Significance
The centromere is essential for chromosome segregation because it is the site at which the kinetochore assembles to control microtubule attachment and chromosome movement. In meiosis, the homologous chromosomes pair, synapse by forming the synaptonemal complex (SC), and segregate. This is associated with the centromeres clustering in the nucleus. Little is known about the function of the integral centromere proteins in these processes because they are essential, and mutations in the genes are lethal. Here, we exploit allelic combinations to show the centromere proteins CENP-C and CAL1 are required in Drosophila meiosis for centromere pairing and clustering, and we find this occurs at the nucleolus. Defects in these proteins affect the SC at the centromere and cause meiosis segregation errors.
Keywords: CENP-A, CID
Abstract
Meiotic chromosome segregation involves pairing and segregation of homologous chromosomes in the first division and segregation of sister chromatids in the second division. Although it is known that the centromere and kinetochore are responsible for chromosome movement in meiosis as in mitosis, potential specialized meiotic functions are being uncovered. Centromere pairing early in meiosis I, even between nonhomologous chromosomes, and clustering of centromeres can promote proper homolog associations in meiosis I in yeast, plants, and Drosophila. It was not known, however, whether centromere proteins are required for this clustering. We exploited Drosophila mutants for the centromere proteins centromere protein-C (CENP-C) and chromosome alignment 1 (CAL1) to demonstrate that a functional centromere is needed for centromere clustering and pairing. The cenp-C and cal1 mutations result in C-terminal truncations, removing the domains through which these two proteins interact. The mutants show striking genetic interactions, failing to complement as double heterozygotes, resulting in disrupted centromere clustering and meiotic nondisjunction. The cluster of meiotic centromeres localizes to the nucleolus, and this association requires centromere function. In Drosophila, synaptonemal complex (SC) formation can initiate from the centromere, and the SC is retained at the centromere after it disassembles from the chromosome arms. Although functional CENP-C and CAL1 are dispensable for assembly of the SC, they are required for subsequent retention of the SC at the centromere. These results show that integral centromere proteins are required for nuclear position and intercentromere associations in meiosis.
Centromeres are the control centers for chromosomes, and thus are essential for accurate segregation in cell division. Centromeres are the DNA regions with a specialized chromatin structure upon which the kinetochore is built. The kinetochore is a complex of at least 100 proteins that contains the proteins to bind microtubules, motors to move on or destabilize microtubules, as well as checkpoint proteins monitoring kinetochore–microtubule attachment (1). The ability of the kinetochore to control microtubule binding and chromosome movement is essential for proper segregation in both mitosis and meiosis. In meiosis, additional constraints are placed on kinetochore function to ensure that homologs segregate in the first division and that segregation of sister chromatids is deferred until the second division (2). Recent studies indicate that in addition, the centromere itself may influence homolog segregation by controlling homolog pairing and formation of the synaptonemal complex (SC) (3).
In prophase of meiosis I, the homologs must pair and ultimately become attached, usually by recombination and crossing-over. By quantifying centromere number through prophase I, it has been observed that centromeres pair in yeast, plants, and Drosophila (3). Perhaps unexpectedly, this pairing can be between nonhomologous centromeres; in yeast, this has been proposed as a mechanism to prevent recombination around the centromere, as centromere pairing resolves from initially being nonhomologous to being homologous (4, 5). Homologous centromere pairing may play a critical role in ensuring segregation of chromosomes that do not undergo crossing-over, possibly by affecting orientation of the kinetochores (3, 6–8).
The centromere also regulates synapsis via the formation of the SC. SC formation initiates at the centromere and sites of cross-over formation in yeast, and the centromere is the first site for SC formation in Drosophila prophase I (9, 10). In addition, the SC persists at the centromere in yeast and Drosophila after the SC present along the chromosome arms has disassembled late in prophase I (7, 9, 11). Although SC assembly does not begin at centromeres in mouse meiosis, it persists at the centromeres and appears to promote proper segregation (12, 13).
Another centromere property has been observed in Drosophila oocytes. In most organisms, the centromeres are clustered together at one site at the onset of meiosis, likely a remnant of their configuration in mitosis, but this clustering breaks down as centromeres arrange in pairs (3, 4). In Drosophila, however, the centromeres remain clustered until exit from prophase I at oocyte maturation (9, 10, 14). Although an essential role for centromere clustering has not been demonstrated, it may facilitate homolog pairing, synapsis, or accurate segregation, particularly given that the homologous telomeres do not pair into a bouquet formation in Drosophila meiosis (15, 16). Components of the SC are necessary for centromere clustering, as is the cohesion protein ORD (9, 14).
The studies on centromere pairing and clustering define centromere geography within the meiotic nucleus, but they did not test whether centromere structure or function was involved. Centromeres have specialized nucleosomes with a histone H3 variant, centromere protein-A (CENP-A) (17). Incorporation of CENP-A into centromere chromatin is regulated precisely, although it occurs at distinct cell cycle times in different cell types, varying between late mitosis and G1 (17). In vertebrates, a complex of 15 proteins, the constitutive centromere-associated network (CCAN), is present on the CENP-A chromatin throughout the cell cycle and is crucial for assembling kinetochore proteins (1). In Drosophila, the entire CCAN complex has not been identified, although the CENP-C protein is present (18). Another Drosophila protein, CAL1, binds to CENP-A (called CID in Drosophila) in a prenucleosomal complex, and CAL1 is required for loading CID (19–22). CAL1 interacts with both CID and CENP-C, and all three proteins show interdependency for centromere localization (21, 23).
Little is known about the activities of these centromere proteins in meiosis. In fission yeast, CENP-C has been demonstrated to be critical for kinetochore–microtubule binding in meiosis and also to control kinetochore orientation in meiosis I (24). The timing of assembly of kinetochore and centromere proteins onto meiotic chromosomes has been examined in mouse spermatocytes (25) and in Drosophila spermatocytes and sperm (26, 27). RNAi studies have shown that CAL1 and CENP-C (the latter to a lesser extent) are needed for CID localization in Drosophila male meiosis, with reduction in the levels of any of these three proteins being associated with meiotic segregation errors (26). Drosophila males differ from most organisms in not undergoing recombination or forming an SC, and centromere clustering does not occur (28). A question of particular interest that has yet to be addressed is whether centromere architecture and function are required for centromere clustering and pairing in meiosis.
Results
CENP-C Function Is Required for Centromere Clustering and Pairing.
The essential function of centromeres has impeded analysis of requirements for centromere function in meiosis. We overcame this by exploiting the viable allele cenp-CZ3-4375 that we identified from a noncomplementation screen with a female-sterile mutant collection (29) (Fig. S1A). It is a missense mutation, but it appears to destabilize the protein because it results in decreased protein levels (Fig. S1 C and D). This allele was used in combination with homozygous lethal alleles of cenp-C and cal1 we recovered from a screen for mutants with mitotic defects during Drosophila embryogenesis (30). These mutations cause C-terminal truncations in the proteins (Fig. S1). Although these genes are essential, we were able to analyze the role of CENP-C in meiosis by using transheterozygotes of the lethal and female-sterile alleles of cenp-C.
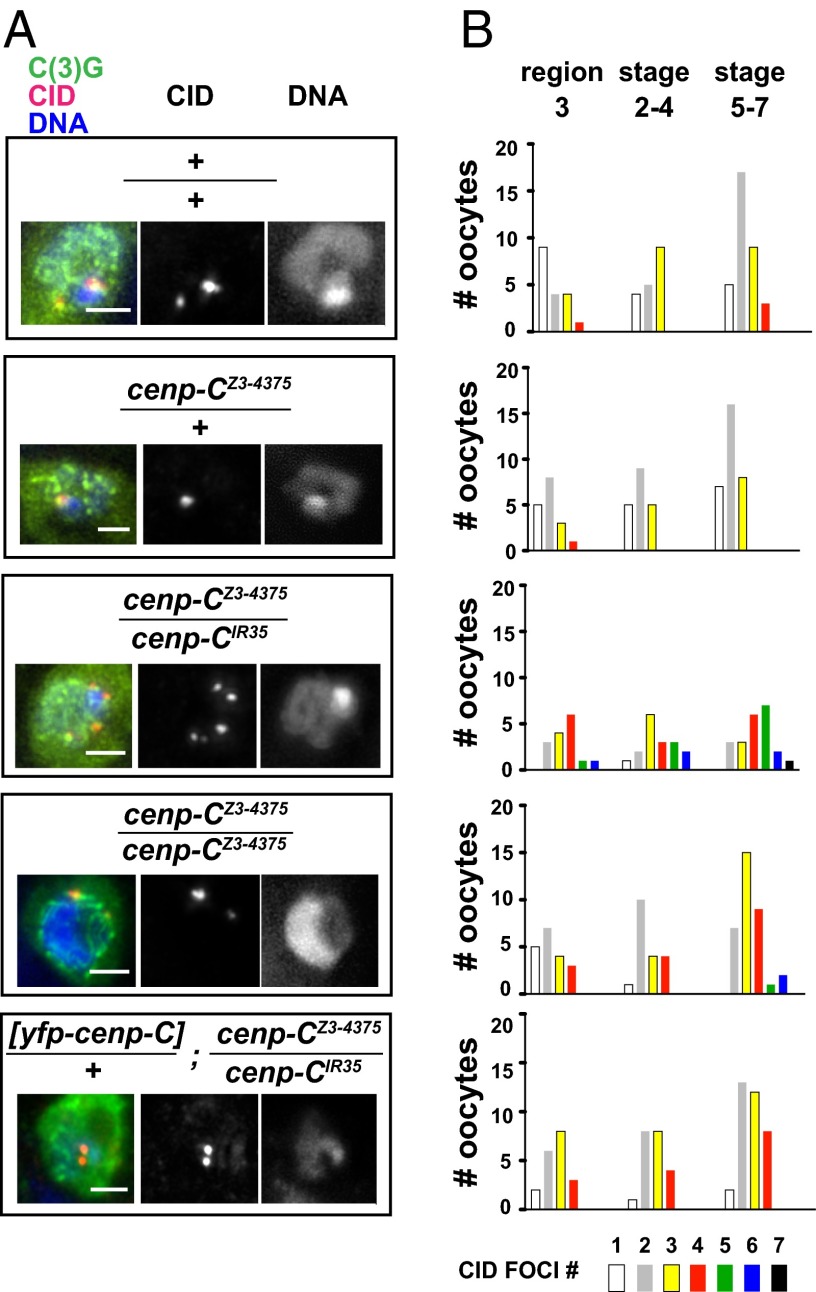
We tested whether a functional centromere structure is required for meiotic centromere clustering. Centromere clustering is detected as less than four CID foci, given that there are four homolog pairs in Drosophila (9, 14) (Fig. 1). The stages of meiosis can be distinguished in the Drosophila ovary by characteristic morphology of egg chambers (the egg chamber is the oocyte plus sister nurse cells and somatic follicle cells) and by staining with a marker for the SC, the transverse filament protein C(3)G (31). The germarium contains oocytes that have initiated meiosis: Region 2a of the germarium corresponds to zygotene and pachytene; from germarium region 2b through egg chamber stage 4, the oocyte remains in pachytene (9, 14). In stages 5–7, the SC disassembles (9, 32). Centromere clustering is not observed in mitotically dividing precursor cells in the germarium, and its initiation occurs in region 2a when meiosis initiates (9). From region 2a on, one or two CID foci are predominantly present, a consequence of centromere clustering (9, 14) (Fig. 1).
Fig. 1.
Centromere clustering and pairing require cenp-C function. (A) CID and DAPI staining of the oocyte from stage 5–7 egg chambers of WT (Oregon R), the indicated cenp-C mutants, and the [yfp::cenp-C]/+; cenp-CZ3-4375/cenp-CIR35 transgenic line. Increased CID foci are observed in the cenp-CZ3-4375/cenp-CIR35 mutants but not in the cenp-CZ3-4375 homozygote shown. The pronounced cenp-CZ3-4375/cenp-CIR35 defect is rescued by the [yfp::cenp-C] transgene. C(3)G, green; CID, red; DAPI, blue. (Right) Separate channels for CID and DAPI staining are shown. (Scale bars, 2 μm.) (B) Quantification of CID foci number in region 3, stages 2–4, and in stages 5–7 for the respective genotypes. The quantification shows that both centromere clustering and centromere pairing are disrupted in cenp-CZ3-4375/cenp-CIR35, whereas cenp-CZ3-4375 females are slightly defective in both. By the Wilcoxon rank sum test, the distribution in cenp-CZ3-4375/cenp-CIR35 is significantly different from WT in all stages (region 3, P = 1.1e-04; stages 2–4, P = 1.9e-03; stages 5–7, P = 1.5e-07). The cenp-CZ3-4375 females were significantly different from WT in stages 5–7 (P = 4.4e-05). The transgene strain was significantly different from cenp-CZ3-4375/cenp-CIR35 in all stages (region 3, P = 0.02; stages 2–4, P = 0.03; stages 5–7, P = 2.5e-05). The cenp-CZ3-4375/+ heterozygote was not significantly different from WT.
Staining of ovaries from cenp-CIR35/cenp-CZ3-4375 transheterozygous females with antibodies against CID revealed defects in centromere clustering. Although CENP-C is required for CID localization in mitosis (21, 23), CID staining was present in these oocytes, permitting us to use it as a centromere marker (Fig. S2). Quantification of the number of CID foci confirmed centromere clustering in WT and in cenp-CZ3-4375/+ controls (Fig. 1). In contrast, there was a marked failure of centromere clustering in the cenp-C transheterozygous mutants, evident in pachytene in region 3 (Fig. 1). The homozygous female-sterile allele also showed centromere clustering defects, although not as pronounced as in the transheterozygotes (Fig. 1). We showed that this effect was due to loss of CENP-C function, because a transgene expressing a functional fusion of YFP–CENP-C (21) restored centromere clustering (Fig. 1). These results demonstrate a requirement for a structurally normal centromere for clustering. Moreover, they highlight the role of CENP-C, given that CID is still detectably localized to centromeres in these cells but clustering is defective nevertheless.
In addition to clustering of the centromeres, the centromeres of each homolog have been demonstrated to be paired until metaphase I in Drosophila oocytes (6, 33). In the cenp-C mutants, we observed more than four CID foci (Fig. 1). This reveals that alteration of centromere function affects centromere pairing as well as centromere clustering. Although this could possibly arise from a complete loss of sister-chromatid cohesion, we think this is unlikely, given that we do not observe the meiotic segregation errors that accompany loss of sister-chromatid cohesion (see below).
We tested whether CENP-C is required for the establishment of centromere clustering by examining earlier meiotic stages by staining with CID and an antibody that recognizes the fusome, a cytoskeletal structure that permits identification of the oocyte (34) (Fig. S3A). In contrast to later stages, centromere clustering was normal in zygotene oocytes of cenp-C transheterozygous females (Fig. S3B). Thus, either CENP-C and proper centromere structure are essential only for maintenance but not establishment of centromere clustering or there is sufficient CENP-C function in the transheterozygous mutant to set up but not to maintain clustering.
cenp-C and cal1 Show Allele-Specific Interactions Causing Defects in Centromere Clustering and Pairing.
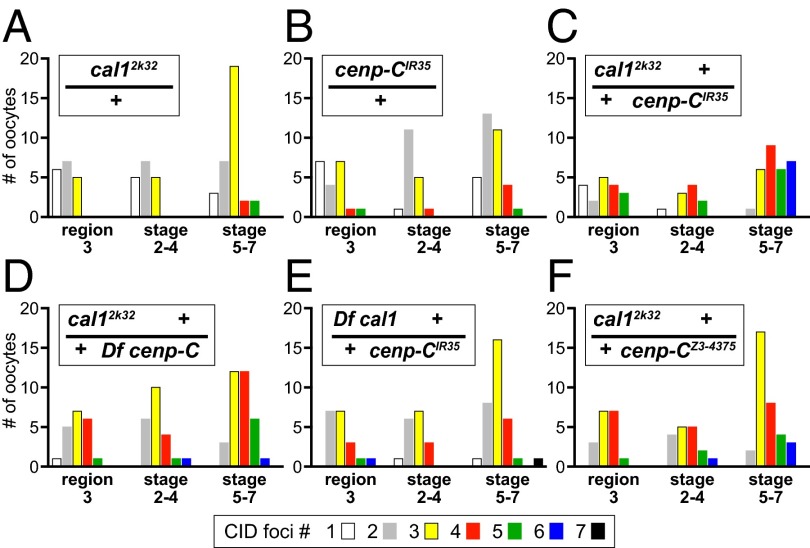
Failure of complementation between two mutations in different genes is diagnostic of a functional interaction between the gene products (35). This can result from a dosage effect, whereby decreasing the levels of the two products causes a phenotype, or it can reflect a disrupted physical interaction between the two proteins. The C terminus of CENP-C mediates its binding to CAL1 and its localization to centromeres (21). CAL1 binds to CENP-C via its C terminus and to CID via its N terminus (21). In mitosis, the three proteins are interdependent for their localization to the centromere (21, 23). Both the cal12k32 allele and the cenp-CIR35 allele (36) truncate the C terminus of the proteins (Fig. S1); thus, they are predicted to weaken or eliminate binding between the two proteins. We wondered whether this would be sufficient to show a complementation failure. The double heterozygotes of these recessive alleles (cal12k32 +/+ cenp-CIR35) retain CID localization (Fig. S2). Remarkably, although cal12k32 alone had only slight defects in centromere clustering, the double heterozygotes (cal12k32 +/+ cenp-CIR35) were defective in centromere clustering (Fig. 2 A and C). In addition, many oocytes showed defective centromere pairing in the double heterozygote, but pairing defects were seen in only one or two oocytes of each of the single heterozygotes (Fig. 2 A–C).
Fig. 2.
Allele-specific noncomplementation between cal1 and cenp-C causes defects in centromere clustering and pairing. Quantification of CID foci number at region 3, stages 2–4, and in stages 5–7 for the respective genotypes is shown. (A and B) cal12k32 causes a slight dominant defect (stages 5–7, P = 0.02), but the cenp-CIR35 stop codon is not significantly different from WT (compare with Fig. 1B). (C) Double heterozygotes cal12k32 +/+ cenp-CIR35 exhibit failure of both centromere clustering and pairing (a significant difference from WT region 3, P = 0.01; stages 2–4, P = 1.0e-03; stages 5–7, P = 8.8e-11). (D–F) These defects are more severe than in females bearing either truncation allele in trans to a deletion of the other gene or with cal12k32 in trans to the cenp-CZ3-4375 allele (significant differences in stages 5–7 between cal12k32 +/+ cenp-CIR35 and cal12k32 +/+ Df cenp-C, P = 0.02; Df cal1 +/+ cenp-CIR35, P = 7.9e-06; cal12k32 +/+ cenp- CZ3-4375, P = 0.01). The deletions were Df(3R)Exel6149 for cenp-C and Df(3R)Exel6176 for cal1. Df, deficiency.
We investigated whether the noncomplementation of cenp-C and cal1 was specific for the C-terminal truncated forms of the proteins by examining each allele in trans to a deficiency for the other gene. These females had defects in centromere clustering, but the severity of the defect was considerably less than in the double heterozygotes with the truncation alleles (Fig. 2 D and E). The double heterozygotes of cal12k32 and cenp-CZ3-4375 also had slight effects that were not as pronounced as in the truncation alleles (Fig. 2F). The cenp-CZ3-4375 allele reduces CENP-C protein levels (Fig. S1D), and it exhibits the same extent of clustering defects as the deficiency when in trans to cal12k32. Centromere pairing defects also occurred at lower frequencies in mutants in which cal12k32 was present in trans to a deficiency of cenp-C or the cenp-CZ3-4375 allele (Fig. 2 D–F).
These results further emphasize the critical requirement for centromere structure in clustering and pairing. The striking noncomplementation between the cal12k32 allele and the cenp-CIR35 allele appears to result from either reduced interaction or a poisonous interaction between the two proteins caused by the C-terminal truncations. There is sensitivity to dosage of the two proteins, however, because a deletion of either gene causes slight centromere clustering defects and rare pairing defects in trans to the C-terminal truncation alleles.
Meiotic Centromere Clustering Is Associated with the Nucleolus.
CAL1 has been demonstrated to localize to the nucleolus as well as the centromere in interphase mitotic cells (19), and it physically associates with the nucleolar protein Modulo, a nucleoplasmin homolog (37). In the absence of CAL1, CENP-C is present in the nucleolus in interphase cells and in meiotic spermatocytes (19, 26). A role for the nucleolus in mitotic centromere formation has been implicated by the observation that RNAi depletion of Modulo diminishes CAL1 at the centromere (37). In addition, depletion of Modulo causes loss of mitotic centromere clustering, changes in heterochromatin, and mitotic segregation defects (38). Given these results and the effect of the cal12k32 +/+ cenp-CIR35 transheterozygote on centromere clustering, we investigated whether the nucleolus could be associated with centromere clustering in meiosis.
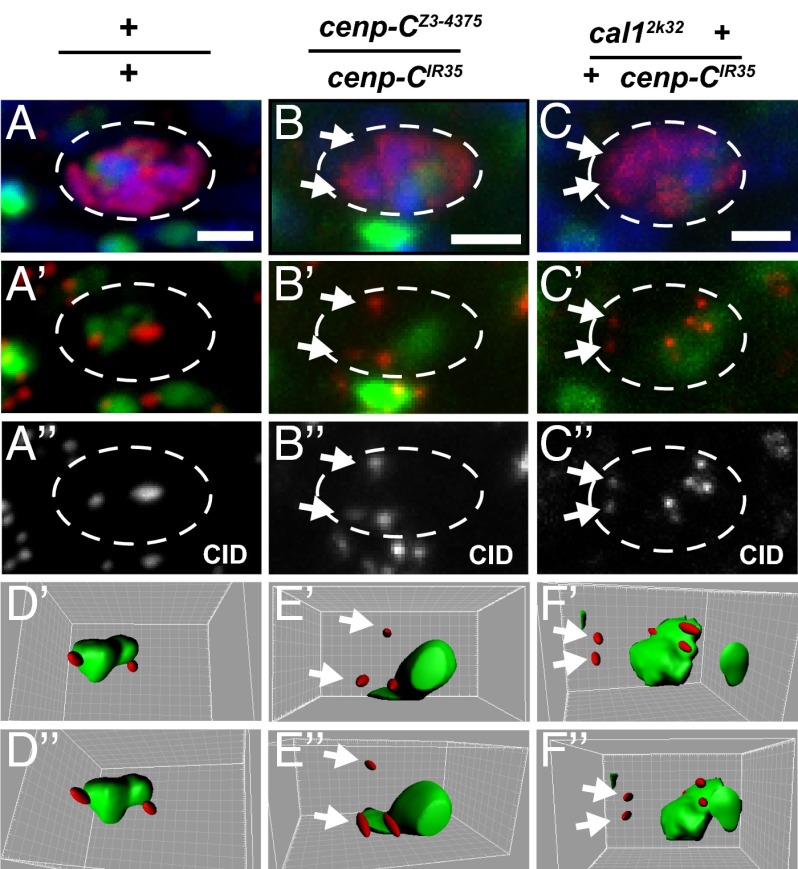
We examined the position of the centromere cluster relative to the nucleolus by staining ovaries with antibodies against CID and the nucleosomal Fibrillarin protein. The distance between the centromeres and the nucleolar surface was measured through 3D reconstruction by Imaris imaging software (www.bitplane.com). Strikingly, in WT, the centromere cluster always was adjacent to the nucleolus (Fig. 3A, quantified in Fig. S4). In contrast, in both the cenp-CIR35/cenp-CZ3-4375 and cal12k32 +/+ cenp-CIR35 mutants, the centromeres were separate from the nucleolus (Fig. 3 B and C and Fig. S4).
Fig. 3.
Centromere clustering is associated with the nucleolus, and this is disrupted in centromere protein mutants. Region 3 germaria of WT (A–A′′), cenp-C Z3-4375/cenp-CIR35 (B–B′′), and cal12k32 +/+ cenp-CIR35 (C–C′′) mutants were examined for centromere–nucleolus association. (A–C) Merged images of C(3)G (red), Fibrillarin (green), and DAPI (blue) staining. C(3)G staining identifies the oocyte nucleus (dotted line). (A′–C′) Merged images of CID (red) and Fibrillarin (green) staining. (A′′–C′′) Isolated CID channels of A′–C′, respectively. (Scale bars, 2 μm.) (D–F′) 3D reconstructions of A′–C′, respectively, via IMARIS software. Centromere (CID, red) and nucleolus (Fibrillarin, green) are shown. The 3D reconstructions are displayed at two different off-center angles to convey positions between the centromere and the nucleolus better. White arrows indicate centromeres not in close proximity with the nucleolus. Quantification is shown in Fig. S4.
These intriguing observations raise the possibility that in meiosis, as in mitosis, the nucleolus is linked to centromere clustering. Furthermore, they suggest that proper centromere structure is required for nucleosomal association.
Centromere Function Affects SC Structure.
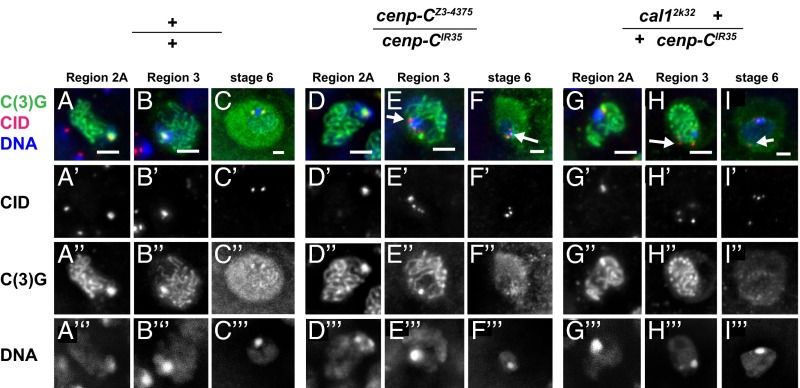
The site of centromere clustering in Drosophila is where synapsis and SC formation initiate in the oocyte (9, 10). Thus, we next tested whether SC formation was affected in the centromere protein mutants that disrupt centromere clustering. SC structure was analyzed by staining with an antibody against the transverse filament protein C(3)G (31). C(3)G was detected in continuous ribbon structures in region 2a of the germaria, reflective of synapsed homologs, in WT, cenp-CIR35/cenp-CZ3-4375, and cal12k32 +/+ cenp-CIR35 (Fig. S5). This indicates that failure of centromere clustering does not impair assembly of the SC.
Although the SC disassembles after pachytene, in stages 5–7, a remnant of the SC persists at the centromere until stage 9, detectable as brightly staining C(3)G foci that overlap with CID foci (9) (Fig. 4 A–C). Despite the lack of detectable effects on SC assembly, we observed that even by region 3 of the germaria, centromere association of C(3)G frequently was absent in centromere protein mutants (Fig. 4 D–I). Thus, in addition to the demonstrated requirement for C(3)G for centromere clustering (9), proper centromere structure is needed for retention of C(3)G at the centromere.
Fig. 4.
cal1 and cenp-C function to maintain the SC at the centromere. CID (red), C(3)G (green), and DAPI (blue) staining of oocyte nuclei from region 2A and region 3 germaria and stage 6 egg chambers from WT (A–C), cenp-CZ3-4375/cenp-CIR35 (D–F), and cal12k32 +/+ cenp-CIR35 (G–I) mutant females are shown. The split single channels are indicated. CID foci colocalize with strong SC staining in region 2A of WT and the centromere protein mutants. In region 3, centromere SC signal is observed in the WT [only 2 of 48 region 3 oocytes have CID foci without coincident C(3)G staining], but centromere SC is not maintained in the centromere protein mutants [15 of 27 region 3 oocytes have CID foci without C(3)G staining in cenp-CZ3-4375/cenp-CIR35, and 8 of 16 have CID foci without C(3)G staining in cal12k32 +/+ cenp-CIR35]. By Fisher’s exact test, the mutants are significantly different from WT with P = 6.8e-07 for cenp-CZ3-4375/cenp-CIR35 and P = 1.0e-04 for cal12k32 +/+ cenp-CIR35. Although the SC persists at WT centromeres in stage 6, it does not localize to centromeres in stage 6 centromere protein mutants. CID foci devoid of C(3)G staining in the mutants are indicated by arrows. (Scale bars, 2 μm.)
Noncomplementation of cenp-C and cal1 Affects Meiotic Chromosome Segregation.
The cenp-CIR35/cenp-CZ3-4375 females are sterile, but the cal12k32 +/+ cenp-CIR35 flies are fertile, permitting us to investigate whether the centromere defects result in meiotic segregation errors. Segregation of the sex chromosomes in females can be quantified in crosses to attached XY males, and the segregation of the 4th chromosome can be assessed by crossing to males with an attached 4th chromosome (39). This permits recovery of exceptional progeny arising from segregation errors, as well as distinction between gametes aberrantly bearing two copies of the chromosome (diplo exceptions) vs. none (nullo exceptions).
We simultaneously measured X and 4th chromosome segregation in WT control, cal12k32/+, cenp-CIR35/+, and cal12k32 +/+ cenp-CIR35 females by crossing to males with attached XY and attached 4 chromosomes. These genetic experiments revealed that both the cal12k32 and cenp-CIR35 alleles are semidominant for X chromosome nondisjunction and cal12k32 is semidominant for 4th chromosome nondisjunction (Table 1). There was a notable noncomplementation and genetic interaction between cal12k32 and cenp-CIR35. Females double-heterozygous for these mutants showed missegregation of both the X and 4th chromosomes. The effects on the two chromosomes were independent; the gamete classes arising from nondisjunction of the X chromosome together with the 4th chromosome were not elevated compared with gametes with missegregation of one of these chromosomes (Table 1). The X chromosome used for these tests was heterozygous for the centromere-linked car mutation. Failure to recover homozygous car mutant diplo exceptional (XX) progeny shows that the nondisjunction events occurred in meiosis I.
Table 1.
Nondisjunction tests in centromere protein mutant females
| Female genotype | +/+ | cal12K32/+ | cenp-CIR35/+ | cal12K32 +/+ cenp-CIR35 |
| Ova | ||||
| X; 4 | 2,214 | 1,650 | 2,226 | 692 |
| X; 0 | 0 | 18 | 0 | 14 |
| X; 44 | 1 | 24 | 11 | 7 |
| 0; 4 | 0 | 5 | 1 | 25 |
| 0; 0 | 0 | 1 | 2 | 1 |
| 0; 44 | 0 | 0 | 0 | 3 |
| XX; 4 | 0 | 1 | 2 | 17 |
| XX; 0 | 0 | 0 | 0 | 1 |
| XX; 44 | 0 | 0 | 0 | 0 |
| Total progeny | 2,215 | 1,700 | 2,242 | 760 |
| Adjusted total | 2,215 | 1,707 | 2,247 | 807 |
| X exceptions, % | 0 | 0.8* | 0.44** | 11.6*** |
| 4 exceptions, % | 0.05 | 2.64**** | 0.58 | 3.4***** |
The percentage nondisjunction for the X and 4th chromosomes are shown in bold. Significantly different from WT, as measured by the X nondisjunction significance test of Zeng et al. (46) (*P = 8.0e-03; **P = 0.025; ***P < 1.0e-04). Significantly different from WT by the Kruskal–Wallis test (****P = 1.5e-03; *****P = 4.9e-03).
To confirm that these genetic interactions were the consequence of mutation of the CAL1 and CENP-C proteins, we tested whether a transgene expressing a functional YFP fusion to CENP-C or a GFP fusion to CAL1 (21) could rescue the nondisjunction phenotype. For each transgene, two copies significantly rescued the nondisjunction resulting from noncomplementation in cal12k32 +/+ cenp-CIR35 (Table S1).
Given that the centromere clustering and pairing defects resulting from the centromere protein mutants showed allele specificity, we explored whether the meiotic nondisjunction was a consequence of genetic interaction between the two truncation alleles. Either a deletion of cal1 in trans to cenp-CIR35 or a deletion of cenp-C in trans to cal12k32 did not cause X chromosome nondisjunction (Table S2). Thus, simply reducing the levels of one of the proteins in the presence of a truncated form of the other is insufficient to cause meiotic segregation defects.
We also analyzed whether cal12k32 +/+ cenp-CIR35 compromised sex chromosome segregation in males by crossing to females with attached X chromosomes. Although there was a significant increase in XY segregation errors (Wilcoxon rank sum probability <0.01), the effect was considerably reduced compared with the defects observed in females (Table S3). Interestingly, the diplo exceptional sperm carried XX rather than XY chromosomes. Thus, meiosis II rather than meiosis I nondisjunction occurred, suggesting that in males, meiosis II segregation may be particularly vulnerable to compromised centromere function. Given the distinct mechanisms used in Drosophila male meiosis (absence of recombination and SC, specific homolog pairing sites and proteins) (28), the effects of the cal1 and cenp-C mutations in males may differ from those observed in females.
Discussion
Localization studies demonstrated centromere pairing in yeast, Drosophila, and plants, and it showed that the centromeres cluster together in Drosophila meiosis I. Here, we establish that centromere function is required for both pairing and clustering. Thus, centromeres are integrally involved in these two processes and not brought together solely by external factors. Because these events occur before assembly of the kinetochore, it is likely that the chromatin and associated proteins at the centromere are critical. The mutations in cenp-C reveal that functional CENP-C is necessary at a minimum for maintenance of centromere pairing and clustering in Drosophila oocytes. The noncomplementation between truncated CENP-C and CAL1 protein forms implicates CAL1 as also being crucial for centromere pairing and clustering. Given the role of CENP-C in recruiting proteins to the centromere (40), the requirement for this protein could reflect a direct role in centromere pairing and clustering or the need for a protein whose localization is dependent on CENP-C and/or CAL1. In the cenp-C mutant and the cenp-C cal1 double-heterozygous mutant, CID is still localized to the centromere, as evidenced by its presence at brightly DAPI-stained heterochromatin at levels that, by immunofluorescence, are not significantly lower than WT (Fig. S2). Thus, CID presence is insufficient for centromere clustering and pairing. The reduced level of CID staining in the double-heterozygous mutant is nearly significant, however; thus, we do not exclude the possibility that reduced CID levels contribute to the mutant defects.
The proteins at the centromere may interact with nuclear structures to promote centromere clustering. This study identifies the nucleolus as a likely candidate. We find that the centromere clusters are associated with the nucleolus in WT oocytes, and this association requires cenp-C and cal1 function. In Drosophila female meiosis, the nucleolus may serve as an anchor site for centromeres throughout prophase I.
The SC also may cluster centromeres. Clustering has been shown to be disrupted in mutants for the SC transverse and central elements (9). Our observation that the SC protein C(3)G fails to be retained at the centromere in cenp-C and cal1 mutants raises the possibility that the failure of clustering in these centromere protein mutants is a consequence of the absence of the SC. The hypothesis of this causality is consistent with the timing of defects; as early as pachytene, both centromere SC and clustering are absent. It remains to be determined how the SC, a structure contained between pairs of homologs, could gather centromeres into a cluster. In c(3)g mutants, more than four CID foci can be observed, indicating that both centromere pairing and clustering can be affected (9). Thus, failure of centromere retention of the SC also could account for the pairing defects in the centromere protein mutants.
The allele-specific noncomplementation [type I second-site noncomplementation (35)] between the mutations causing C-terminal truncations of CENP-C and CAL1 is unusual and informative. Such mutations that alter protein structure rather than simply reducing protein levels provide the opportunity to investigate genetic interactions. This allele-specific noncomplementation affects all the processes we analyzed: centromere pairing, centromere clustering and nucleolar association, SC retention at the centromere, and meiotic segregation. The antagonistic genetic interaction requires the truncated protein forms, because deficiencies for each of the genes complement the truncation allele of the other for meiotic segregation and cause only slight defects in centromere pairing and clustering. This is also true for the cenp-CZ3-4375 allele that reduces protein levels. Thus, simply decreasing the levels of the proteins does not perturb these processes. The C-terminal region of CAL1 binds to CENP-C, whereas the N terminus binds to CID; thus, the truncated form could have a dominant negative effect by binding CID and blocking its link to CENP-C (21). The C terminus of CENP-C is required for its localization to the centromere as well as binding to CAL1, whereas it binds the KNL-1/Mis12 complex/Ndc80 complex (KMN) kinetochore network via its N terminus (41). Thus, C-terminal truncated CENP-C also could act as a dominant negative to uncouple the KMN complex from a functional centromere association, particularly given that the N terminus alone can bind to kinetochore proteins but not to the centromere (41, 42). Expression of the N terminus alone also can disrupt the spindle assembly checkpoint (43). The truncation alleles of cenp-C and cal1 each alone have slight semidominant effects on centromere pairing, clustering, and meiotic segregation, consistent with dominant negative activities. The combination of the two dominant negative effects could account for perturbation of the meiotic processes. We cannot exclude, however, the possibility that these truncation alleles act as recessive neomorphs, conferring novel properties on the proteins.
A critical question is whether centromere clustering is required for proper meiotic segregation. It remains to be determined whether the meiotic nondisjunction that occurs in these centromere protein mutants is linked to the failure of centromere clustering and/or centromere pairing. The meiotic segregation errors in oocytes affect both the X chromosome, which undergoes recombination, and the 4th chromosome, which is achiasmate and lacks SC (44, 45). One way that meiotic segregation of both types of chromosomes could be dependent on clustering would be if association with the nucleolus is necessary for proper assembly of the kinetochore later in prophase I. It is notable, however, that the meiotic segregation errors in oocytes assayed for the X chromosome occurred exclusively in meiosis I; thus, a defect in kinetochore function necessary for both meiosis I and II was not evident. There are known meiosis I-specific requirements of the kinetochore, such as the need for the two sister kinetochores to co-orient in meiosis I, and establishment of these may require centromere clustering and/or nucleolar association. This proposal is consistent with the demonstrated effects of cenp-C mutants in meiosis in Saccharomyces pombe (24).
An alternative possibility is that the centromere mutations have independent effects on centromere clustering and subsequent segregation. For example, the centromere clustering defects could result from failure to retain the SC at the centromere and the meiotic nondisjunction could be an independent consequence of improperly assembled kinetochores later in meiosis I. The centromere mutations clearly can affect meiotic segregation independent of centromere pairing and clustering, given the meiotic nondisjunction in males double-heterozygous for the cenp-C and cal1 alleles. In Drosophila male meiosis, centromere clustering, SC formation, and recombination do not occur.
Although observed in yeast, plants, and Drosophila, a role for intrinsic centromere function in the nuclear localization of centromeres and associations between centromeres in meiosis has not yet been defined. The demonstration that proper centromere architecture is necessary for these interactions opens a path to define the molecular basis of centromere pairing and clustering across these species in meiosis.
Materials and Methods
Ovary Immunostaining.
Fixation and staining protocols are provided in SI Materials and Methods. Antibodies used were rat anti-CID (Claudio Sunkel, Instituto de Biologia Molecular e Celular, Porto, Portugal) at 1:1,000, rabbit anti-C(3)G (Mary Lilly, National Institutes of Health, Bethesda, MD) at 1:1,000, mouse anti-1B1 (Developmental Studies Hybridoma Bank) at 1:5, and mouse anti-Fibrillarin (Cytoskeleton, Inc.) at 1:200. Signals were detected using Alexa Fluor dye-conjugated secondary antibodies (1:400; Molecular Probes) and stained with 0.5 μg/mL DAPI.
Microscopy.
The images in Figs. 1 and 4 were taken with a Nikon Eclipse Ti microscope with a Hamamatsu camera and a Nikon Apo TIRF 100× oil objective. Confocal images were acquired with a Zeiss Axio LSM 700 or LSM710 microscope and a Zeiss LSM-TPMT camera. Objectives used for confocal images were a Zeiss 25× apochromat and a Zeiss 40× Plan-apochromat. Wide-field fluorescence image acquisition and deconvolution were carried out using NIS elements software (Nikon Instruments). Confocal image acquisition was carried out using Zen software (Carl Zeiss). IMARIS software (BITPLANE Scientific Solutions) was used to measure the distance between the nucleolus and CID foci in the oocyte from images acquired on a Zeiss LSM 710 confocal microscope.
Nondisjunction Tests.
Nondisjunction tests for the X and 4th chromosomes in the female and the sex chromosomes in the male were carried out, and the frequency of nondisjunction was calculated as in the study by Kerrebrock et al. (39).
Three tests were used to compare nondisjunction frequencies between genotypes statistically: the nonparametric Kruskal–Wallis test, the Wilcoxon rank sum test, and the method detailed by Zeng et al. (46) for the X chromosome frequencies.
Supplementary Material
Acknowledgments
We thank Helena Kashevsky for providing technical assistance in the nondisjunction tests and statistical analysis. George Bell also provided help with the significance tests. We thank Wendy Salmon for help with image acquisition and quantification, and Wendy Salmon and Tom Dicesare for assistance with figures. We are grateful to Claudio Sunkel, Christian Lehner, Aaron Straight, and Mary Lilly for antibodies. Cathleen Lake, Scott Hawley, Iain Cheeseman, Mina Kojima, Belinda Pinto, Jared Nordman, and Laura Frawley gave helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM39341 and by the Mathers Charitable Foundation. T.L.O.-W. is an American Cancer Society Research Professor.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320074110/-/DCSupplemental.
References
- 1.Gascoigne KE, Cheeseman IM. Kinetochore assembly: If you build it, they will come. Curr Opin Cell Biol. 2011;23(1):102–108. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe Y. Geometry and force behind kinetochore orientation: Lessons from meiosis. Nat Rev Mol Cell Biol. 2012;13(6):370–382. doi: 10.1038/nrm3349. [DOI] [PubMed] [Google Scholar]
- 3.Stewart MN, Dawson DS. Changing partners: Moving from non-homologous to homologous centromere pairing in meiosis. Trends Genet. 2008;24(11):564–573. doi: 10.1016/j.tig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obeso D, Dawson DS. Temporal characterization of homology-independent centromere coupling in meiotic prophase. PLoS ONE. 2010;5(4):e10336. doi: 10.1371/journal.pone.0010336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsubouchi T, Roeder GS. A synaptonemal complex protein promotes homology-independent centromere coupling. Science. 2005;308(5723):870–873. doi: 10.1126/science.1108283. [DOI] [PubMed] [Google Scholar]
- 6.Dernburg AF, Sedat JW, Hawley RS. Direct evidence of a role for heterochromatin in meiotic chromosome segregation. Cell. 1996;86(1):135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 7.Gladstone MN, Obeso D, Chuong H, Dawson DS. The synaptonemal complex protein Zip1 promotes bi-orientation of centromeres at meiosis I. PLoS Genet. 2009;5(12):e1000771. doi: 10.1371/journal.pgen.1000771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpen GH, Le M-H, Le H. Centric heterochromatin and the efficiency of achiasmate disjunction in Drosophila female meiosis. Science. 1996;273(5271):118–122. doi: 10.1126/science.273.5271.118. [DOI] [PubMed] [Google Scholar]
- 9.Takeo S, Lake CM, Morais-de-Sá E, Sunkel CE, Hawley RS. Synaptonemal complex-dependent centromeric clustering and the initiation of synapsis in Drosophila oocytes. Curr Biol. 2011;21(21):1845–1851. doi: 10.1016/j.cub.2011.09.044. [DOI] [PubMed] [Google Scholar]
- 10.Tanneti NS, Landy K, Joyce EF, McKim KS. A pathway for synapsis initiation during zygotene in Drosophila oocytes. Curr Biol. 2011;21(21):1852–1857. doi: 10.1016/j.cub.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newnham L, Jordan P, Rockmill B, Roeder GS, Hoffmann E. The synaptonemal complex protein, Zip1, promotes the segregation of nonexchange chromosomes at meiosis I. Proc Natl Acad Sci USA. 2010;107(2):781–785. doi: 10.1073/pnas.0913435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisig CG, et al. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet. 2012;8(6):e1002701. doi: 10.1371/journal.pgen.1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiao H, et al. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet. 2012;8(6):e1002790. doi: 10.1371/journal.pgen.1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Webber HA, Howard L, Bickel SE. The cohesion protein ORD is required for homologue bias during meiotic recombination. J Cell Biol. 2004;164(6):819–829. doi: 10.1083/jcb.200310077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 16.Carpenter ATC. Electron microscopy of meiosis in Drosophila melanogaster females. I. Structure, arrangement, and temporal change of the synaptonemal complex in wild-type. Chromosoma. 1975;51(2):157–182. doi: 10.1007/BF00319833. [DOI] [PubMed] [Google Scholar]
- 17.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120(5):425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 18.Orr B, Sunkel CE. Drosophila CENP-C is essential for centromere identity. Chromosoma. 2011;120(1):83–96. doi: 10.1007/s00412-010-0293-6. [DOI] [PubMed] [Google Scholar]
- 19.Erhardt S, et al. Genome-wide analysis reveals a cell cycle-dependent mechanism controlling centromere propagation. J Cell Biol. 2008;183(5):805–818. doi: 10.1083/jcb.200806038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goshima G, et al. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316(5823):417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schittenhelm RB, Althoff F, Heidmann S, Lehner CF. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. J Cell Sci. 2010;123(Pt 21):3768–3779. doi: 10.1242/jcs.067934. [DOI] [PubMed] [Google Scholar]
- 22.Schuh M, Lehner CF, Heidmann S. Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Curr Biol. 2007;17(3):237–243. doi: 10.1016/j.cub.2006.11.051. [DOI] [PubMed] [Google Scholar]
- 23.Mellone BG, et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7(5):e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17(3):334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Parra MT, et al. Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet. 2009;5(3):e1000417. doi: 10.1371/journal.pgen.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunleavy EM, et al. The cell cycle timing of centromeric chromatin assembly in Drosophila meiosis is distinct from mitosis yet requires CAL1 and CENP-C. PLoS Biol. 2012;10(12):e1001460. doi: 10.1371/journal.pbio.1001460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri N, et al. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 2012;10(12):e1001434. doi: 10.1371/journal.pbio.1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hawley RS. Meiosis: How male flies do meiosis. Curr Biol. 2002;12(19):R660–R662. doi: 10.1016/s0960-9822(02)01161-2. [DOI] [PubMed] [Google Scholar]
- 29.Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: A resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167(1):203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unhavaithaya Y, Park EA, Royzman I, Orr-Weaver TL. Drosophila embryonic cell cycle mutants. G3 (Bethesda) 2013;3(10):1875–1880. doi: 10.1534/g3.113.007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15(23):3130–3143. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Resnick TD, et al. Mutations in the chromosomal passenger complex and the condensin complex differentially affect synaptonemal complex disassembly and metaphase I configuration in Drosophila female meiosis. Genetics. 2009;181(3):875–887. doi: 10.1534/genetics.108.097741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilliland WD, Hughes SF, Vietti DR, Hawley RS. Congression of achiasmate chromosomes to the metaphase plate in Drosophila melanogaster oocytes. Dev Biol. 2009;325(1):122–128. doi: 10.1016/j.ydbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 34.de Cuevas M, Spradling AC. Morphogenesis of the Drosophila fusome and its implications for oocyte specification. Development. 1998;125(15):2781–2789. doi: 10.1242/dev.125.15.2781. [DOI] [PubMed] [Google Scholar]
- 35.Hawley RS, Gilliland WD. Sometimes the result is not the answer: The truths and the lies that come from using the complementation test. Genetics. 2006;174(1):5–15. doi: 10.1534/genetics.106.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heeger S, et al. Genetic interactions of separase regulatory subunits reveal the diverged Drosophila Cenp-C homolog. Genes Dev. 2005;19(17):2041–2053. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen CC, Greene E, Bowers SR, Mellone BG. A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila. PLoS ONE. 2012;7(9):e45094. doi: 10.1371/journal.pone.0045094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padeken J, et al. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell. 2013;50(2):236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 39.Kerrebrock AW, Miyazaki WY, Birnby D, Orr-Weaver TL. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 1992;130(4):827–841. doi: 10.1093/genetics/130.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145(3):410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21(5):399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 42.Trazzi S, et al. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS ONE. 2009;4(6):e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21(5):391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lake CM, Hawley RS. The molecular control of meiotic chromosomal behavior: Events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol. 2012;74:425–451. doi: 10.1146/annurev-physiol-020911-153342. [DOI] [PubMed] [Google Scholar]
- 45.Walker MY, Hawley RS. Hanging on to your homolog: The roles of pairing, synapsis and recombination in the maintenance of homolog adhesion. Chromosoma. 2000;109(1-2):3–9. doi: 10.1007/s004120050407. [DOI] [PubMed] [Google Scholar]
- 46.Zeng Y, Li H, Schweppe NM, Hawley RS, Gilliland WD. Statistical analysis of nondisjunction assays in Drosophila. Genetics. 2010;186(2):505–513. doi: 10.1534/genetics.110.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.