Significance
Populations of the African malaria vector, Anopheles gambiae, are structured into M and S forms. All current work assumes the two rarely hybridize. Here we show this assumption is false. We demonstrate (i) significant exchange of genes between the two forms, even though (ii) hybrids have reduced fitness and (iii) the gene exchange process is spatially and temporally dynamic. For malaria, it is important to determine if genes for traits like insecticide resistance are shared between forms. For evolutionary biologists, this work confirms that this mosquito is a good model for studying if and how species may evolve in cases where there is ongoing gene flow.
Keywords: reproductive isolation, population structure, Anopheles coluzzii
Abstract
The M and S forms of Anopheles gambiae have been the focus of intense study by malaria researchers and evolutionary biologists interested in ecological speciation. Divergence occurs at three discrete islands in genomes that are otherwise nearly identical. An “islands of speciation” model proposes that diverged regions contain genes that are maintained by selection in the face of gene flow. An alternative “incidental island” model maintains that gene flow between M and S is effectively zero and that divergence islands are unrelated to speciation. A “divergence island SNP” assay was used to explore the spatial and temporal distributions of hybrid genotypes. Results revealed that hybrid individuals occur at frequencies ranging between 5% and 97% in every population examined. A temporal analysis revealed that assortative mating is unstable and periodically breaks down, resulting in extensive hybridization. Results suggest that hybrids suffer a fitness disadvantage, but at least some hybrid genotypes are viable. Stable introgression of the 2L speciation island occurred at one site following a hybridization event.
The M and S forms of the African malaria vector Anopheles gambiae have been the subject of intense study over the past decade. The focus has centered on models of the evolution and maintenance of genetic divergence between the two forms in relation to speciation (reviewed in ref. 1). A. gambiae has become a model, described in a number of recent reviews on speciation (2–5).
The two forms occur in sympatry throughout West and Central Africa (6). They were initially described on the basis of several single nucleotide polymorphisms (SNPs) in the X-linked ribosomal DNA locus (7, 8). Heterozygotes were rarely found in nature and studies of reproductive isolation (RI) confirmed strong assortative mating with interform matings occurring at a frequency of ∼1% (9). Progeny of laboratory crosses and backcrosses show no signs of reduced fitness (10). However, it is widely held that, in nature, some degree of ecologically dependent postzygotic isolation, in addition to assortative mating, contributes to divergence between the two forms (11, 12).
Studies of the genetic structure of M and S based on microsatellite markers revealed little between-form differentiation outside the centromeric region of the X chromosome and a few regions associated with inversions (13–15). This overall lack of divergence was attributed to the homogenizing effect of gene flow between the forms (16).
In 2008, several reports of much higher frequencies of M/S hybrids, as high as 24%, appeared (17–19). These were all observed in populations in coastal West Africa, an area now thought to represent a zone of secondary contact (20, 21). These reports resulted in the emergence of this species as the focus of research aimed at exploring the evolution and maintenance of genetic divergence with gene flow (11, 12, 22, 23).
The first genome-wide comparison of the M and S forms by Turner et al. (24) was consistent with earlier observations that divergence overall is low, but there are small, discrete regions of divergence representing about 3% of the genome. They identified three diverged regions: one near the centromere on the X chromosome, one on the left arm of chromosome 2 (2L), and one on the right arm of chromosome 2 (2R). A number of genome-wide scans comparing M and S have been conducted since. These have applied several methods, including the same microarray used by Turner et al. (25, 26), high-density SNP arrays (12, 21, 23, 27), and whole-genome sequencing (28). These studies likewise revealed little divergence except in a few discrete regions of the genome characterized by high levels of differentiation (islands of divergence).
This body of work has culminated in two opposing models aimed at describing the evolution of M and S (1, 11). The “islands of speciation” model supposes that (i) small regions of divergence contain genes responsible for RI because they are directly associated with assortative mating and/or are under strong ecologically dependent divergent selection, and (ii) the rest of the genome is either neutral with respect to differentiation or close enough to neutral so that contemporary gene flow overwhelms selection. The alternative “incidental island” model recognizes the presence of islands of divergence, but suggests that (i) these are not related to RI and the remainder of the genome is less differentiated due to segregating ancestral polymorphism, not gene flow, and (ii) F1 hybrids are effectively sterile, therefore the amount of “realized” gene flow between M and S is near zero and that M and S are in fact “good species.” Indeed, the M form has recently been elevated to species status and provided the formal species name Anopheles coluzzii (29). We continue to refer to M and S forms to facilitate discussion with reference to the recent literature.
Limitations in the genome-wide scans described above may have contributed to disparate views of the evolution of divergence between A. gambiae subgroups. All comparisons of natural populations to date used single-locus, X-linked genotypes (7, 30–32) to identify M and S form specimens used in downstream analyses. These single-locus assays misidentify a significant proportion of backcross individuals. In addition, assessment of hybridization frequencies and comprehensive tests for introgression are precluded from these studies because DNA pools were used (21, 23, 27), sample sizes were too small (24, 26), or such assessments may be irrelevant because M and S laboratory colony mosquitoes were used (28). Finally, there are strong limitations in relying on genome scans alone for detecting selection, gene flow, and recombination that occurs during speciation (33, 34).
In this study, we used a simple multilocus SNP genotype approach to distinguish M, S, F1 hybrids, and backcross individuals (35), which we call the “divergence island SNP” (DIS) assay (SI Appendix). We describe the spatial and temporal distribution of hybridization frequencies, the extent to which hybridization results in introgression, and the fitness of hybrid genotypes in nature. Assumptions concerning these between-form interactions have played a central role in the interpretation of comparative genomics data as applied to understanding speciation in this system. The results we report challenge a number of these assumptions and they provide unique information about temporal dynamics in the way in which M and S forms interact that suggests new avenues for future research.
Results
Multilocus, Single-Copy DIS Assay.
A few studies have used multiple restriction fragment length polymorphism assays and/or mismatch primer-based SNP assays to genotype one SNP in each of the three (X, 2L, and 3L) islands of divergence (26, 36). We extended this approach with the development of the DIS assay (35) to describe A. gambiae populations throughout West Africa (Fig. 1 and SI Appendix, Table S3). The DIS assay includes 7 SNPs spanning 4.4 Mbp on the X chromosome, 5 spanning 2.2 Mbp on chromosome 2L, and 3 spanning 117 kb on chromosome 3L (35). Genotypes at these 15 SNPs were used to designate individuals as M form, S form (homozygous for form-specific alleles at a minimum of 13 of the 15 SNPs), F1 hybrids (heterozygous at all 15 SNPs), and backcross (individuals with mixed M, S, and heterozygous genotypes at 3 or more SNPs). The GOUNDRY and ENDO populations were defined on the basis of genotypes at eight microsatellite loci as described previously (37). We used the method described in ref. 35 to illustrate the distribution of DIS genotypes among each study population, using what we refer to as “DIS maps” (Figs. 2 and 3 and SI Appendix, Figs. S3–S7).
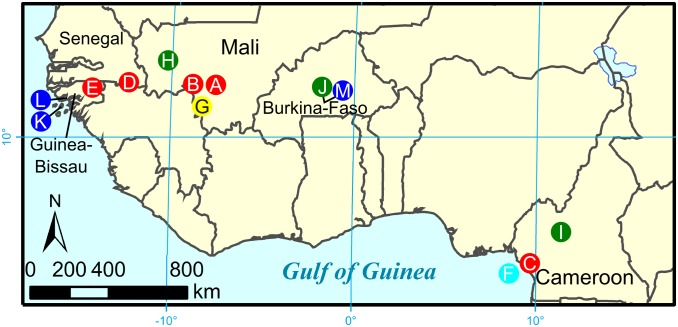
Fig. 1.
Collection site map. Site index follows Table 1. Sites are marked with colors that group populations by genetic structure as follows: red, past introgression lacking evidence for ongoing gene flow; cyan, past introgression and loss of divergence on 2L; green, ongoing gene flow and introgression; blue, complete or nearly complete introgression; and yellow, Selinkenyi where a longitudinal study was conducted.
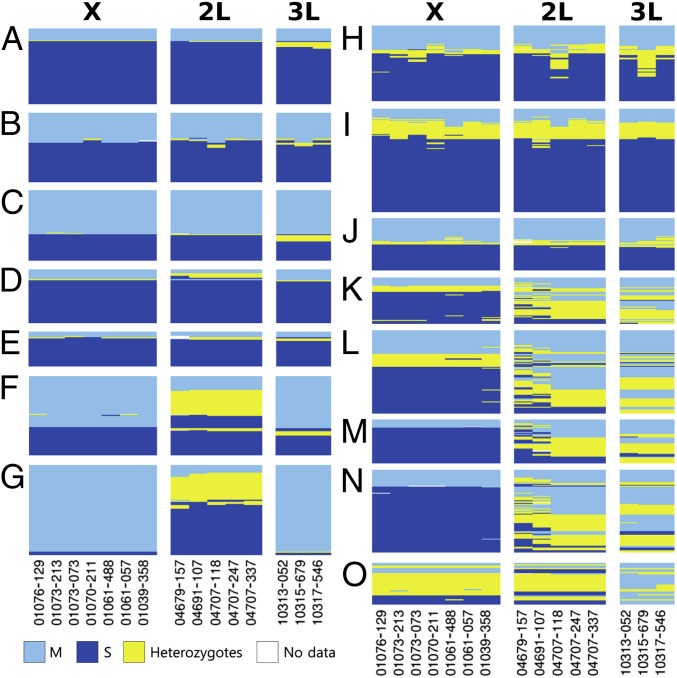
Fig. 2.
DIS maps for sympatric populations. Site index follows Table 1. A, Banambani; B, Founia; C, Tiko; D, Nathia; E, Canjufa; F, Bioko; G, Selinkenyi; H, Bantinngoungou; I, Njigalap; J, ENDO; K, Abu females; L, Prabis females; M, Abu males; N, Prabis males; O, GOUNDRY.
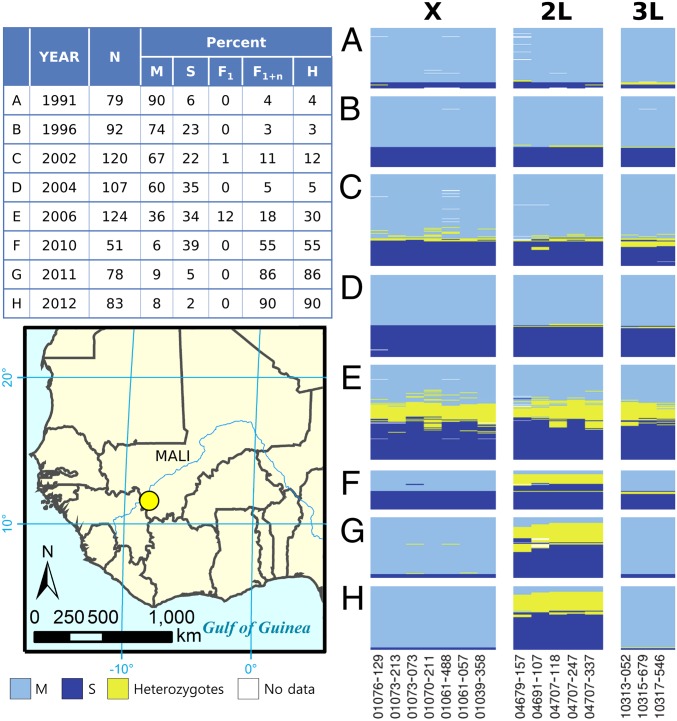
Fig. 3.
DIS maps for sympatric populations from the town of Selinkenyi, Mali collected at eight time points over a 21-y period, 1991–2012 (A–H).
Analysis of Allopatric M and S Populations.
Five allopatric populations, two “pure” M forms and three pure S forms (pure = predominant form occurs at frequency >0.96), were analyzed to establish that molecular-form–specific SNPs are not polymorphic within forms. These sites covered a broad geographic area, from central Mali to southern Cameroon (SI Appendix, Fig. S3). Only 12 of 466 carried mixed genotypes. Heterozygotes occurred at SNPs distributed over all three chromosomes. Ten of the 14 individuals were heterozygous at multiple, physically linked, SNPs. Four were heterozygous at a single SNP, but each of these was only observed in a single population, suggesting these were due to rare past migration events and do not represent shared ancestral polymorphisms.
Analysis of Sympatric M and S Populations.
Thirteen populations in which the M and S forms occur in sympatry were analyzed (Fig. 1). These included the ENDO and GOUNDRY populations from Burkina Faso (37), four populations from Mali, three from Guinea-Bissau, two from Cameroon, one population from Equatorial Guinea, and one from Senegal. A total of 825 individuals were sampled (Table 1). In every population studied, individuals with hybrid genotypes occurred at a frequency in excess of 5%, ranging from 5.2% to 96.9%. The genetic structure of these populations formed a continuum, from populations with evidence of past introgression, but clear boundaries between M and S, to those in which the boundaries between M and S are completely absent.
Table 1.
M, S, F1, and backcross (F1+n) frequencies
| Index | Site | Year | N | %M | %S | %F1 | %F1+n |
| Populations with past introgression and no ongoing gene flow | |||||||
| A | Banambani | 2005 | 87 | 15 | 73 | 0 | 12 |
| B | Founia | 2006 | 77 | 39 | 56 | 0 | 5 |
| C | Tiko | 2003 | 81 | 16 | 2 | 0 | 82 |
| D | Nathia | 2009 | 41 | 7 | 78 | 0 | 15 |
| E | Canjufa | 2009 | 26 | 15 | 73 | 0 | 12 |
| Populations with past introgression and loss of divergence on 2L | |||||||
| F | Bioko | 2002 | 95 | 0 | 96 | 0 | 4 |
| G | Selinkenyi | 2012 | 83 | 8 | 2 | 0 | 90 |
| Populations with ongoing gene flow and introgression | |||||||
| H | Bantinngoungou | 2006 | 87 | 24 | 46 | 5 | 25 |
| I | Njigalap | 2006 | 119 | 10 | 70 | 12 | 8 |
| J | ENDO | 2006 | 60 | 42 | 45 | 3 | 10 |
| Populations with complete or nearly complete introgression | |||||||
| K | Abu (♀♀) | 2009 | 50 | 98 | 0 | 0 | 2 |
| L | Prabis (♀♀) | 2009 | 92 | 25 | 0 | 1 | 74 |
| M | Abu (♂♂) | 2009 | 48 | 19 | 0 | 19 | 62 |
| N | Prabis (♂♂) | 2009 | 90 | 13 | 0 | 15 | 72 |
| O | Goundry | 2006 | 32 | 3 | 0 | 0 | 97 |
Populations with past introgression and no ongoing gene flow.
Populations at Banambani and Founia, Tiko, Nathia, and Canjufa included backcross individuals at frequencies ranging from 5% to 15%, but of the 312 individuals genotyped, no F1 hybrids were detected (Fig. 2 A–E and SI Appendix, Fig. S4). Genotype frequencies at all SNPs departed significantly from Hardy–Weinberg equilibrium (HWE) with a deficiency of heterozygotes in every case. Association among all three unlinked islands of divergence was strong, but nowhere near complete (mean r2 = 0.83; SI Appendix, Table S4). Recombination was more common in autosomal islands than for X chromosome islands (SI Appendix, Table S4). Introgressed SNPs in both the homozygous and heterozygous states were present, suggesting backcrossing for multiple generations. Excluding the possibility of ancestral polymorphism (as justified above), the distribution of genotype frequencies can be explained by hybridization, either ongoing at low frequencies (too low for the detection of F1 genotypes) or in the recent past, followed by the reestablishment of strong premating barriers. In either case, introgression has occurred with backcross genotypes maintained at nonequilibrium frequencies, likely due to incomplete assortative mating and/or selection too weak to eliminate them (at least between the time of their formation by hybridization and the time they were sampled).
Populations with past introgression and loss of divergence on 2L.
Populations at Bioko and Selinkenyi collected in 2012 were composed of a large number of backcross individuals, 54.9–89.2% (Table 1, Fig. 2 F and G, and SI Appendix, Fig. S5). As in populations described above, the absence of F1 hybrids suggests a lack of ongoing gene flow in these populations. Presence of S form chromosome 2 SNPs in otherwise typical M form genomes accounted for nearly all introgression (110 of 116 recombinant genotypes). Genotype frequencies at all SNPs on both 3L and X departed significantly from HWE, with a deficiency of heterozygotes in all cases and strong association of form-specific SNPs across these two islands of divergence. SNPs on 3L and X are associated (mean r2 > 0.81; SI Appendix, Table S4), whereas SNPs on 2L were not associated with X or 3L (r2 < 0.23). Genotype frequencies for all five SNPs on 2L, in both populations, were in HWE, suggesting stable introgression of the 2L island. Introgression of X-linked SNPs was absent (0/116).
Populations with ongoing gene flow and introgression.
Populations at Bantinngoungou, Njigalap, and the ENDO population included F1 hybrids at frequencies ranging from 3.3% to 11.8% and backcross genotypes from 10.0% to 25.3% (Table 1, Fig. 2 H–J, and SI Appendix, Fig. S6). Of 45 tests for compliance of genotype frequencies to HWE, all but 2 departed from expectations, with a deficiency of heterozygotes in every case. The proportion of backcross genotypes relative to F1s is quite low (in Njigalap the frequency of F1 genotypes exceeds backcrosses by 50%) and in nearly every case the introgressed SNP is heterozygous (56/62), indicating these are the product of first generation backcrosses (F1 × parental). This suggests that hybridization is consistently high in this population with strong selection against later generation backcross genotypes or that at the time these populations were sampled, they were experiencing a very recent breakdown in assortative mating (isolate breaking) (38). Overall form-specific islands remained associated (mean r2 = 0.82).
Populations with complete or nearly complete introgression.
Populations at Abu and Prabis consist of backcross individuals at frequencies of 82.0% and 73.9%, respectively (Fig. 2 K–N and SI Appendix, Fig. S7). Introgression is strongly asymmetric, with the M form introgressed into the S (22). Our analysis supports this observation and illustrates that introgression into the S form has proceeded to the extent that pure S form genotypes are completely absent, whereas pure M form individuals persist. Genotype frequencies for all eight SNPs on chromosomes 2 and 3 are in HWE; however, strong linkage disequilibrium (LD) is maintained for all SNPs on the X chromosome (r2 > 0.93), with a significant deficiency of heterozygotes at all seven SNPs. A separate analysis of males reveals the strength of the asymmetric introgression (Fig. 2 M and N and SI Appendix, Fig. S7 C and D). Males that were heterozygous for all eight autosomal SNPs (males are hemizygous for X-linked SNPs) were presumed to be F1 hybrids. Of 19 F1 hybrid males, 18 were hemizygous for S-specific SNPs on the X chromosome, suggesting S form females exhibit weak assortative mating.
The GOUNDRY population exhibits the most extreme level of introgression, with a frequency of backcross genotypes of 96.9% (Fig. 2O and SI Appendix, Fig. S7E). Only a single individual of 32 sampled was homozygous at all SNPs. Genotype frequencies at all 15 SNPs are in HWE. Within each island, all pairs of SNPs were in significant LD (r2 > 0.60, P = 0.00). No association among form-specific islands was evident (r2 < 0.17, P > 0.05).
Temporal Variation in Hybridization.
Patterns of hybridization and introgression vary spatially, probably due to differences among local environments (39–41). These differences may include environmental cues used in mate selection and/or factors that influence hybrid fitness. If true, it is likely that hybridization and introgression also have a temporal dynamic in places that experience environmental fluctuations over time. To examine this possibility we conducted a longitudinal study of sympatric M and S populations spanning a period of 21 y at Selinkenyi. Rainfall in this area is controlled by movement of the intertropical conversion zone (ITCZ), which oscillates between the northern and southern tropics annually. Variations in the latitudinal movement of the ITCZ from year to year cause large interannual variability in wet-season (June–October) rainfall (42).
The distribution of DISs in populations at Selinkenyi between 1991 and 2012 are illustrated in Fig. 3. All collections were made during the rainy season (July–October). In 1991, M was the predominant form, representing 89.9% of individuals sampled. A total of 3.8% of individuals had mixed, backcross genotypes, but no F1 hybrids were observed (Fig. 3A). The relative proportion of forms changed in 1996 with S form increasing to 22.8%, but the proportion of backcross hybrids remained roughly as it was in 1991 (3.3%, Fig. 3B). As in 1991, no F1 hybrids were collected. Temporal changes in the relative abundance of the two forms have been reported in the past (43, 44). The presence of backcross individuals indicates that some degree of hybridization had occurred in the past, but a lack of F1 hybrids suggests that there was no ongoing hybridization at the time these samples were collected.
A change occurred in 2002 when the relative abundance of the two forms remained similar to 1996 (66.7% M and 21.7% S); however, the frequency of backcross hybrids increased to 10.8% and F1 hybrids were observed at a frequency of 0.8% (Fig. 3C). The occurrence of F1 hybrids and accompanying increase in backcross frequencies suggests this sample was taken at a time when hybridization between the two forms was occurring and that hybrid individuals were actively mating, resulting in introgression of the two forms. Two years later (2004) near complete assortative mating appears to have been reestablished, evident from an absence of F1 hybrids and a decrease in the frequency of backcross hybrids (from 11.6% in 2002 to 5.6% in 2004, Fig. 3D).
A dramatic change occurred in 2006 when the frequency of F1 hybrids jumped to 12.1% and backcross hybrids increased to 18.5% (Fig. 3E). An apparent breakdown of assortative mating in 2006 precipitated a profound change in the genetic structure of this population such that the frequency of hybrids in samples collected in 2010, 2011, and 2012 increased to 54.9%, 85.9%, and 89.2%, respectively (Fig. 3 F–H). Nearly all backcross individuals in the 2010–2012 populations had mixed genotypes due to introgression of SNPs within the 2L island, resulting in the complete loss of 2L association with the X and 3L islands. The X and 3L SNPs remained in tight LD (r2 > 0.83, SI Appendix, Table S4). Genotype frequencies for all five SNPs on 2L remained at similar frequencies subsequently (2010–2012), so it appears that divergence in this region of the genome has been permanently lost.
The nearly complete disappearance of heterozygotes at all SNPs following the episode of hybridization that occurred in 2002 (Fig. 3C), and for heterozygotes on 3L and X following the hybridization event in 2006 (Fig. 3E), provides strong evidence for selection against hybrids in nature.
Patterns of Introgression Among Islands of Divergence.
White et al. (26) reported nearly complete LD between the three islands of divergence in natural populations. Hahn et al. (36) used laboratory crosses to demonstrate a lack of intrinsic incompatibilities that might explain this association. Based on these observations, it was proposed that the association among the three islands is maintained by near-complete RI between M and S forms in nature. RI is presumed to be the consequence of strong premating isolation and that those few hybrids that may be formed rarely survive to reproduce (26). This idea is central to the incidental islands model described above. The notion that association is maintained by strong selection against mixed genotypes is weakened by reports of linkage equilibrium in coastal populations where hybridization rates are high (20). We included two coastal populations in this study, the villages of Abu and Prabis in Guinea-Bissau, and we likewise found low LD among the three divergence islands (r2 < 0.17). The pattern we observed is complicated by the fact that the extent of introgression among the three islands is not equivalent. Genotypes for all eight SNPs on chromosomes 2 and 3 are at equilibrium frequencies so that introgression is complete. This suggests that selection against mixed genotypes at these SNPs is very low.
The pattern is quite different for the X chromosome where LD among the seven SNPs is quite strong (r2 > 0.93) and genotype frequencies depart significantly from HWE with a deficiency of heterozygotes at all seven SNPs. A pattern of lower introgression on the autosomes relative to the X chromosome was generally observed in all populations where M and S are sympatric (SI Appendix, Table S4). It has, however, been suggested that coastal populations represent unique environments and are therefore unlikely to be instructive about the genomics of RI in more “typical” areas (23). Results from the longitudinal survey conducted at Selinkenyi and the “snapshot” data for Goundry and Njigalap, however, demonstrate that hybridization and introgression do occur in typical A. gambiae habitats.
Within and Between Divergence Island Recombination.
Recombination within islands was lowest (r2 = 0.95) for chromosome 3 and highest (r2 = 0.90) for chromosome 2 (SI Appendix, Table S4). Recombination within the X chromosome was similar (r2 = 0.94) to the level observed in chromosome 3, despite covering a larger region of the genome (37 times larger).
Recombination between islands was highest for the 2L island (r2 = 0.87 vs. r2 = 0.94 for X and r2 = 0.93 for 3L) with a mean proportion of mismatched islands = 20% (SI Appendix, Table S4). Collections that displayed high levels of 2L island mismatch (P > 10%) include Nathia, Bioko, and 2010–2012 Selinkenyi. The wide geographic range of this 2L island mismatched genome, as well as the loss of 2L divergence post-2006 in Selinkenyi, suggest that this particular genome combination is stable and likely to be present in other geographic regions.
Discussion
The results presented here provide the basis for a critical reassessment of the relationship between the M and S forms of A. gambiae. A long-standing debate has centered on the level of gene flow between forms. On one side many authors consider that RI between the two is complete or nearly complete (7, 8, 45, 46) and that the two forms represent distinct species (29). On the other side are those who argued that RI, even if recent, cannot explain the “mosaic” pattern of genome divergence, and that it is selection operating on a few genomic regions in the face of gene flow that shapes this pattern (13, 16, 47). Here we have demonstrated that M/S hybridization occurs at frequencies well above the ∼1% frequently cited and at locations throughout West and Central Africa (Figs. 2 and 3 and SI Appendix, Figs. S3–S7), outside the coastal areas where high levels of hybridization are well known, but have been dismissed as “anomalous” (23).
Genome-wide scans have identified genomic islands of divergence, consistent with the mosaic divergence identified in earlier work. These were initially interpreted as representing islands of speciation containing genes responsible, directly or indirectly, for partial RI between forms and that the remaining, roughly 97% of the genome remains undifferentiated due to gene flow between the two forms (24). An opposing view recognizes islands of divergence as standing out, but argues this divergence is due to the location of the islands near centromeres where recombination is low and that the surrounding genome is relatively undifferentiated because complete RI evolved only recently (27). In support of this incidental islands model, it has been argued that the infrequent hybrids observed in nature are largely F1 hybrids and that these are effectively sterile, therefore their presence does not represent “effective” gene flow (26). The longitudinal study at Selinkenyi does suggest reduced fitness of hybrids; however, the loss of divergence on the chromosome 2L island after 2006 and the persistence of backcross genotypes in nearly all populations surveyed, often at high frequencies, reveals that hybridization does in fact represent gene flow and at levels far exceeding what has heretofore been recognized.
The existence of temporal variation in hybridization rates represents a unique and unanticipated feature in the relationship between M and S. The longitudinal study conducted over a period of 21 y at Selinkenyi reveals that RI is unstable (Fig. 3). Sustained maintenance of M and S populations, presumably by strong assortative mating, is periodically disrupted by episodes of hybridization, which in 2006 resulted in 12% of the population consisting of F1 hybrids. Remarkably this episode resulted in the complete loss of the 2L island of divergence present before 2006. In samples taken in 2010, 2011, and 2012, 2L SNP genotypes were at equilibrium frequencies.
This longitudinal survey also provided evidence for selection against hybrids. A smaller episode of hybridization occurred in 2002 with a frequency of F1 hybrids = 0.08, and backcrossed genotype frequencies = 0.11. Strong assortative mating appears to have been reestablished in 2004 with no F1s present and a decline in backcross genotypes to a frequency of 0.05. Likewise, following the 2006 episode, the frequency of backcrossed genotypes for the X and 3L islands declined from 13.7% (X) and 2.4% (3L) in 2006 to 2.0% (X) and 0% (3L) in 2010. This decline in hybrids was presumably the result of selection. Loss of the 2L island after 2006 is likely the consequence of relatively weak selection being overwhelmed by gene flow that year [but see discussion by Weetman et al. concerning selection on the kdr locus on chromosome 2L (12)], whereas selection on the 3L and X islands appears to have been strong enough to maintain divergence, despite this relatively high level of gene flow.
Overall, it is the X chromosome island that remains the most highly diverged. This observation supports the suggestion that the X island contains genes responsible for RI (26, 48). This idea is further supported by the identification of high levels of divergence in the same region of the X between the Bamako and Savanna chromosomal forms of A. gambiae, which exhibit a high degree of RI, despite the fact that both are S form (49). Reduced introgression and recombination on the sex chromosome shown here accords well with other studies on flycatchers (50–52), Drosophila simulans group (53), and Heliconius butterflies (54) that demonstrated that sex chromosomes display a more advanced stage of speciation compared with autosomes.
Overall the picture that emerges is consistent with the islands of speciation model (24) as the best fit to describe the relationship between the M and S forms. However, in a larger sense these results challenge the more fundamental view of the relationship between M and S. The extent of gene flow between the two forms is far greater than presumed and the frequency of hybridization is dynamic, fluctuating in both time and space. This observation is consistent with a genic model of speciation where populations may diverge even in the face of considerable gene flow (4, 55). This process has been described as proceeding along a speciation continuum (56, 57) and it is possible for divergent populations to lie somewhere midway on this continuum (58, 59). Movement on the speciation continuum may be toward increasing divergence and speciation; however, the reverse is also possible (60, 61). Our data suggest the possibility that M and S forms represent diverged populations that sporadically move “forward” (diverging) and “backward” (introgressing).
The M form is generally believed to be the more recently evolved (62, 63), presumably via niche expansion into marginal habitats in parapatry with the S form (64). It is in these marginal habitats that they are believed to have diverged via adaptation to the peculiar ecological conditions that occur there and ultimately by the acquisition of premating mechanisms reducing the possibility of mating with the S form. Range expansion has increasingly brought M and S in contact, so that today they occur largely in sympatry throughout their range in West and Central Africa (1, 6). We provide evidence that pre- and postzygotic isolation between the two forms is relatively weak, such that varying degrees of introgression of the two forms has occurred throughout their range and that this is an ongoing phenomenon. The GOUNDRY population in Burkina Faso is the most extreme example where all form-specific SNP genotypes are in HWE (Fig. 2O and SI Appendix, Fig. S7E). Coastal populations in Guinea-Bissau (Abu and Prabis) have complete introgression of the autosomal islands of divergence; however, the X chromosome remains largely diverged (Fig. 2 K–N and SI Appendix, Fig. S7 A–D), presumably due to a strong asymmetry in assortative mating (22). Loss of the chromosome 2L island of divergence was observed in populations at Bioko and Selinkenyi (Fig. 2 F and G and SI Appendix, Fig. S5). Evidence that this is a dynamic and ongoing process is provided by the longitudinal study conducted at Selinkenyi where periodic episodes of increased hybridization have recently culminated in the loss of the 2L island of divergence in this population (Fig. 3).
The results presented here confirm the importance of the M and S designation as a proxy for reproductively isolated populations (in most, but not all locations analyzed). However, it seems plausible that the M and S forms lie at an unstable position, midway on a species continuum, where forces promoting divergence (selection and assortative mating) are frequently overwhelmed by recurrent gene flow leading to introgression. It is likely that divergence occurs on a very long time scale, over hundreds to millions of generations (65). Our longitudinal study at Selinkenyi spans 21 y (∼250 A. gambiae generations). In this regard, our data are still just a snapshot of a much longer process. In addition, our analysis covers a tiny fraction of the M and S genomes. Genome-wide surveys will provide a much better picture of the extent of M–S introgression and should provide insights into the importance of hybridization as an adaptive force in these mosquitoes. It is clear that A. gambiae remains an intriguing system for the exploration of mechanisms underlying the speciation process.
Methods
Mosquito Collections and Species Identification.
A. gambiae sensu lato were collected from inside human dwellings using mouth aspirators during the rainy season from various sites in Burkina Faso (ENDO), Cameroon (three sites), Equatorial Guinea (one site), Guinea-Bissau (three sites), Mali (seven sites) and Senegal (two sites) (SI Appendix, Table S3). The GOUNDRY, Burkina Faso collections were larvae from natural breading sites. A. gambiae sensu stricto were identified using a diagnostic PCR developed by Scott et al. (66).
SNP Genotyping.
We used the DIS method (35) to genotype A. gambiae individuals. The DIS method includes 15 SNPs occurring on all three chromosomes; 7 on the X, 5 on 2L, 3 on 3L. Full details of the DIS method are provided in SI Appendix. Genotyping was conducted using the Sequenom iPLEXGold assay platform at the Veterinary Genetics Laboratory at the University of California, Davis. Mass spectrogram visualization and genotype calls were done using TyperAnalyzer version 4.0 (Sequenom).
Data Analysis.
We plotted DIS maps using R heatmap plotting. We calculated LD using the EMLD program that implements the maximum-likelihood method described in ref. 67. Arlequin version 3.5 (68) was used to test departure from HWE and to determine significance of LD.
Supplementary Material
Acknowledgments
We thank Drs. K. Vernick (Pasteur Institute) and M. Riehle (University of Minnesota) for the GOUNDRY and ENDO specimens, Dr. B. Foy (Colorado State University) for the Senegal specimens, C. Neiman and A. Weakley (Vector Genetics Laboratory) for technical assistance, J. Malvick (Veterinary Genetics Laboratory) for assistance in SNP genotyping, and Dr. M. Sanford (Harris County Institute of Forensic Sciences) for advice in developing this project. This research was supported by National Institutes of Health Grants R01AI 078183, R21AI062929, D43TW007390, and T32AI074550.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The raw genotype data reported in this paper have been deposited in the online individual level population genomic database PopI, with OpenProject id AgDIS, http://popi.ucdavis.edu.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316851110/-/DCSupplemental.
References
- 1.Lanzaro GC, Lee Y. Speciation in Anopheles gambiae—The distribution of genetic polymorphism and patterns of reproductive isolation among natural populations. In: Manguin S, editor. Anopheles Mosquitoes: New Insights into Malaria Vector. Rijeka, Croatia: InTech; 2013. [Google Scholar]
- 2.Pinho C, Hey J. Divergence with gene flow: Models and data. Annu Rev Ecol Evol Syst. 2010;41(41):215–230. [Google Scholar]
- 3.Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28(7):342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Hey J. Recent advances in assessing gene flow between diverging populations and species. Curr Opin Genet Dev. 2006;16(6):592–596. doi: 10.1016/j.gde.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Nosil P. Ecological Speciation. Oxford: Oxford Univ Press; 2012. p. xvii. [Google Scholar]
- 6.della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35(7):755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Favia G, Lanfrancotti A, Spanos L, Sidén-Kiamos I, Louis C. Molecular characterization of ribosomal DNA polymorphisms discriminating among chromosomal forms of Anopheles gambiae s.s. Insect Mol Biol. 2001;10(1):19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 8.della Torre A, et al. Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001;10(1):9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 9.Tripet F, et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol Ecol. 2001;10(7):1725–1732. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- 10.Diabaté A, Dabire RK, Millogo N, Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae) J Med Entomol. 2007;44(1):60–64. doi: 10.1603/0022-2585(2007)44[60:eteopi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 11.Turner TL, Hahn MW. Genomic islands of speciation or genomic islands and speciation? Mol Ecol. 2010;19(5):848–850. doi: 10.1111/j.1365-294X.2010.04532.x. [DOI] [PubMed] [Google Scholar]
- 12.Weetman D, Wilding CS, Steen K, Pinto J, Donnelly MJ. Gene flow-dependent genomic divergence between Anopheles gambiae M and S forms. Mol Biol Evol. 2012;29(1):279–291. doi: 10.1093/molbev/msr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanzaro GC, et al. Complexities in the genetic structure of Anopheles gambiae populations in west Africa as revealed by microsatellite DNA analysis. Proc Natl Acad Sci USA. 1998;95(24):14260–14265. doi: 10.1073/pnas.95.24.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wondji C, Simard F, Fontenille D. Evidence for genetic differentiation between the molecular forms M and S within the Forest chromosomal form of Anopheles gambiae in an area of sympatry. Insect Mol Biol. 2002;11(1):11–19. doi: 10.1046/j.0962-1075.2001.00306.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang R, Zheng L, Touré YT, Dandekar T, Kafatos FC. When genetic distance matters: Measuring genetic differentiation at microsatellite loci in whole-genome scans of recent and incipient mosquito species. Proc Natl Acad Sci USA. 2001;98(19):10769–10774. doi: 10.1073/pnas.191003598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor C, et al. Gene flow among populations of the malaria vector, Anopheles gambiae, in Mali, West Africa. Genetics. 2001;157(2):743–750. doi: 10.1093/genetics/157.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ndiath MO, et al. Dynamics of transmission of Plasmodium falciparum by Anopheles arabiensis and the molecular forms M and S of Anopheles gambiae in Dielmo, Senegal. Malar J. 2008;7:136. doi: 10.1186/1475-2875-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caputo B, et al. Anopheles gambiae complex along The Gambia river, with particular reference to the molecular forms of An. gambiae s.s. Malar J. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveira E, et al. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45(6):1057–1063. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Caputo B, et al. The “far-west” of Anopheles gambiae molecular forms. PLoS ONE. 2011;6(2):e16415. doi: 10.1371/journal.pone.0016415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nwakanma DC, et al. Breakdown in the process of incipient speciation in Anopheles gambiae. Genetics. 2013;193(4):1221–1231. doi: 10.1534/genetics.112.148718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsden CD, et al. Asymmetric introgression between the M and S forms of the malaria vector, Anopheles gambiae, maintains divergence despite extensive hybridization. Mol Ecol. 2011;20(23):4983–4994. doi: 10.1111/j.1365-294X.2011.05339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reidenbach KR, et al. Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae in West and Central Africa. Genome Biol Evol. 2012;4(12):1202–1212. doi: 10.1093/gbe/evs095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3(9):e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White BJ, et al. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009;183(1):275–288. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19(5):925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neafsey DE, et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science. 2010;330(6003):514–517. doi: 10.1126/science.1193036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawniczak MK, et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science. 2010;330(6003):512–514. doi: 10.1126/science.1195755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coetzee M, et al. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa. 2013;3619(2):246–274. [PubMed] [Google Scholar]
- 30.Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16(4):461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- 31.Santolamazza F, Della Torre A, Caccone A. Short report: A new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am J Trop Med Hyg. 2004;70(6):604–606. [PubMed] [Google Scholar]
- 32.Santolamazza F, et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar J. 2008;7:163. doi: 10.1186/1475-2875-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22(8):437–446. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 34.Sousa V, Hey J. Understanding the origin of species with genome-scale data: modelling gene flow. Nat Rev Genet. 2013;14(6):404–414. doi: 10.1038/nrg3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee Y, Marsden CD, Nieman C, Lanzaro GC (2013) A new multiplex SNP genotyping assay for detecting hybridization and introgression between the M and S molecular forms of Anopheles gambiae. Mol Ecol Resour, 10.1111/1755-0998.12181. [DOI] [PMC free article] [PubMed]
- 36.Hahn MW, White BJ, Muir CD, Besansky NJ. No evidence for biased co-transmission of speciation islands in Anopheles gambiae. Philos Trans R Soc Lond B Biol Sci. 2012;367(1587):374–384. doi: 10.1098/rstb.2011.0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riehle MM, et al. A cryptic subgroup of Anopheles gambiae is highly susceptible to human malaria parasites. Science. 2011;331(6017):596–598. doi: 10.1126/science.1196759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elseth GD, Baumgardner KD. Population Biology. New York: D. Van Nostrand; 1981. p. xvi. [Google Scholar]
- 39.Aldridge G. Variation in frequency of hybrids and spatial structure among Ipomopsis (Polemoniaceae) contact sites. New Phytol. 2005;167(1):279–288. doi: 10.1111/j.1469-8137.2005.01413.x. [DOI] [PubMed] [Google Scholar]
- 40.Rieseberg LH, Whitton J, Gardner K. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics. 1999;152(2):713–727. doi: 10.1093/genetics/152.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teeter KC, et al. The variable genomic architecture of isolation between hybridizing species of house mice. Evolution. 2010;64(2):472–485. doi: 10.1111/j.1558-5646.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- 42.McSweeney C, New M, Lizcano G, Lu X. UNDP Climate Change Country Profiles (University of Oxford, Oxford) 2010. Available at http://country-profiles.geog.ox.ac.uk/. Accessed September 5, 2013.
- 43.Lehmann T, et al. Aestivation of the African malaria mosquito, Anopheles gambiae in the Sahel. Am J Trop Med Hyg. 2010;83(3):601–606. doi: 10.4269/ajtmh.2010.09-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Touré YT, et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998;40(4):477–511. [PubMed] [Google Scholar]
- 45.della Torre A, et al. Speciation within Anopheles gambiae—the glass is half full. Science. 2002;298(5591):115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- 46.Mukabayire O, et al. Patterns of DNA sequence variation in chromosomally recognized taxa of Anopheles gambiae: Evidence from rDNA and single-copy loci. Insect Mol Biol. 2001;10(1):33–46. doi: 10.1046/j.1365-2583.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- 47.Lehmann T, et al. Population structure of Anopheles gambiae in Africa. J Hered. 2003;94(2):133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- 48.Slotman MA, et al. Reduced recombination rate and genetic differentiation between the M and S forms of Anopheles gambiae s.s. Genetics. 2006;174(4):2081–2093. doi: 10.1534/genetics.106.059949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee Y, et al. Chromosome inversions, genomic differentiation and speciation in the African malaria mosquito Anopheles gambiae. PLoS ONE. 2013;8(3):e57887. doi: 10.1371/journal.pone.0057887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491(7426):756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 51.Harr B, Price T. Speciation: Clash of the genomes. Curr Biol. 2012;22(24):R1044–R1046. doi: 10.1016/j.cub.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 52.Saetre GP, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc Biol Sci. 2003;270(1510):53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrigan D, et al. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Res. 2012;22(8):1499–1511. doi: 10.1101/gr.130922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin SH, et al. Genome-wide evidence for speciation with gene flow in Heliconius butterflies. Genome Res. 2013;23(11):1817–1828. doi: 10.1101/gr.159426.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5(2):114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- 56.Mallet J, Beltrán M, Neukirchen W, Linares M. Natural hybridization in heliconiine butterflies: The species boundary as a continuum. BMC Evol Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18(3):375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 58.Peccoud J, Ollivier A, Plantegenest M, Simon JC. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci USA. 2009;106(18):7495–7500. doi: 10.1073/pnas.0811117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merrill RM, et al. Mate preference across the speciation continuum in a clade of mimetic butterflies. Evolution. 2011;65(5):1489–1500. doi: 10.1111/j.1558-5646.2010.01216.x. [DOI] [PubMed] [Google Scholar]
- 60.Seehausen O, Takimoto G, Roy D, Jokela J. Speciation reversal and biodiversity dynamics with hybridization in changing environments. Mol Ecol. 2008;17(1):30–44. doi: 10.1111/j.1365-294X.2007.03529.x. [DOI] [PubMed] [Google Scholar]
- 61.Taylor EB, et al. Speciation in reverse: Morphological and genetic evidence of the collapse of a three-spined stickleback (Gasterosteus aculeatus) species pair. Mol Ecol. 2006;15(2):343–355. doi: 10.1111/j.1365-294X.2005.02794.x. [DOI] [PubMed] [Google Scholar]
- 62.Ayala FJ, Coluzzi M. Chromosome speciation: Humans, Drosophila, and mosquitoes. Proc Natl Acad Sci USA. 2005;102(Suppl 1):6535–6542. doi: 10.1073/pnas.0501847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298(5597):1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- 64.Costantini C, et al. Living at the edge: Biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26(2):229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 66.Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49(4):520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 67.Huang Q, Shete S, Swartz M, Amos CI. Examining the effect of linkage disequilibrium on multipoint linkage analysis. BMC Genet. 2005;6(Suppl 1):S83. doi: 10.1186/1471-2156-6-S1-S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Excoffier L, Lischer HE. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.