Significance
Toll/interleukin-1 receptor (TIR) homology domains mediate the downstream signaling of Toll-like receptors (TLRs), but the molecular mechanism of the signal transduction is elusive on the structural basis. Here, we determined the structures of TIR domain-containing adaptor molecule (TICAM) 1 and TICAM-2 TIR domains and demonstrated their homotypic and heterotypic interaction surfaces. Both TICAM-1 and TICAM-2 TIR domains form a BB-loop–mediated homodimer. The dimerization of TICAM-2 TIR presents an interaction surface for TICAM-1 TIR. The present result is consistent with the notion that the BB-loop mutant is dominant negative and that dimerization of upstream TIRs is crucial for recruiting downstream TIRs.
Keywords: innate immunity, TLR signaling, MyD88-independent pathway, TRAM, TRIF
Abstract
Homotypic and heterotypic interactions between Toll/interleukin-1 receptor (TIR) domains in Toll-like receptors (TLRs) and downstream adaptors are essential to evoke innate immune responses. However, such oligomerization properties present intrinsic difficulties in structural studies of TIR domains. Here, using BB-loop mutations that disrupt homotypic interactions, we determined the structures of the monomeric TIR domain-containing adaptor molecule (TICAM)-1 and TICAM-2 TIR domains. Docking of the monomeric structures, together with yeast two hybrid-based mutagenesis assays, reveals that the homotypic interaction between TICAM-2 TIR is indispensable to present a scaffold for recruiting the monomeric moiety of the TICAM-1 TIR dimer. This result proposes a unique idea that oligomerization of upstream TIR domains is crucial for binding of downstream TIR domains. Furthermore, the bivalent nature of each TIR domain dimer can generate a large signaling complex under the activated TLRs, which would recruit downstream signaling molecules efficiently. This model is consistent with previous reports that BB-loop mutants completely abrogate downstream signaling.
The extracellular domain of toll-like receptor 4 (TLR4) specifically binds lipopolysaccharides (LPSs) from Gram-negative bacteria, inducing dimerization and leading to the dimerization of cytosolic Toll/interleukin-1 receptor (TIR) domains. This activated conformation of TLR4 recruits the TIR domain of a downstream adaptor molecule, TIR domain-containing adaptor molecule-2 (TICAM-2) [also known as TRIF-related adaptor molecule (TRAM)], that subsequently recruits the TIR domain of another adaptor molecule, TIR domain-containing adaptor molecule-1 (TICAM-1) [also known as TIR domain-containing adaptor inducing IFN-β (TRIF)] (1–3) at endosomes. Eventually this process activates IFN response factors and generates type-I interferons (IFNs) (4–7). Elucidation of the homotypic and heterotypic interactions between TICAM-1 and TICAM-2 is essential for understanding of TLR4-mediated type-I IFN generation (8).
A large number of TIR domain structures, including receptors and adaptors, have been determined by X-ray crystallography and NMR. The receptors include TLR1 (9), TLR2 (10), and IL-1R accessory protein-like (IL-1RAPL) (11). Adaptors include myeloid differentiation factor 88 (MyD88) (12) and MyD88 adaptor-like (Mal) (13, 14). In addition, AtTIR (15, 16) derived from Arabidopsis thaliana and PdTIR (17) from bacteria have been solved. Each of these TIR domain structures has a ferredoxin fold with five β-strands (βA–βE), five α-helices (αA–αE), and loops connecting β-strands and α-helices (9). Although homotypic interactions of the TIR domains have been proposed based on the crystal structures, most proposed models have small interacting surfaces, possibly due to crystal contacts. Recently, however, a crystal structure of the TLR10 TIR domain was reported that forms a homotypic dimer mediated by the loop connecting βB and αB (designated “BB-loop”) (18). Interestingly, BB-loop mutations in TLR4 were reported to be dominant-negative and abrogated downstream signaling (19). TICAM-1 and TICAM-2 harboring BB-loop mutations are also dominant-negative and unable to form homotypic interactions (1, 2), reinforcing the importance of BB-loop–mediated homotypic dimer formation in signal propagation.
Despite extensive structural studies, it is not known why homotypic interactions are essential for downstream signaling (20–27). To address this issue, it is necessary to discriminate residues required for homotypic and those required for heterotypic interactions. Here, we first determine the structures of the monomeric BB-loop mutants of the TICAM-1 and TICAM-2 TIR domains using NMR. Then, based on the solution structures of the BB-loop mutants, coupled mutagenesis/yeast two-hybrid experiments, and restrained docking calculations, we show that the homotypic interaction of TICAM-2 TIR is essential to form a scaffold for recruiting the TICAM-1 TIR domain.
Results
Monomerization of the TICAM-1 and TICAM-2 TIR Domains by BB-Loop Mutations.
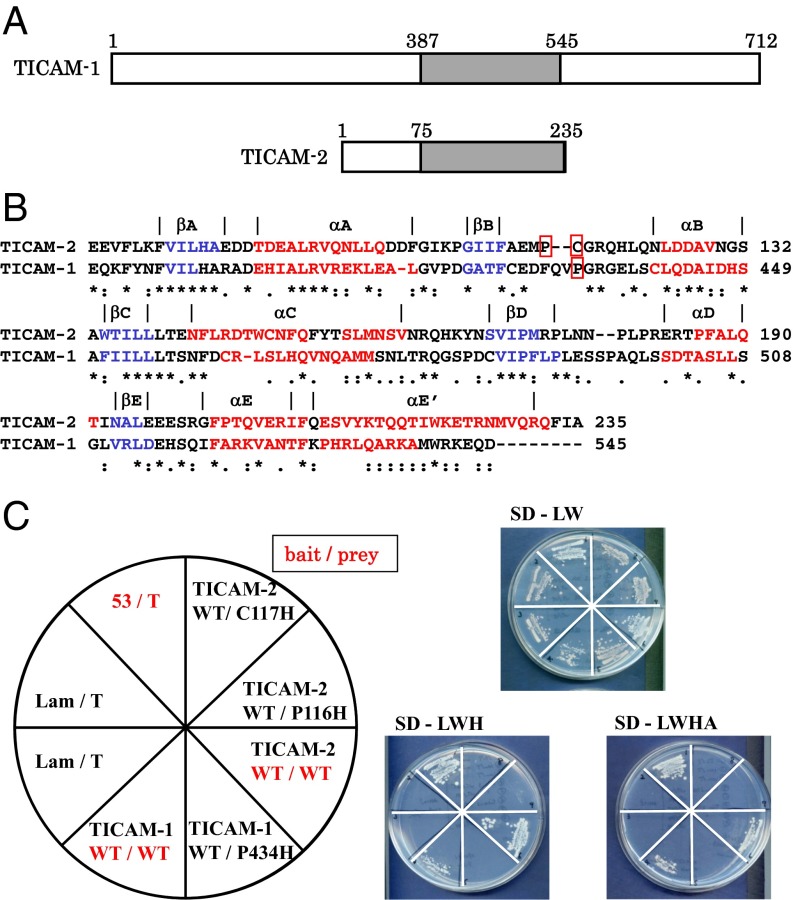
The TIR domains of TICAM-1 (387–545) and TICAM-2 (75–235) (Fig. 1A) oligomerized and precipitated in aqueous solution at ∼200 μM concentration, so monomerization was indispensable for structure determination by NMR. BB-loop mutants of TICAM-1 and TICAM-2 are known to be dominant-negative and unable to form homotypic interactions (1, 2). Thus, we prepared P434H and C117H (P116H) mutants of the TICAM-1 and TICAM-2 TIR domains, respectively (Fig. 1B) [hereafter designated TICAM-1 P434H and TICAM-2 C117H (TICAM-2 P116H)] and analyzed their homotypic interactions using yeast two-hybrid experiments. Yeast two-hybrid experiments showed that the homotypic interaction is disrupted by the BB-loop mutation (Fig. 1C), consistent with previous reports (1, 2) and NMR observation of both wild types and BB-loop mutants (SI Text and Fig. S1).
Fig. 1.
Homotypic interaction of TICAM-1 and TICAM-2 TIR domains. (A) TIR domains of TICAM-1 and TICAM-2. The gray boxes represent the TIR domains of TICAM-1 and TICAM-2. (B) Sequence alignment of the TIR domains of human TICAM-1 and human TICAM-2. Amino acid residues involved in the β-sheet and α-helix are shown in blue and red, respectively. The residues on the BB-loop enclosed by a red line were substituted by His for the solution structure determination in this study. Numbers at the right side of the sequences correspond to the residue number in human TICAM-1 and TICAM-2. (C) Yeast two-hybrid analysis of homotypic interaction in TICAM-1 and TICAM-2. Pro434 of TICAM-1, Pro116 and Cys117 of TICAM-2 in the BB-loop were substituted by His residue. These mutants disrupted homotypic interaction of the TIR domain in TICAM-1 and TICAM-2. p53/T-antigen and Lamine/T-antigen were used as positive and negative controls in the yeast two-hybrid assay, respectively. SD-LW, SD-LWH, and SD-LWHA indicate a synthetic dropout medium lacking Leu and Trp, lacking Leu, Trp, and His, and that further lacking Ade, respectively.
NMR Structures of the TICAM-1 P434H and TICAM-2 C117H Mutants.
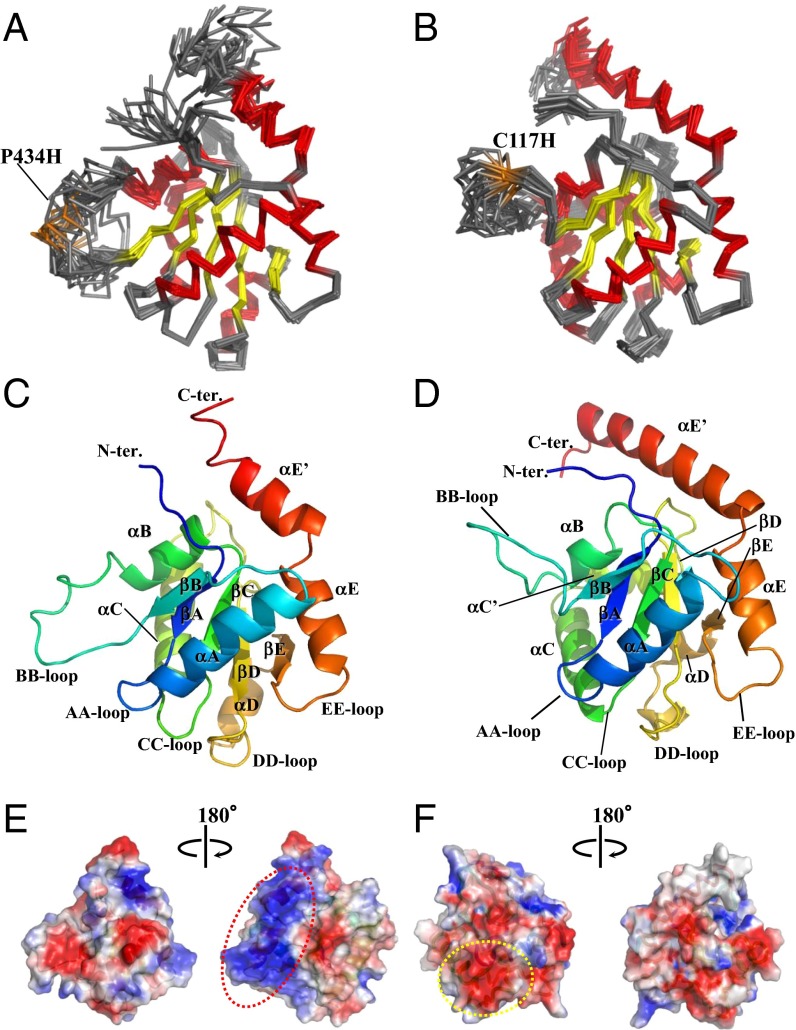
The solution structures of the TICAM-1 P434H and TICAM-2 C117H mutants were determined based on distance and dihedral angle constraints. Structural statistics for the final 20 conformers of each protein are summarized in Table S1. The core structures, consisting of residues other than the BB-loops and N-terminal and C-terminal regions, were well defined. The root-mean-square-deviation (rmsd) of the core backbone atoms (Cα, N, C′) of TICAM-1 P434H (395–427 and 442–527) and TICAM-2 C117H (83–110, 132–215) were 0.45 Å and 0.50 Å, respectively (Fig. 2 A and B). The global structures of both mutants were comprised of five parallel β-strands surrounded by six or seven α-helices and loops that connect β-strands and α-helices. Following the conventional nomenclature for TIR domains, the five strands in TICAM-1 were designated βA(397–400), βB(424–427), βC(451–455), βD(486–491), and βE(511–514), and the six helices in TICAM-1 were designated αA(406–419), αB(441–449), αC(462–474), αD(501–507), αE(520–528), and αE′(530–538) with a kink at residue 529 (Fig. 2C). Similarly, TICAM-2 C117H also contained five strands designated βA(81–85), βB(109–112), βC(134–138), βD(169–173), and βE(193–195) and seven α-helices, designated αA(90–101), αB(125–129), αC(142–152), αC′(156–161), αD(186–191), αE(202–210), and αE′(212–232) (Fig. 2D). In both structures, the conformation of the BB-loop was not well defined due to broadening of the NMR signals, resulting in insufficient NOE distance restraints. (Fig. 2 A and B).
Fig. 2.
Structures of the TICAM-1 and TICAM-2 TIR domains. (A and B) The overlay of the 20 lowest energy structures of the TIR domains of TICAM-1 and TICAM-2 determined by NMR. The backbone atoms (N, Cα, C′) of polypeptides are drawn in wire model; β-sheets in yellow; α-helices in red. Pro434 of TICAM-1 and Cys117 of TICAM-2 in BB-loop are substituted with His, which are shown in orange and labeled. (C and D) The TIR domain of TICAM-1 (C) and TICAM-2 (D) in ribbon model. β-strands, α-helices, and the connecting loops are labeled after the conventional nomenclature of the TIR domain (9). (E and F) Electrostatic surface potential of the TIR domains of TICAM-1 (E) and TICAM-2 (F). Positive, negative, and neutral electrostatic surface potentials are presented in blue, red, and white, respectively. The electrostatic surface potential in E and F, Left is presented as the same orientation as shown in A (C) and B (D). The opposite surfaces are shown in E and F, Right. The basic surface in TICAM-1 TIR and the acidic surface in TICAM-2 TIR are enclosed by red and yellow dotted lines, respectively.
Electrostatic surface potentials of TICAM-1 P434H and TICAM-2 C117H are shown in Fig. 2 E and F, respectively. TICAM-1 P434H has an extensive basic surface comprised of αE (Arg522, Lys523) and αE′ (Lys529, Arg532, Arg536, Lys537, Arg541, Lys542). In contrast, TICAM-2 C117H has an extensive acidic surface comprised of the AA-loop (Glu87, Asp88, Asp89) and the αA-helix (Asp91, Glu92, Asp102, Asp103).
A Dali search (28) revealed that the structure of the TIR domain of TICAM-1 is most similar to that of TICAM-2, with a z-score of 9.6 and an rmsd of 3.8 Å for the structured region (Cα 122 atoms), followed by the TIR domains of TLR2 (z-score 9.2), TLR1 (z-score 8.8), TLR10 (z-score 8.7), IL-1RAPL (z-score 8.5), and MyD88 (z-score 7.5). A structural superposition was made to align the secondary structures and functionally important residues in TICAM-1, TICAM-2, and other TIR domains using the MATRAS program (29, 30) (Fig. S2). Intriguingly, the residues that form an extensive acidic surface in the TICAM-2 TIR domain and an extensive basic surface in the TICAM-1 TIR domain are not conserved in other TIR domains, suggesting that these residues might be responsible for specific interaction between TICAM-1 and TICAM-2.
Acidic Region of TICAM-2 and Basic Region of TICAM-1 Are Essential for Heterotypic Interaction.
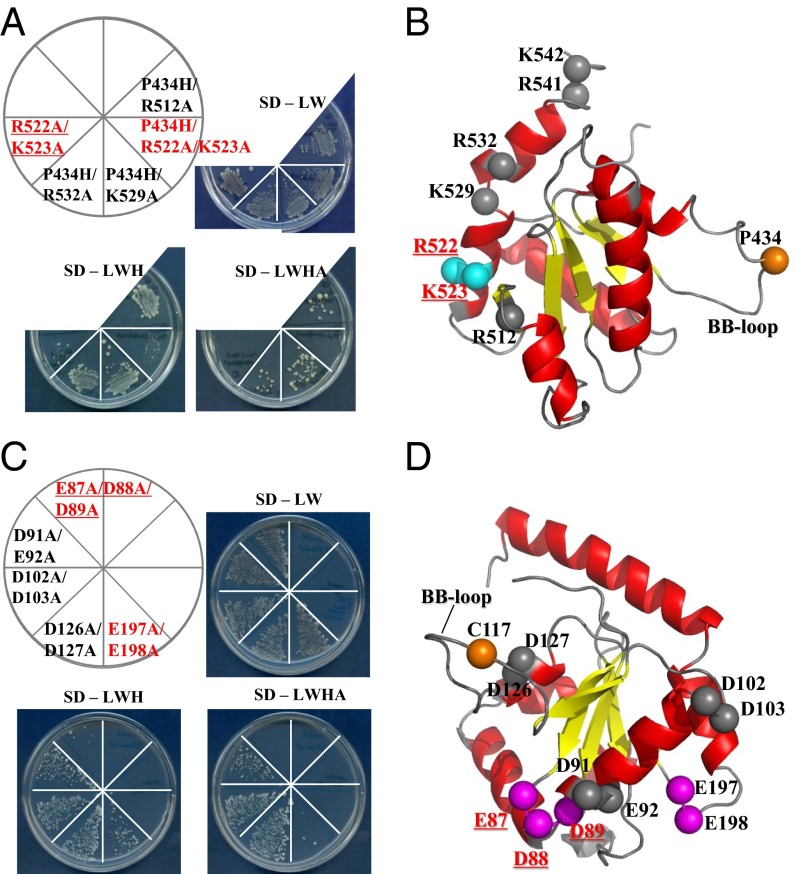
To investigate heterotypic interactions between the TICAM-1 and TICAM-2 TIR domains, further yeast two-hybrid experiments were carried out (31). Because previous studies showed that oligomerization of the TICAM-2 TIR domain is essential for its interaction with the TICAM-1 TIR domain (1, 2, 27), the wild-type TICAM-2 TIR domain was used as bait, and the TICAM-1 TIR domain mutants were used as prey. To search for residues that are essential for the interaction with TICAM-2, basic residues within TICAM-1 αE and αE′-helices were selected and mutated to alanine in the first round of two-hybrid experiments. The TICAM-1 mutants harboring the BB-loop mutation, P434H/R512A, P434H/K529A, and P434H/R532A, could interact with TICAM-2, but the mutants R522A/K523A and P434H/R522A/K523A could not (Fig. 3A). These results indicate that Arg522 and Lys523 of TICAM-1, but not Pro434, Arg512, Lys529, and Arg532, are crucial for direct interaction with TICAM-2, consistent with the observation that TICAM-1 oligomerization is not required for interaction with the TICAM-2 TIR domain. We designated the region involving Arg522 and Lys523 the “RK site.” Actually, Pro434 is located on the opposite side of the RK site in TICAM-1, indicating that the homotypic and heterotypic interaction sites in the TICAM-1 TIR domain are distinct (Fig. 3B). Interestingly, the BB-loop mutant could still interact with wild-type TICAM-2, implying that monomeric TICAM-1 retains its ability to bind to the TIR domain of the TICAM-2 wild-type dimer.
Fig. 3.
Heterotypic interaction between TICAM-1 and TICAM-2 TIRs revealed by yeast two-hybrid assays (1). (A) Yeast two-hybrid assays using the wild type of the TICAM-2 TIR domain as bait and the mutants of the TICAM-1 TIR domain as prey. (B) The basic residues on the EE-loop and αE- and αE′-helices in the TICAM-1 TIR domain (shown as spheres) are mutated to Ala. The residues that disrupt the heterotypic interaction are displayed in cyan. The position of the BB-loop mutation in TICAM-1 TIR is shown in orange. (C) Yeast two-hybrid assays using the wild type of the TICAM-1 TIR domain as bait and the mutants of the TICAM-2 TIR domain harboring no BB-loop mutation as prey. (D) The acidic residues in the TICAM-2 TIR domain (shown as spheres) are mutated to Ala. The residues that disrupt the heterotypic interaction are displayed in magenta. The position of the BB-loop mutation in TICAM-2 TIR is shown in orange.
After finding two basic residues in the TICAM-2 binding surface of TICAM-1, we searched for acidic residues in TICAM-2 that complemented the interaction. Arrays of two or three acidic residues from the TIR domain of TICAM-2 (E87/D88/D89 in the AA-loop, D91/E92 in the N-terminal side of the αA-helix, D102 /D103 in the C-terminal side of the αA-helix, D126/D127 in the αB-helix, and E197/E198 in the EE-loop) were substituted with alanine. Yeast two-hybrid analyses showed that TICAM-2 mutants bearing E87A/D88A/D89A and E197A/E198A could not grow completely on SD-LWHA medium due to disruption of their interaction with wild-type TICAM-1 (Fig. 3C), suggesting that the acidic cluster consisting of residues Glu87/Asp88/Asp89 in the AA-loop and Glu197/Glu198 in the EE-loop of the TICAM-2 TIR domain might be responsible for association with the TICAM-1 TIR domain (Fig. 3D, in magenta). However, in yeast two-hybrid experiments, it is generally difficult to distinguish mutations that disrupt the protein interaction surface from those that disrupt the tertiary structure. To discriminate these two scenarios, the mutant proteins were expressed and subjected to gel-filtration chromatography. We showed the results in SI Text and Fig. S3.
Coupled Mutations Identify Additional Binding Sites.
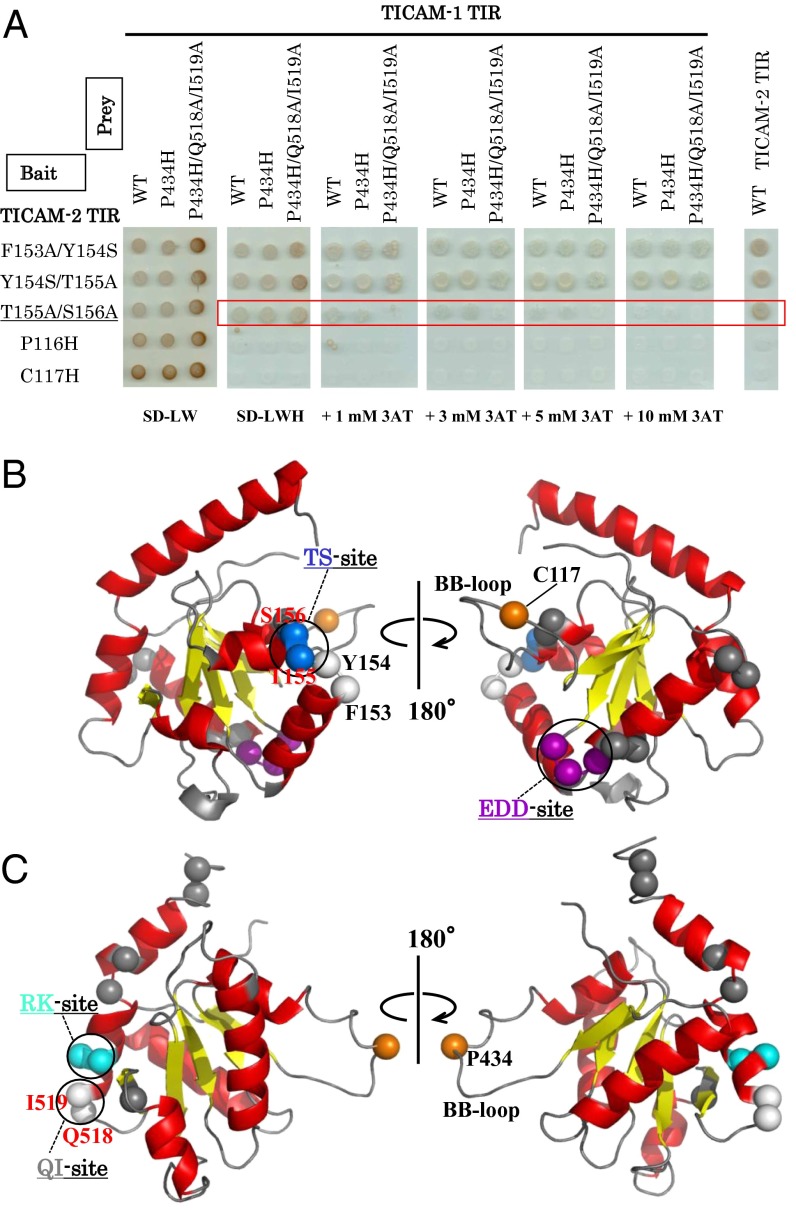
Because dimer formation of the TICAM-2 TIR domain is indispensable for interaction with the TICAM-1 TIR domain, we used TICAM-2 TIR domain mutants that harbored no BB-loop mutations in the following yeast two-hybrid experiments. Because the β-strands formed a core structure surrounded by α-helices and loop regions, the residues in the β-strands would be expected to stabilize the structure of the TIR domain. Thus, we applied mutations to the exposed surface residues located on α-helices or loops, based on the NMR structure of TICAM-2 C117H (Fig. S4) and studied further interaction sites between the TICAM-2 and TICAM-1 TIR domains using yeast two-hybrid experiments. The heterotypic interaction between the TICAM-2 wild type and the TICAM-1 mutants was studied using an SD-LWH medium where the 3AT concentration is successively increased (Fig. 4A). Tables S2 and S3 list the results of the yeast two-hybrid experiments. Only TICAM-2 mutants involving residues on the CC′-loop between the αC and the αC′ helices showed reduced affinity for TICAM-1 as is summarized in Fig. 4A. First, the interaction of the F153A/Y154S, Y154S/T155A, and T155A/S156A mutants of the TICAM-2 TIR domain was studied with the wild type of the TICAM-2 TIR domain, showing that these mutants retain the homotypic interaction with the TICAM-2 wild type (Fig. 4A, right lane). Next, these mutants were applied to the yeast two-hybrid analyses to study heterotypic interactions with wild-type TICAM-1 and TICAM-1 P434H. Considering that monomeric TICAM-1 P434H can interact with the TICAM-2 dimer, the contact residues should be located near the RK site. Thus, we selected Gln518 and Ile519 on the EE-loop and αE-helix as further candidates for interaction with the TICAM-2 TIR domain.
Fig. 4.
Heterotypic interaction between TICAM-2 and TICAM-1 TIRs revealed by yeast two-hybrid assays (2). (A) Yeast two-hybrid assays using the mutants of TICAM-2 TIR as bait and the wild type and mutants of TICAM-1 TIR as prey. The wild-type TICAM-2 TIR is also used as prey as shown in Right. Yeast growth was analyzed with SD-LWH medium supplemented with 3-AT. (B) Residue mapping on the structure of TICAM-2 TIR. Thr155 and Ser156 are shown in blue (TS site), and Phe153 and Y154 are shown in white. Acidic residues, Glu87, Asp88, and Asp89 (EDD site), which are critical for interaction with TICAM-1, are shown in magenta. The residue C117 substituted by His is shown in orange. (C) Residue mapping on the structure of TICAM-1 TIR. Arg522 and Lys523 (RK site) are shown in cyan, and Gln518 and Ile519 (QI site) are shown in white. The Pro434 substituted by His in BB-loop of TICAM-1 is shown in orange.
Among the TICAM-2 TIR domain mutants F153A/Y154S, Y154S/T155A, and T155A/S156A, only the T155A/S156A mutant on the CC′-loop showed reduced interaction with wild type, P434H, and P434H/Q518A/I519A TICAM-1 mutants in a 3AT dose-dependent manner, with higher growth inhibition for the P434H/Q518A/I519A mutant. This result implied that Thr155 and Ser156 in TICAM-2 (designated the “TS site”) and Gln518 and Ile519 in TICAM-1 (designated the “QI site”) are also involved in the heterotypic interaction (Fig. 4A). Intriguingly, the TS site in blue and the EDD site in magenta are located on opposite sides of the TICAM-2 TIR domain (Fig. 4B). In TICAM-1, the interacting residues with the TICAM-2 TIR domain are located on the RK site (in cyan) and QI site (in white), which form a wedge-like shape on the TICAM-1 TIR domain surface (Fig. 4C).
Restrained Docking of Trimeric TICAM-2/TICAM-1 Complex.
The solution structures of the TICAM-1 P434H and TICAM-2 C117H mutants, along with the results of the mutagenesis and yeast two-hybrid assays, permitted docking of the complex formed by two TICAM-2 TIR domains and a single TICAM-1 TIR domain (SI Text, Fig. S5).
As shown in Fig. 5, the heterotypic interaction sites on TICAM-2 in the top-ranked model involve the acidic surface of the EDD site on the first TICAM-2 chain and the TS site on the second TICAM-2 chain, located on the opposite side (Fig. 5A). Significantly, the TICAM-2 homo-dimer positions the EDD and TS sites next to each other (Fig. 5A), forming a concave surface that can accommodate the TICAM-1 TIR domain. The interaction site on TICAM-1 includes the RK site on the αE-helix and the QI site on the EE-loop, which are located on the top of the wedge (Fig. 5A). In the top-scoring model, all 7 mutations that abrogated TICAM-2/TICAM-1 binding and 30 out of the 31 mutations that did not affect TICAM-2/TICAM-1 binding were recapitulated in the binding energy calculations (Table S4). The TICAM-2 dimer is symmetrically related by a twofold axis along the BB-loop and is maintained by the BB-loop and αC-helix interactions, respectively, consistent with the TLR10 dimer structure (Fig. 5B).
Fig. 5.
Docking model of the TIR domains of TICAM-2 and TICAM-1. (A) The docking structure comprised of TICAM-2 dimer and TICAM-1 monomer. Homotypic and heterotypic interfaces are enclosed by blue and orange dotted lines, respectively. (B) TICAM-2 dimer presenting the binding surface with TICAM-1 TIR. The EDD and TS sites are located at the front surface (also at the back surface) that interact with the wedge of the TICAM-1 TIR (the RK and QI sites). (C) The front view of the interacting surface of TICAM-2 is shown in electrostatic surface potential presentation whereas the wedge of the TICAM-1 is presented in ribbon model. The residues on the RK and QI sites are shown in wire model and labeled.
The top view of the interaction surface between the TICAM-2 dimer and the TICAM-1 monomer is shown in Fig. 5C, where the binding surface of TICAM-2 is represented by the electrostatic surface potential and TICAM-1 by a ribbon model. As shown in the figure, the RK site interacts with the acidic surface of the EDD site, and the QI site with the TS site (Fig. 5C).
Reporter Gene and Binding Assays Using Full-Length TICAM-1 and TICAM-2 Mutants.
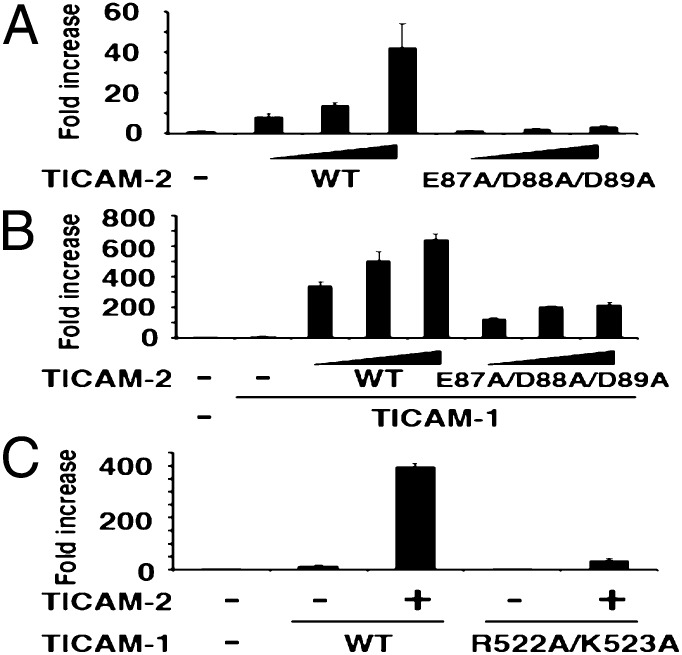
Based on the yeast two-hybrid analysis, we constructed mammalian expression vectors coding the wild type and various mutants of TICAM-2 and TICAM-1 and measured their IFN-β promoter activation abilities by reporter gene assays. Forcedly expressed wild type TICAM-2 activated the IFN-β promoter in HEK293FT cells in a dose-dependent manner (Fig. 6A). In contrast, the TICAM-2 EDD-site mutant E87A/D88A/D89A failed to activate the IFN-β promoter (Fig. 6A). Overexpressed TICAM-2 undergoes homo-dimerization, which in turn recruits TICAM-1, resulting in activation of the IFN-β promoter. Therefore, we next analyzed TICAM-2–TICAM-1-dependent IFN-β promoter activation in the presence of limiting amounts of TICAM-1. A marked enhancement of TICAM-1-mediated IFN-β promoter activation was observed with wild-type TICAM-2, and to a lesser extent with the EDD-site TICAM-2 mutant (Fig. 6B). In the case of TICAM-1, the RK-site mutant (R522A/K523A) only weakly activated the IFN-β promoter (Fig. 6C), consistent with the predicted electrostatic interaction between the acidic surface of the TICAM-2 TIR domain and the basic surface of TICAM-1 observed in the yeast two-hybrid experiments and the docking model. We also measured the NF-κB activation abilities of the EDD-site mutant of TICAM-2 (Fig. S6). Both IFN-β and NF-κB promoter assays demonstrated that the TICAM-2 EDD-site mutant suppressed both signals.
Fig. 6.
Functional assays of TICAM-1 and TICAM-2 mutants in mammalian cells. (A) TICAM-2–dependent IFN-β promoter activation. IFN-β promoter activity was reduced by the E87A/D88A/D89A mutation in TICAM-2. (B) TICAM-2-TICAM-1–dependent IFN-β promoter activation. IFN-β promoter activity was reduced by the E87A/D88A/D89A mutation in TICAM-2. (C) TICAM-2-TICAM-1–dependent IFN-β promoter activation. The mutation of R522/K523 in TICAM-1 reduced IFN-β promoter activation.
To confirm that the TICAM-2 EDD site is involved in the binding of TICAM-1 TIR domain, a coimmunoprecipitation assay was performed in HEK293FT cells. Although the expression level of TICAM-1, the TICAM-2 wild type, and the TICAM-2 EDD mutant in HEK293FT cells was similar (Fig. S7, Lower), the affinity between the TICAM-2 mutant and TICAM-1 was much reduced compared with that between the TICAM-2 wild type and TICAM-1 (Fig. S7, Upper). All of the data suggest that the reduced IFN-β promoter activity of the TICAM-2 EDD mutant was due to its reduced affinity to TICAM-1.
Discussion
The oligomerization properties of TIR domains are closely related to their biological functions. This study presents challenges for structural analysis by NMR. Here, we applied dominant-negative BB-loop mutations to the TICAM-1 and TICAM-2 TIR domains that disrupted the oligomerization. To study the interaction between TICAM-1 and TICAM-2 TIR domains, we used yeast two-hybrid experiments in combination with monomeric structural information. Although the yeast two-hybrid method can produce false positives, we could eliminate such false positives by structural analysis of the BB-loop mutants together with gel-filtration studies of the expressed proteins. Using this approach, we identified two binding regions, the EDD site and the TS site, located on the opposite sides of TICAM-2. Our docking calculations, based on yeast two-hybrid data, revealed a TICAM-2 TIR homo-dimer that assumed a twofold axis of symmetry around the BB-loop, similar to the TLR10 TIR domain structure. Importantly, the two binding regions revealed by the yeast two-hybrid experiments are spatially close in this TICAM-2 homo-dimer structure (Fig. 5A). Heterotypic interaction sites on the TICAM-1 TIR domain could also be elucidated by our approach, which indicated that the RK and QI sites are responsible for interaction with TICAM-2. It is notable that this heterotypic interaction does not require dimer formation of the TICAM-1 TIR domain. In our restraint-driven docked model, the wedge-shaped surface containing the RK and QI sites binds to the concave surface formed by the EDD and TS sites belonging to different TICAM-2 TIR domains (Fig. 5 A and B).
An important conclusion derived from the present study is that the homo-dimerization of the TICAM-2 TIR domains presents a surface that recruits the monomeric moiety of the TICAM-1 TIR domain. In our previous reports, we showed that TLR4 binds the TICAM-2 wild type, but the interaction is disrupted by the BB-loop mutation of TLR4, whereas the BB-loop mutant of TICAM-2 still binds to TLR4 wild type based on the yeast two-hybrid experiments (1). We also showed that the TICAM-1 BB-loop mutant still interacts with the TIR domain of TLR3 wild type (27). Taken together, these results suggest a unique paradigm in which dimer formation of the upstream TIR domain is essential for recruitment of the monomeric moiety of the downstream TIR dimer. Consistent with this paradigm, BB-loop mutants of TLR3 and TLR4 as well as TICAM-1 and TICAM-2 completely abrogate downstream signaling.
It is notable that the myristoylation of TICAM-2 at the N terminus is essential for its localization on the plasma membrane or endosome, and colocalization with TLR4 (32). Interestingly, according to the present TICAM-2 TIR dimer model, the N termini of both TICAM-2 TIR domains are oriented in the same direction, permitting anchoring of the TICAM-2 dimer to the membrane.
A current consensus is that LPS on the surface of Gram-negative bacteria induces clustering of TLR4, leading to formation of the active TLR4 TIR dimer, which triggers activation of the MyD88 and TICAM-1 pathways. Recently, the structure of the Myddosome, a molecular complex mediated by the death domains of MyD88, IRAK4, and IRAK1/2, was determined (33–35). This TICAM-1/2 structural study, on the other hand, would allow us to speculate a model that the TLR4 TIR dimer bridges the TICAM-2 TIR dimer, which further couples with the TICAM-1 TIR, generating an extended signaling network downstream of TLR4. The present work proposes a key for future analysis about an IFN-inducing signaling complex in the context of TLR4-mediated LPS signaling. Recent studies showed the importance of lateral TLR3 clustering mediated by TICAM-1 for downstream signaling (36), consistent with our model.
Our reconstitution study revealed that the TICAM1/2 heterodimer formation is reproducible in HEK293FT cells and that TICAM-2 EDD mutant has less ability to recruit TICAM-1 than wild type (Fig. S7). Reporter activity reflecting IFN induction is accordingly decreased (Fig. 6). Thus, the proposed model is at least right in the formation of the two-adaptor complex. However, herein we only abstracted the complex of TICAM-1 and TICAM-2 from the IFN-inducing axis of LPS signaling of TLR4, which consists of an array of many different molecules. Reconstituting the optimal LPS–IFN signal axis in human TICAM-2 knockout cells will be required to test physiological importance of the TICAM-2 EDD domain.
Materials and Methods
For details, see SI Materials and Methods.
Protein Expression and Purification.
The human TICAM-1 gene encoding the TIR domain (387–545) with mutation of Pro434 to His was cloned into the pET22b (Novagen) vector. The human TICAM-2 gene encoding the TIR domain (75–235) with mutation of Cys117 to His was cloned into the pGEX6p-1(GE healthcare) vector.
NMR Measurements and Structure Calculation.
NMR data for chemical shift assignments for TICAM-1 P434H and TICAM-2 C117H were collected using a suite of triple resonance experiments on Varian UNITY INOVA 600 and 800 spectrometers.
Yeast Two-Hybrid Analysis.
The TIR domains were constructed by direct cloning of two-step PCR products using mutant oligonucleotide primers and subcloned into pGBKT7 and pGADT7 plasmid (Clontech).
Restrained Docking Calculations.
Two docking calculations were made to generate representative TICAM-2 TIR homo-dimer models, from which trimeric TICAM-2/TICAM-1 models were then constructed.
Luciferase Reporter Gene Assay.
Mutants of TICAM-1(1–566) and TICAM-2(1–235) were generated using PCR and subcloned into pEF-BOS vector.
Immunoprecipitation and Immunoblot Analysis.
FLAG-tagged TICAM-2 were immunoprecipitated using anti-FLAG mAb (2.5 µg per sample) and Protein G Sepharose (GE healthcare).
Supplementary Material
Acknowledgments
This research is supported by the Japan Society for the Promotion of Science through its “Funding Program for World-Leading Innovative R&D on Science and Technology.”
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The chemical shift assignments and NOE and dihedral restraint data have been deposited in the BioMagResBank, www.bmrb.wisc.edu [accession nos. 18883 (TICAM-1) and 18882 (TICAM-2)]. The atomic coordinates for the ensemble have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 2mlx (TICAM-1) and 2mlw (TICAM-2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222811110/-/DCSupplemental.
References
- 1.Oshiumi H, et al. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278(50):49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 2.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat Immunol. 2003;4(2):161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4(11):1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto M, Takeda K, Akira S. TIR domain-containing adaptors define the specificity of TLR signaling. Mol Immunol. 2004;40(12):861–868. doi: 10.1016/j.molimm.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Gay NJ, Gangloff M, Weber AN. Toll-like receptors as molecular switches. Nat Rev Immunol. 2006;6(9):693–698. doi: 10.1038/nri1916. [DOI] [PubMed] [Google Scholar]
- 7.Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu Rev Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald KA, et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198(7):1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408(6808):111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 10.Tao X, Xu Y, Zheng Y, Beg AA, Tong L. An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem Biophys Res Commun. 2002;299(2):216–221. doi: 10.1016/s0006-291x(02)02581-0. [DOI] [PubMed] [Google Scholar]
- 11.Khan JA, Brint EK, O’Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J Biol Chem. 2004;279(30):31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- 12.Ohnishi H, et al. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc Natl Acad Sci USA. 2009;106(25):10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valkov E, et al. Crystal structure of Toll-like receptor adaptor MAL/TIRAP reveals the molecular basis for signal transduction and disease protection. Proc Natl Acad Sci USA. 2011;108(36):14879–14884. doi: 10.1073/pnas.1104780108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Lu J, Zhou W, Shen Y. Structural insights into TIR domain specificity of the bridging adaptor Mal in TLR4 signaling. PLoS ONE. 2012;7(4):e34202. doi: 10.1371/journal.pone.0034202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19(1):155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernoux M, et al. Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe. 2011;9(3):200–211. doi: 10.1016/j.chom.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan SL, et al. Molecular mimicry in innate immunity: Crystal structure of a bacterial TIR domain. J Biol Chem. 2009;284(32):21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nyman T, et al. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J Biol Chem. 2008;283(18):11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- 19.Poltorak A, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282(5396):2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 20.Gautam JK, Ashish, Comeau LD, Krueger JK, Smith MF., Jr Structural and functional evidence for the role of the TLR2 DD loop in TLR1/TLR2 heterodimerization and signaling. J Biol Chem. 2006;281(40):30132–30142. doi: 10.1074/jbc.M602057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunne A, Ejdeback M, Ludidi PL, O’Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J Biol Chem. 2003;278(42):41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Zienkiewicz J, Hawiger J. Interactive sites in the MyD88 Toll/interleukin (IL) 1 receptor domain responsible for coupling to the IL1β signaling pathway. J Biol Chem. 2005;280(28):26152–26159. doi: 10.1074/jbc.M503262200. [DOI] [PubMed] [Google Scholar]
- 23.Radons J, et al. The interleukin 1 (IL-1) receptor accessory protein Toll/IL-1 receptor domain: analysis of putative interaction sites in vitro mutagenesis and molecular modeling. J Biol Chem. 2003;278(49):49145–49153. doi: 10.1074/jbc.M306077200. [DOI] [PubMed] [Google Scholar]
- 24.Bovijn C, et al. Identification of interaction sites for dimerization and adapter recruitment in Toll/interleukin-1 receptor (TIR) domain of Toll-like receptor 4. J Biol Chem. 2012;287(6):4088–4098. doi: 10.1074/jbc.M111.282350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Núñez Miguel R, et al. A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signalling adaptor proteins. PLoS ONE. 2007;2(8):e788. doi: 10.1371/journal.pone.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seya T, Oshiumi H, Sasai M, Akazawa T, Matsumoto M. TICAM-1 and TICAM-2: Toll-like receptor adapters that participate in induction of type 1 interferons. Int J Biochem Cell Biol. 2005;37(3):524–529. doi: 10.1016/j.biocel.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain-containing adaptor molecule-1-mediated NF-kappaB and interferon regulatory factor-3 activation. J Biol Chem. 2008;283(26):18283–18291. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16(6):566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 29.Kawabata T. MATRAS: A program for protein 3D structure comparison. Nucleic Acids Res. 2003;31(13):3367–3369. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawabata T, Nishikawa K. Protein structure comparison using the markov transition model of evolution. Proteins. 2000;41(1):108–122. [PubMed] [Google Scholar]
- 31.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: A method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88(21):9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowe DC, et al. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. Proc Natl Acad Sci USA. 2006;103(16):6299–6304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gay NJ, Gangloff M, O’Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011;32(3):104–109. doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Kersse K, Verspurten J, Vanden Berghe T, Vandenabeele P. The death-fold superfamily of homotypic interaction motifs. Trends Biochem Sci. 2011;36(10):541–552. doi: 10.1016/j.tibs.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Luo J, et al. Lateral clustering of TLR3:dsRNA signaling units revealed by TLR3ecd:3Fabs quaternary structure. J Mol Biol. 2012;421(1):112–124. doi: 10.1016/j.jmb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.