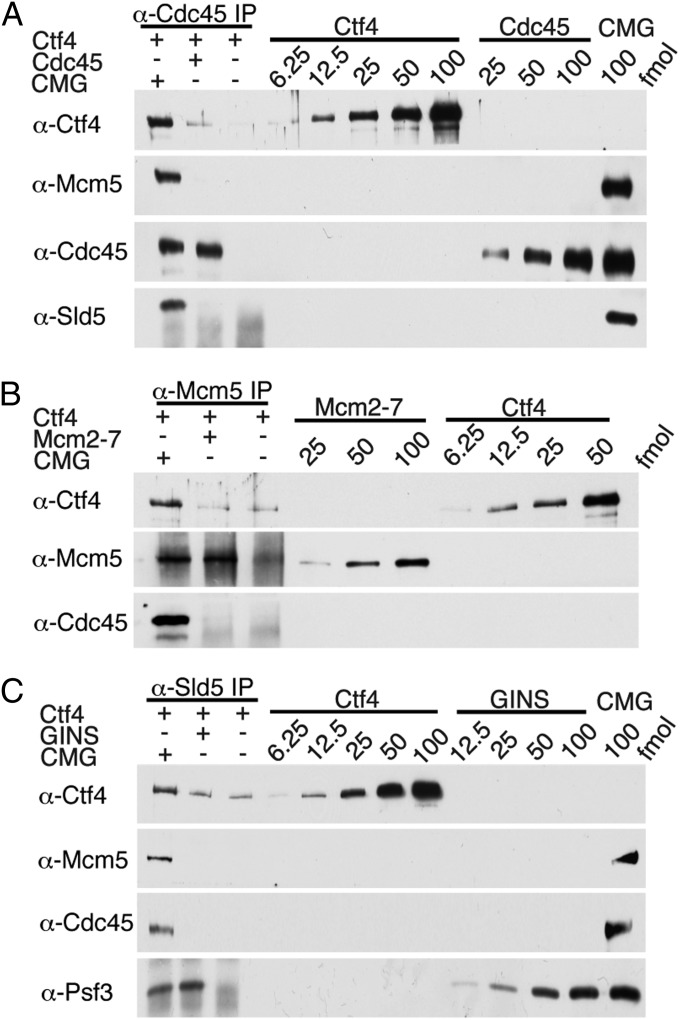
Interactions between hCtf4 and the CMG complex, as well as with each component of the complex, were examined. Sf9 cells were infected with viruses expressing HF-hCtf4 or untagged hCtf4 with human Cde45 (hCdc45), HF–human Mcm2-7 (hMcm2-7), or human GINS (hGINS) (GST-tagged Sld5), and extracts were prepared (using less stringent 150 mM potassium acetate rather than 420 mM potassium acetate, which is used in the isolation of hCtf4–CMG). Western blot analyses revealed relatively low levels of interactions (Fig. S5 A–C). Glycerol gradient analyses of the products formed indicated that Cdc45 and Mcm2-7 were not stably complexed to hCtf4 (Fig. S5 A, Right and B, Right). Coexpression of HF-hCtf4 and hGINS (containing GST-Sld5), followed by FLAG precipitation, yielded a complex of hGINS associated with excess hCtf4 (Fig. S5C, Left). SDS/PAGE separation following binding to and elution from GST beads revealed the removal of the excess hCtf4 and the presence of an hCtf4–GINS complex at a molar ratio of 1:8, respectively. Glycerol gradient separation of this complex resulted in the detection of relatively low levels of stable hCtf4–GINS complex, as well as a shift in the sedimentation properties of the hGINS complex [compared with free GINS (Fig. S5C, Right)]. These results indicate that hCtf4 forms a complex with hGINS, as reported previously in yeast (22), but the human complex is less stable than the yeast Ctf4–GINS complex.
The stability of the hCtf4–GINS and hCtf4–CMG complexes were compared during their isolation from virally infected Sf9 cells. Following elution of these complexes from glutathione beads, significantly lower levels of hCtf4 were associated with hGINS than with the hCMG complex (Fig. S5D). Glycerol gradient separation of the complexes revealed a stable hCtf4–CMG complex (Fig. S5E, Left), but not an hCtf4–GINS complex (Fig. S5E, Right). However, the hGINS distribution in both gradients (compared with the sedimentation of free hGINS) suggested that it may have associated with hCtf4 (and other hCMG components) unstably. It should be noted that these complexes were extracted from infected cells with higher salt (420 mM potassium acetate) than described in Fig. S5 A–C. This difference may have contributed to the discrepancy in stability of the hCtf4–GINS complex described in Fig. S5 C, Right, and E, Right. These findings indicate that the presence of hMcm2-7, hCdc45, and hGINS in the hCMG complex increased the stable association (and possibly binding) of hCtf4, suggesting cooperative interactions between these proteins.
The in vitro interactions of hCtf4 with individual hCMG components and the hCMG complex were compared. The hCdc45, hMcm2-7, and hGINS were isolated from baculovirus-infected cells as described in SI Materials and Methods (Fig. S6 A and B). The hCdc45 used in these experiments sedimented as a monomer (66 kDa) in glycerol gradients (Fig. S6B). Interactions between hCtf4 and the hCMG complex were carried out with 100 fmol of the hCMG complex or hCMG components in the presence of 400 fmol of hCtf4. Immunoprecipitations of hCdc45 + hCtf4 by α-Cdc45, hMcm2-7 + hCtf4 by α-Mcm5, and hGINS + hCtf4 with α-Sld5 resulted in precipitations of 39%, <1%, and 19% of the level of hCMG + hCtf4 immunoprecipitated by these antibodies, respectively (Fig. 5 A–C). Furthermore, the level of each protein that interacted with hCtf4 was unaffected by mixtures containing two or all three of the purified proteins of the hCMG complex. Collectively, these findings indicate that the coassociation of the proteins in the hCMG complex contribute to its more efficient and stable interaction with hCtf4 rather than a selective interaction of hCtf4 with a single protein of the complex.
Fig. 5.
Comparison of interactions of hCtf4 with the hCMG complex and hMcm2-7, hGINS, or hCdc45. (A) hCMG or hCdc45 (100 fmol) was incubated with hCtf4 (400 fmol), and the mixture was immunoprecipitated with 1 μg of α-Cdc45. The precipitated material and various levels of input proteins were separated by 10% (wt/vol) SDS/PAGE and immunoblotted against the indicated proteins. (B) hCMG or hMcm2-7 (100 fmol) was incubated with hCtf4 (400 fmol) and immunoprecipitated with 2.5 μg of α-Mcm5. (C) hCMG or hGINS (100 fmol) was incubated with hCtf4 (400 fmol) and immunoprecipitated with 2 μg of α-Sld5. IP, immunoprecipitation.
hCMG Complex Associates with hCtf4 on Chromatin in HeLa Cells.
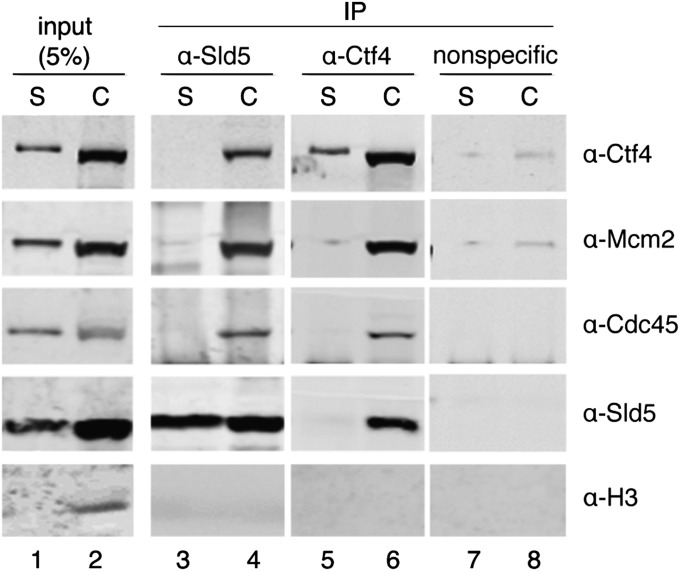
We investigated the interaction of hCtf4 with the hCMG complex in mammalian cells. For this purpose, soluble and chromatin fractions were isolated from HeLa cells as described in SI Materials and Methods and immunoprecipitated with Sld5 or Ctf4 antibodies (Fig. 6). As shown by Western blot analyses, the subunits of the hCMG complex and hCtf4 coimmunoprecipitated only from the chromatin fraction (Fig. 6, lanes 3–6). Previous studies in HeLa cells showed that the hCMG complex was found stably associated only in the chromatin fraction (14). Histone H3, which is localized mostly on chromatin, was used as a quality control of the lysate fractionation (Fig. 6, compare lanes 1 and 2). These findings indicate that the hCMG complex interacts with hCtf4 only on chromatin, although all of these proteins are abundantly present in chromatin-free fractions.
Fig. 6.
hCMG complex associates with hCtf4 on chromatin in HeLa cells. Soluble (S) or chromatin (C) fractions (500 μg) of protein isolated from HeLa cells were incubated with Sld5 antibodies (lanes 3 and 4), Ctf4 (lanes 5 and 6), or nonspecific (GST) antibodies (lanes 7 and 8), as indicated above the immunoblots. Specific interactions were detected by Western blotting using antibodies to Ctf4, Mcm2, Cdc45, Sld5, or Histone H3. Input represents 5% of the lysate used for immunoprecipitation (lanes 1 and 2).
Discussion
Ctf4 is a member of a conserved family of proteins required for multiple chromosome transactions (17). Members of this family contain either two or three motifs, including the N-terminal WD40 domain (of variable copies of conserved repeats), followed by three highly conserved SepB boxes. Higher eukaryotes include a C-terminal HMG AT hook DNA binding domain that is missing in lower eukaryotes. All family members harbor a SepB domain (∼300 aa) within their central region that can form a four-bladed β-propeller (25). Thus, hCtf4, budding yeast Ctf4, Aspergillus nidulans SepB, and Mcl1 family members contain multiple β-propeller domains likely to mediate protein interactions. In keeping with this notion, interactions between Ctf4 and many proteins involved in DNA replication have been detected (7, 19–22). The data reported here indicate direct interaction of hCtf4 with the hCMG complex.
Ctf4 was identified as a Pol α-binding protein by affinity chromatography (20), and recent results show that it is required for the association of Pol α with the RPC (21, 22). In higher eukaryotes, Zhu et al. (7) reported that both full-length hCtf4 and its derivative containing the SepB and HMG domains interacted with the p180 subunit of Pol α in vivo. In vitro studies demonstrated that hCtf4 stimulated reactions catalyzed by Pol α and Pol ε and weakly interacted with these Pols (19). Furthermore, recent studies on the identification of replisome proteins revealed the association of hCtf4 with nascent DNA chains (26).
In this report, we show that hCtf4 interacts with the hCMG complex following viral expression of these proteins in insect cells and in vitro with purified components. The isolation of the hCtf4–CMG complex from HeLa chromatin fractions indicates that the complex is formed in humans as well as in budding yeast (22). However, formation of the complex in higher eukaryotes requires the SepB and HMG domains (C domain), whereas the budding yeast Ctf4, which lacks the HMG domain, depends only on the SepB domain for its interactions with the RPC (22). Although interactions between hCtf4 and the individual components of the hCMG complex (Mcm2-7, GINS, or Cdc45) were observed (measured by coimmunoprecipitation), the complexes formed were poorly stable (measured by glycerol gradient sedimentation). In contrast, in vitro interaction of hCtf4 with the hCMG complex was more efficient and yielded a stable and more stoichiometric complex following glycerol gradient separation. These findings suggest that the stable binding of hCtf4 to hCMG occurs through its interaction with multiple components of the hCMG complex.
Previous hydrodynamic studies revealed that hCtf4 is a homodimer and the region responsible for its dimerization resides within the SepB domain (19). The in vitro interaction and stimulation of the helicase activity of the hCMG complex required both the SepB and HMG domains. Our findings indicate that hCtf4 present in the hCtf4–CMG complex is a homodimer, whereas the hCMG complex is present as a monomer. It should be noted that the SepB domain used here (and in our previous study) contains not only the SepB homology domain but an additional ∼140-aa region at its C terminus (19). This corresponding region in budding yeast Ctf4 was reported to contain a helix–loop–helix motif that is essential for Ctf4 function (27). It was postulated that this region mediated the dimerization of Ctf4. Future studies will be required to evaluate this proposal and to determine whether the dimeric structure of Ctf4 is responsible for its interaction with proteins and contributes to its biological activity.
The dimeric structure of Ctf4 raises the possibility that Ctf4 acts as a platform by which the Pol α–primase complex is associated with the CMG complex. Previous studies with budding yeast RPC revealed that Pol ε is required for CMG complex formation and is linked to CMG through the interaction of the p59 subunit of Pol ε with the C terminus of the GINS Psf1 subunit (28). These findings support the notion proposed by Labib’s group (10) that the CMG complex acts as the replisome core structure. Many of the proteins associated with budding yeast RPC appear to be bound directly to the CMG complex (which was isolated from yeast cells following benzonase digestion and immunoprecipitation of the RPC). The mechanism by which proteins interact with the CMG complex can now be explored in vitro. As shown here, hCtf4 interacts directly with the hCMG complex in the absence of chromatin. Future studies will be required to determine which proteins associated with the replisome interact with the CMG complex (directly or indirectly through other protein–CMG derivatives) and which interact with the CMG complex as a CMG–chromatin complex. Future studies directed at this problem may lead to the in vitro reconstitution of the replisome.
Materials and Methods
Expression and Purification of Proteins.
The isolation of hCMG, hCtf4–CMG, hCdc45, hMcm2-7, and GINS is described in SI Materials and Methods. Untagged full-length hCtf4, HF-tagged derivatives of full-length hCtf4, and various hCtf4 domains were isolated as described previously (19).
Helicase Assay.
Helicase assays were carried out in reactions (20 μL) containing 25 mM Hepes⋅NaOH (pH 7.5), NaCl (5 or 50 mM), 0.5 mM ATP, 10 mM magnesium acetate, 1 mM DTT, 0.1 mg/mL BSA, and 2.5 fmol of DNA substrate. Reactions were incubated at 37 °C for 30 min, halted with 4 μL of 6× stop solution [50 mM EDTA (pH 8.0), 40% (vol/vol) glycerol, 2% (wt/vol) SDS, 0.8% xylene cyanol, and 0.3% bromophenol blue], subjected to 10% (wt/vol) PAGE separation at 150 V in 1× TBE (89 mM Tris⋅base, 89 mM boric acid, and 2 mM EDTA), and dried on DEAE-cellulose paper, followed by autoradiography. The resolved DNA products were quantified using a PhosphorImager (Fujifilm).
Supplementary Material
Acknowledgments
This study was supported partly by National Institutes of Health Grant GMS R01 GM034559 and by institutional funds from Memorial Sloan–Kettering Cancer Center.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320202110/-/DCSupplemental.
References
- 1.Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87(1):53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- 2.Evrin C, et al. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106(48):20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Remus D, et al. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139(4):719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labib K. How do Cdc7 and cyclin-dependent kinases trigger the initiation of chromosome replication in eukaryotic cells? Genes Dev. 2010;24(12):1208–1219. doi: 10.1101/gad.1933010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araki H. Initiation of chromosomal DNA replication in eukaryotic cells; Contribution of yeast genetics to the elucidation. Genes Genet Syst. 2011;86(3):141–149. doi: 10.1266/ggs.86.141. [DOI] [PubMed] [Google Scholar]
- 6.Thu YM, Bielinsky AK. Enigmatic roles of Mcm10 in DNA replication. Trends Biochem Sci. 2013;38(4):184–194. doi: 10.1016/j.tibs.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu W, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21(18):2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boos D, Yekezare M, Diffley JF. Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science. 2013;340(6135):981–984. doi: 10.1126/science.1237448. [DOI] [PubMed] [Google Scholar]
- 9.Im JS, et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci USA. 2009;106(37):15628–15632. doi: 10.1073/pnas.0908039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambus A, et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8(4):358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- 11.Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21(4):581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 12.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103(27):10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37(2):247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang YH, Galal WC, Farina A, Tappin I, Hurwitz J. Properties of the human Cdc45/Mcm2-7/GINS helicase complex and its action with DNA polymerase epsilon in rolling circle DNA synthesis. Proc Natl Acad Sci USA. 2012;109(16):6042–6047. doi: 10.1073/pnas.1203734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124(2):237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miles J, Formosa T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Mol Cell Biol. 1992;12(12):5724–5735. doi: 10.1128/mcb.12.12.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams DR, McIntosh JR. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryot Cell. 2002;1(5):758–773. doi: 10.1128/EC.1.5.758-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui Y, et al. Genetic and physical interactions between Schizosaccharomyces pombe Mcl1 and Rad2, Dna2 and DNA polymerase alpha: Evidence for a multifunctional role of Mcl1 in DNA replication and repair. Curr Genet. 2005;48(1):34–43. doi: 10.1007/s00294-005-0584-2. [DOI] [PubMed] [Google Scholar]
- 19.Bermudez VP, Farina A, Tappin I, Hurwitz J. Influence of the human cohesion establishment factor Ctf4/AND-1 on DNA replication. J Biol Chem. 2010;285(13):9493–9505. doi: 10.1074/jbc.M109.093609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miles J, Formosa T. Protein affinity chromatography with purified yeast DNA polymerase alpha detects proteins that bind to DNA polymerase. Proc Natl Acad Sci USA. 1992;89(4):1276–1280. doi: 10.1073/pnas.89.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka H, et al. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes Cells. 2009;14(7):807–820. doi: 10.1111/j.1365-2443.2009.01310.x. [DOI] [PubMed] [Google Scholar]
- 22.Gambus A, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28(19):2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Köhler A, Schmidt-Zachmann MS, Franke WW. AND-1, a natural chimeric DNA-binding protein, combines an HMG-box with regulatory WD-repeats. J Cell Sci. 1997;110(Pt 9):1051–1062. doi: 10.1242/jcs.110.9.1051. [DOI] [PubMed] [Google Scholar]
- 24.Gosnell JA, Christensen TW. Drosophila Ctf4 is essential for efficient DNA replication and normal cell cycle progression. BMC Mol Biol. 2011;12:13. doi: 10.1186/1471-2199-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris SD, Hamer JE. sepB: An Aspergillus nidulans gene involved in chromosome segregation and the initiation of cytokinesis. EMBO J. 1995;14(21):5244–5257. doi: 10.1002/j.1460-2075.1995.tb00209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Contreras AJ, et al. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 2013;3(4):1105–1116. doi: 10.1016/j.celrep.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouprina N, et al. CTF4 (CHL15) mutants exhibit defective DNA metabolism in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1992;12(12):5736–5747. doi: 10.1128/mcb.12.12.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sengupta S, van Deursen F, de Piccoli G, Labib K. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr Biol. 2013;23(7):543–552. doi: 10.1016/j.cub.2013.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.