Significance
High hydrostatic pressure facilitates the characterization of functionally relevant, but sparsely populated, excited conformational states of proteins by reversibly increasing their equilibrium population. Here, high-pressure instrumentation for circular dichroism and developments in high-pressure site-directed spin-labeling EPR are reported, and a combination of EPR and circular dichroism is used to map pressure-populated structural changes in various states of myoglobin. The data reveal that the high-pressure molten globule (MG) of apomyoglobin at neutral pH retains native-like helical content despite a fluctuating tertiary fold, an MG state of holomyoglobin populated at low pH and high-pressure retains ligand-binding capacity, and a transient folding intermediate of apomyoglobin is populated under similar conditions at equilibrium.
Abstract
Excited states of proteins may play important roles in function, yet are difficult to study spectroscopically because of their sparse population. High hydrostatic pressure increases the equilibrium population of excited states, enabling their characterization [Akasaka K (2003) Biochemistry 42:10875–85]. High-pressure site-directed spin-labeling EPR (SDSL-EPR) was developed recently to map the site-specific structure and dynamics of excited states populated by pressure. To monitor global secondary structure content by circular dichroism (CD) at high pressure, a modified optical cell using a custom MgF2 window with a reduced aperture is introduced. Here, a combination of SDSL-EPR and CD is used to map reversible structural transitions in holomyoglobin and apomyoglobin (apoMb) as a function of applied pressure up to 2 kbar. CD shows that the high-pressure excited state of apoMb at pH 6 has helical content identical to that of native apoMb, but reversible changes reflecting the appearance of a conformational ensemble are observed by SDSL-EPR, suggesting a helical topology that fluctuates slowly on the EPR time scale. Although the high-pressure state of apoMb at pH 6 has been referred to as a molten globule, the data presented here reveal significant differences from the well-characterized pH 4.1 molten globule of apoMb. Pressure-populated states of both holomyoglobin and apoMb at pH 4.1 have significantly less helical structure, and for the latter, that may correspond to a transient folding intermediate.
Proteins in solution are dynamic molecules, exhibiting conformational flexibility across a range of time and length scales (1). In addition to a well-ordered native state, conformational excursions to low-lying “excited states” may be required in protein function (2). For example, on a funnel-shaped energy landscape (3, 4), excited states have increased configurational entropy that may give rise to the promiscuous protein–protein interactions that define a protein interactome (5, 6). Structural changes involved in formation of the excited state may take a variety of forms, from rigid body motions of helices (7) to local unfolding of secondary structural elements (8). Despite their functional relevance, excited states are sparsely populated and may escape detection by standard spectroscopic techniques. Hydrostatic pressure apparently offers a solution to this problem by reversibly populating excited states of proteins, allowing for spectroscopic characterization (9–14). Pressure application is particularly useful because it shifts the relative population of preexisting conformational states while minimally affecting the conformational landscape itself (15).
Assuming that pressure can populate excited states for study, the increased configurational entropy of such states presents a challenge to spectroscopic methods that aim to describe structure; depending on the intrinsic time scale of the method, either a population-weighted average structure or a heterogeneous ensemble is observed. The intrinsic time scale of continuous-wave EPR spectroscopy (0.1–100 ns) is fast compared with protein conformational fluctuations (typically microseconds to milliseconds), and site-directed spin-labeling EPR (SDSL-EPR) may provide a snapshot of a conformational equilibrium frozen in time (16–18). In the most common implementation of SDSL, a nitroxide side chain (designated R1) is attached to a site-specifically introduced cysteine using a methanethiosulfonate reagent (19), although other side chains and strategies are possible (20). The EPR spectrum of R1 in a protein encodes information on local structure (21–23), topology (24), and picosecond-to-nanosecond backbone dynamics (18, 25), and thereby may serve to identify conformational heterogeneity in equilibrium states (18), including those populated by pressure. Recently, an EPR system capable of pressures up to 4 kbar and optimized for SDSL was reported, making it possible to explore the structure and dynamics of high-pressure states of proteins using SDSL-EPR (26). An improved system with a programmable pressure intensifier and ceramic sample cell is introduced here.
Although SDSL-EPR provides site-specific information on tertiary contact sites and topology in a protein, it does not provide the global information on secondary structure that is needed to reliably interpret SDSL-EPR data in terms of conformational changes due to pressure application. Far-UV circular dichroism (CD) spectroscopy may provide the required information on secondary structure (27, 28), but high-pressure CD spectroscopy has not been developed because of the lack of a viable window material for a high-pressure sample cell (29); materials used in other optical spectroscopies for high-pressure experiments (quartz and sapphire) are unsuitable for use with circularly polarized light (Supporting Information). As a result, CD studies have been limited to low pressures (0.2 kbar) (30), or for measuring pressure-populated changes after pressure has been released (31–36). Here, we introduce a high-pressure optical cell for CD studies up to 2.4 kbar, using a custom MgF2 window with a reduced aperture. Design and evaluation of the modified optical cell are presented in Supporting Information. This development sets the stage for combined application of SDSL-EPR and CD to explore the structure of pressure-populated excited states of proteins.
An example of an excited protein state is the molten globule (MG), a compact but dynamic state with native-like secondary structure but without the close side-chain packing in the core that is typical of the native state (37). MGs typically are stabilized at low pH or in the presence of denaturants and are believed to represent intermediates in protein folding (38, 39) and, in some cases, may mediate protein function (40). The MG of apomyoglobin (apoMb) stabilized at pH 4.1 is an ideal subject for exploring the structure and dynamics of such states because it can be populated under conditions suitable for study by high-resolution NMR (41). At pH 4.1, NMR 13Cα chemical shift data showed that the native A, G, and H helices are largely retained in the MG state, but reduced values of the chemical shift relative to the native state of apoMb suggest that nonhelical states are sampled. Moreover, heteronuclear NOE data reveal gradients of fast backbone fluctuations in these helices not observed in native apoMb (42). Relaxation dispersion NMR showed the MG to be in pH-dependent equilibrium with other partially folded forms, indicating that the MG state also will be present in an equilibrium mixture at neutral pH, although with a small population relative to the native structure (i.e., an excited state) (39). Furthermore, this equilibrium MG was found to be structurally similar to the second of two intermediates identified during kinetic refolding by ultrafast hydrogen/deuterium (H/D) exchange coupled with 2D NMR (43).
Application of pressure is expected to increase the equilibrium population of an MG state at neutral pH to levels that can be observed spectroscopically (15). Indeed, a state of apoMb, populated at 2 kbar, pH 6, was designated as an MG based on the similarity of its intrinsic fluorescence properties with that of the pH 4.1 state of apoMb (44), although it does not bind the dye 1-anilino-8-naphthalenesulfonic acid, as is typical of MG states in general (45, 46). This pressure-populated state has been examined with high-resolution NMR (47, 48), in which the integral intensities of most cross-peaks in the 1H-15N heteronuclear single quantum coherence (HSQC) spectrum disappear as a result of broadening, suggesting that the entire polypeptide chain is transformed into a heterogeneously disordered conformation fluctuating on the millisecond time scale (48). Based on this behavior, the high-pressure state also was designated as an MG, but the lack of spectral information precluded any information regarding details of the structures present. Clearly, this high-pressure MG state differs in detail from the pH 4.1 MG, for which high-resolution information was obtained (42). Limited proteolysis and H/D exchange experiments in the same pressure range also reported increased flexibility, particularly in the B and E helices (49), and fluorescence and absorption spectroscopy revealed pressure-dependent changes in both apoMb and holomyoglobin (holoMb) at pH 4.1, but again without specific information regarding the structures present (44–46, 50).
Thus, there is little information on the secondary or tertiary structure of the pressure-populated MG and how it compares with the well-characterized pH 4.1 MG. In the present study, high-pressure CD and SDSL-EPR are used to explore the secondary structure and tertiary fold of high-pressure states of both apo- and holoMb. Results show that unlike the pH 4.1 MG of apoMb, the high-pressure state formed at pH 6 at 2 kbar retains the full secondary structure content of native apoMb, despite a heterogeneous tertiary fold revealed by SDSL-EPR. At pH 4.1, an MG-like state of holoMb is reversibly populated at 2 kbar but with a reduced helical content. Finally, at pH 4.1 and 2 kbar, a state of apoMb is formed that retains a significant population of secondary structure and may correspond to a transient folding intermediate. In each case, EPR provides site-specific information regarding the location and local dynamics associated with the transition to these states.
Results
At pH 6.0 and atmospheric pressure, apoMb retains a structure largely similar to the holo protein, except for local unfolding in the region of helix F that caps the heme pocket (51, 52); at pH 4.1, the well-characterized MG state is populated (42). In contrast, holoMb retains the WT structure under the same conditions (53). In the present study, far-UV CD spectra were acquired on holoMb and apoMb at pH 6.0 and pH 4.1 in the pressure range 0–2.4 kbar (Figs. 1 and 2). These data were analyzed to determine the changes in secondary structural content as a function of pressure in each state (Table 1), and the apparent change in partial molar volume (Δ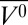 ) and free energy (ΔG0) for transitions observed in the low pH states (Figs. 1 and 2, Insets). UV-visible (UV-Vis) spectra were acquired on holoMb at pH 6.0 and pH 4.1 to monitor the Soret band as an indicator of structural changes in the heme-binding pocket in the pressure range 0–2 kbar (Fig. S1). The reversibility of all pressure-populated changes in CD and UV-Vis spectra was verified by acquiring spectra at 0 bar after maximum pressurization; these were universally superimposable upon prepressurization spectra taken at 0 bar and, for clarity of presentation, are not included.
) and free energy (ΔG0) for transitions observed in the low pH states (Figs. 1 and 2, Insets). UV-visible (UV-Vis) spectra were acquired on holoMb at pH 6.0 and pH 4.1 to monitor the Soret band as an indicator of structural changes in the heme-binding pocket in the pressure range 0–2 kbar (Fig. S1). The reversibility of all pressure-populated changes in CD and UV-Vis spectra was verified by acquiring spectra at 0 bar after maximum pressurization; these were universally superimposable upon prepressurization spectra taken at 0 bar and, for clarity of presentation, are not included.
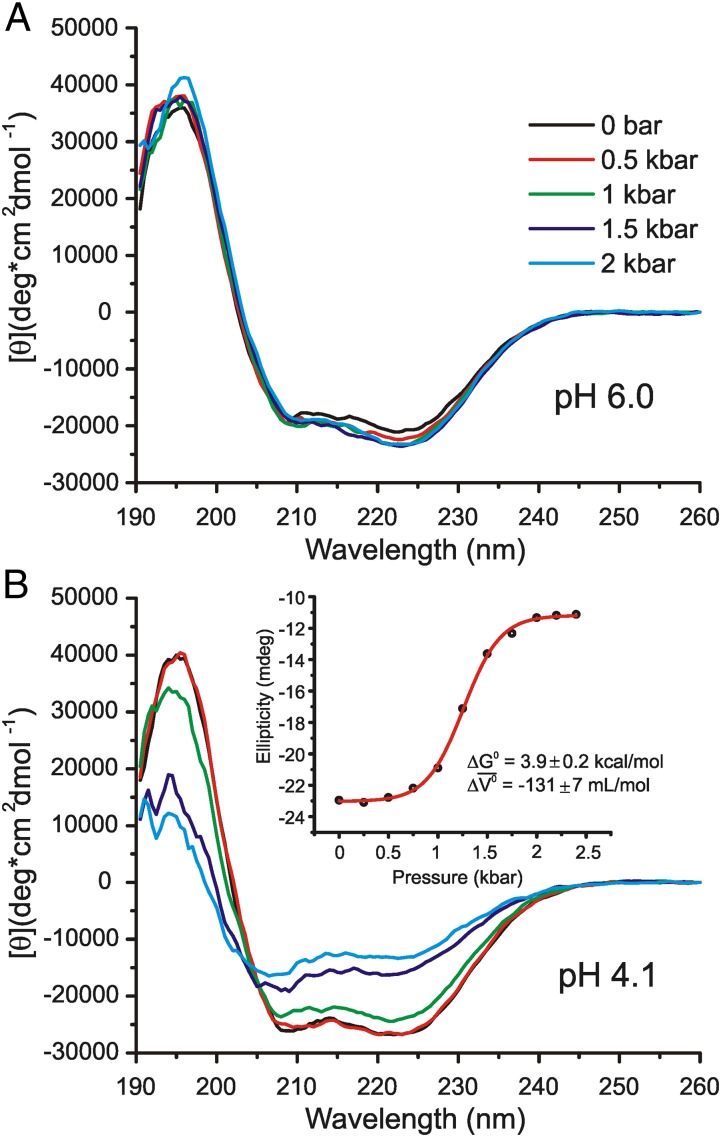
Fig. 1.
Variable-pressure CD spectra of holoMb. Each spectrum is the average of nine scans. (A) Far-UV CD spectra of 14.1 μM holoMb in 5 mM MES at pH 6.0, without salts, from 0 to 2 kbar in 0.5-kbar increments. (B) Far-UV CD spectra of 11.2 μM holoMb in 5 mM sodium acetate at pH 4.1, without salts. (Inset) Plot of ellipticity vs. pressure and the best fit to a two-state model (red trace) to give the indicated ΔG0 and Δ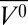 of transition (Methods).
of transition (Methods).
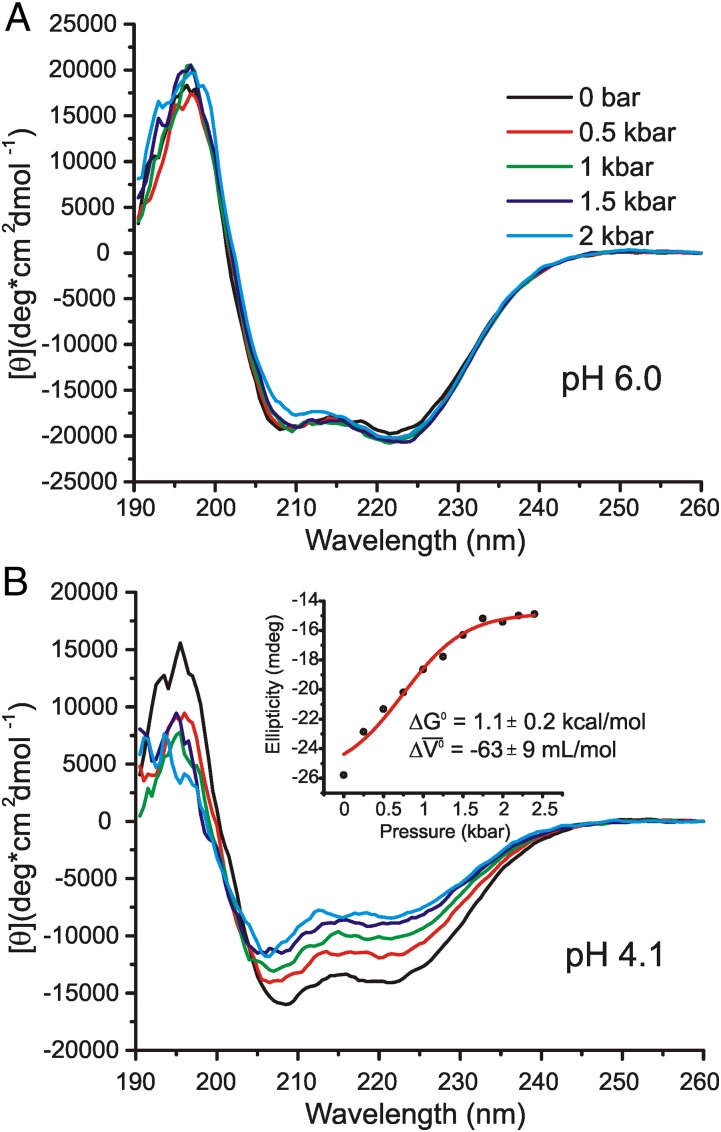
Fig. 2.
Variable-pressure CD spectra of apoMb. Each spectrum is the average of nine scans. (A) Far-UV CD spectra of 24 μM apoMb in 5 mM MES at pH 6.0, without salts, from 0 to 2 kbar in 0.5-kbar increments. (B) Far-UV CD spectra of 24 μM apoMb in 5 mM MES at pH 4.1, without salts. (Inset) Plot of ellipticity vs. pressure and the best fit (red trace) to a two-state model of the transition to give the indicated ΔG0 and Δ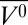 (Methods).
(Methods).
Table 1.
Pressure-dependent secondary structure content estimation from far-UV CD
| Protein | pH | Pressure* | α-Helix† | β-Sheet | Turn | Unordered |
| HoloMb | 6.0 | 0 | 0.67 | 0.02 | 0.13 | 0.18 |
| 2 | 0.67 | 0.02 | 0.10 | 0.21 | ||
| 2.4 | 0.68 | 0.01 | 0.11 | 0.20 | ||
| 4.1 | 0 | 0.72 | 0.01 | 0.10 | 0.17 | |
| 2 | 0.43 | 0.05 | 0.22 | 0.30 | ||
| 2.4 | 0.42 | 0.06 | 0.22 | 0.30 | ||
| ApoMb | 6.0 | 0 | 0.57 | 0.02 | 0.20 | 0.21 |
| 2 | 0.56 | 0.02 | 0.19 | 0.23 | ||
| 2.4 | 0.56 | 0.02 | 0.19 | 0.23 | ||
| 4.1 | 0 | 0.48 | 0.08 | 0.22 | 0.22 | |
| 2 | 0.28 | 0.19 | 0.22 | 0.31 | ||
| 2.4 | 0.26 | 0.23 | 0.22 | 0.29 |
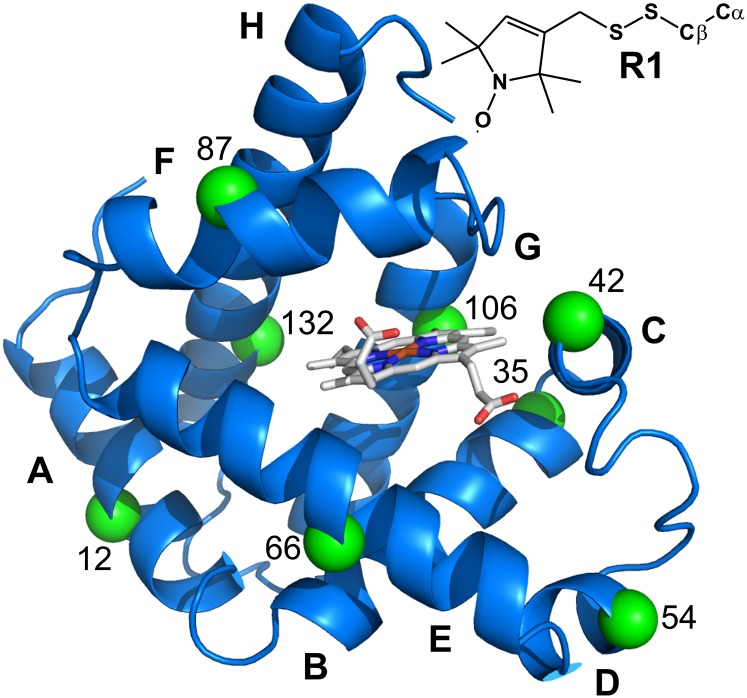
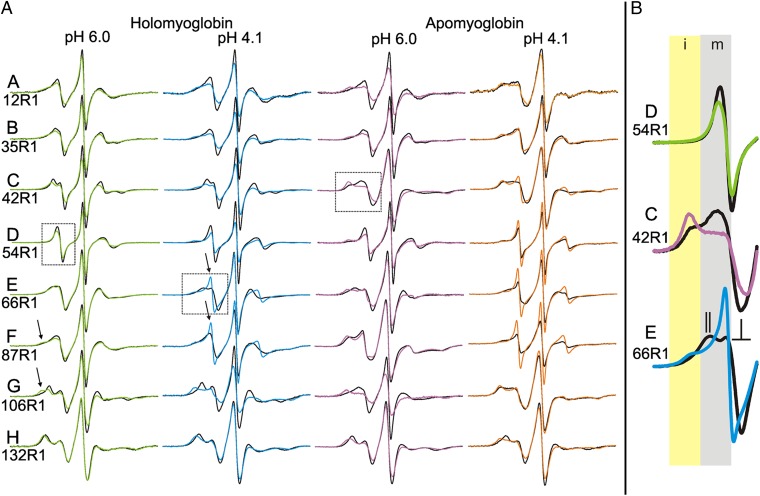
To monitor site-specific changes in structure and dynamics due to pressure in holoMb and apoMb, a single R1 residue was introduced in each of the eight native helices, one at a time, at the sites shown in Fig. 3, and the EPR spectra were recorded for each state investigated at 0 and 2 kbar (Fig. 4A). Although this is a sparse sampling of the structure, the data serve to illustrate the capability of the combined CD/SDSL approach and to identify salient features of each of the myoglobin states under pressure; a more extensive SDSL-EPR dataset as a function of pressure will be published elsewhere. Atmospheric-pressure EPR spectra for each of these mutants were reported previously for holoMb and apoMb at pH 6.0 and for apoMb at pH 4.1 (18); these spectra are reproduced here for comparative purposes. The atmospheric-pressure EPR spectra of holoMb at pH 4.1 are not previously reported, as are the high-pressure EPR spectra for all states. As for the CD and UV-Vis data, EPR spectral changes at high pressure are reversible (Fig. S2), ensuring thermodynamic equilibrium between the conformational states observed over the pressure range investigated.
Fig. 3.
Ribbon model of the holoMb structure (Protein Data Bank ID code 2MBW) (79). The green spheres at the Cα positions identify the residues where the R1 side chain was introduced, one at a time, for EPR experiments. The heme group is shown in stick representation. (Inset) Structure of R1.
Fig. 4.
EPR spectra of R1 in the various states of myoglobin as a function of pressure. (A) EPR spectra at 0 bar (black trace) and 2 kbar (colored trace) are shown for R1 at the indicated sites and states of myoglobin. The arrows highlight examples of spectral components corresponding to immobilization of the nitroxide in the holoMb pH 6.0 state, and to rapid isotropic motion of the nitroxide in the holo pH 4.1 state. (B) The different classes of pressure-dependent spectral change are illustrated using enlargements of the low-field regions of the spectra identified by the dotted boxes. Shaded areas indicate the portions of the spectra that display intensity corresponding to mobile (m, gray) and immobile (i, yellow) motional states of the nitroxide. In response to an increase in pressure, the spectrum of residue 54R1 (Top) exhibits only subtle changes in the single spectral component; the spectrum of 42R1 (Middle) exhibits a shift in the population toward the component corresponding to an immobilized state, whereas that for 66R1 (Bottom) exhibits a shift toward a spectral component representing rapid isotropic motion of the nitroxide. The 0-bar spectrum of 66R1 (black trace) illustrates the characteristic lineshape for a nitroxide undergoing rapid anisotropic motion, where parallel and perpendicular hyperfine components are resolved (25), as indicated. This feature may be recognized in several of the spectra for holoMb in column 1 in A. The component corresponding to the more mobile state of 54R1 and 42R1 (black traces) also reflects anisotropic motion, but the order is too weak to resolve the parallel and perpendicular components.
HoloMb at pH 6.0 Is Rigid and Relatively Pressure Insensitive to 2 kbar.
Analysis of the far-UV CD spectrum of holoMb (Fig. 1A) using the Contin algorithm (27) estimates 67% helical content at 0 bar (gauge pressure; equivalent to atmospheric pressure), consistent with previously reported CD measurements of holoMb under similar conditions (54). Upon pressurization, there is little change in the fractional helicity, with a final value of 68% at 2.4 kbar. The Soret band intensity remains constant from 0 to 2 kbar, with only a small shift in the wavelength of maximum intensity, from 408 to 409 nm (Fig. S1), suggesting little change in the structure surrounding the heme-binding pocket.
Most of the EPR spectra for holoMb at 0 bar (black traces, column 1, Fig. 4A) have a single component reflecting a weakly anisotropic motion of the nitroxide, typical of noninteracting helix surface residues (55). The lineshape variation among these spectra reflects, in part, residue-to-residue differences in the nanosecond time scale dynamics of the backbone (18). Exceptions are residues 87R1 and 106R1, which have two resolved components, one of which corresponds to a small population of an immobilized state of the nitroxide (arrows, column 1, Fig. 4A). In the case of 87R1, it was shown previously that the origin of the immobile state was flexibility of the F helix, in which the residue is located, where some conformations allow contact of the nitroxide with the nearby protein environment (18). Flexibility in the adjacent C helix (18, 56) similarly may account for the immobile state in residue 106R1, although this remains to be established.
Upon pressurization, two classes of behavior are observed. In most residues, there are only small lineshape changes arising from a decrease in the nitroxide mobility; this is evidenced by the reduction in intensity of the central line (compare black and green traces, column 1, Fig. 4A). A similar effect was observed for R1 at sites in T4 lysozyme under pressure and was interpreted in terms of an activation volume for rotational diffusion of the nitroxide (26). The second behavior exhibited is a shift in the relative population of spectral components toward the more immobile component, illustrated in Fig. 4B (center panel), and observed on a smaller scale for 87R1 and 106R1. This pressure-dependent shift in population for 87R1 and 106R1 presumably is a reflection of local compressibility of the protein. For 87R1, this interpretation is consistent with crystallographic and near-IR/Raman spectroscopic studies of holoMb at high pressure, which revealed flexibility in helix F (57, 58).
ApoMb at pH 6.0 Retains the Full Complement of Helical Structure to 2 kbar but Adopts a Heterogeneous Tertiary Fold.
ApoMb is 57% helical at 0 bar, according to the far-UV CD spectrum. This is ∼10% less than that for holoMb at pH 6.0 and 0 bar (Table 1), in agreement with previously published values from CD (52, 54). 13Cα and 1H-15N HSQC NMR has shown this loss is the result of conformational exchange involving nonhelical states, primarily in the F helix, the N-terminal end of the G helix, and the C-terminal end of helix H (51). Upon pressurization of apoMb, there is very little change in the far-UV CD spectrum (Fig. 2). Remarkably, at 2 kbar, where apoMb is known to form an MG-like state, the total helicity is 56%, essentially the same as that at 0 bar.
EPR spectra of spin-labeled residues 12R1, 54R1, and 132R1 in apoMb at pH 6.0 and 0 bar are identical to their holoMb counterparts, indicating that the structure and backbone dynamics are unchanged at these locations upon removal of the heme group (compare black traces, columns 1 and 3, Fig. 4A). The spectrum of 66R1 is single component, and the change in the lineshape between the holo- and apoMb states reflects differences in the local dynamics of helix E. On the other hand, residues 35R1, 42R1, 87R1, and 106R1 have well-resolved two-component spectra. For residues 42R1, 87R1, and 106R1, the two-component spectra previously were shown to arise from conformational exchange involving helices C, F, and G, respectively; conformational exchange also was identified at the N-terminus of E and the C-terminal of H (18).
Pressurization to 2 kbar, at which the putative MG state essentially is completely populated (44, 48), results in well-resolved spectral changes in most residues, unlike the holoMb samples at pH 6.0 (Fig. 4A). For 54R1 and 66R1 in the D and E helices, respectively, the single-component spectra at 0 bar give rise to two-component spectra signaling the presence of alternative conformations of the protein; one component closely resembles that of the 0-bar state, and the other reflects immobilization due to tertiary contact with the protein. For 12R1, 35R1, 42R1, and 106R1 in the A, B, C, and G helices, respectively, pressure induces a strong shift toward a more immobile state. For 87R1 and 132R1, the changes are relatively small and indicate a general decrease in mobility, as would be expected from local compression. The two-component EPR spectra found throughout the protein at 2 kbar are consistent with widespread conformational exchange, slow compared with the intrinsic time scale of EPR, involving at least two states (Discussion). Based on the CD data, the conformational substates must differ in the relative positions of helical segments.
HoloMb Reversibly Loses Helicity upon Pressurization at pH 4.1.
HoloMb has 72% helical content at 0 bar (Table 1), and the Soret band intensity is similar to that at pH 6.0 and atmospheric pressure (Fig. S1). This agrees with previously reported data and indicates a native-like conformation at pH 4.1 and 0 bar (50, 53, 59). The helical content drops as a function of pressure, beginning at 0.5 kbar and leveling off at 43% at 2 kbar. The Soret band intensity showed significant pressure dependence as well, in agreement with previous high-pressure UV-Vis absorbance measurements (50). The ellipticity at 222 nm was used to monitor the fractional helicity during the pressure-populated transition observed in this state. The sigmoidal character of the pressure dependence of the ellipticity is well-fit using a two-state model to give a ΔG0 and Δ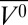 of 3.9 ± 0.2 kcal/mol and −131 ± 7 mL/mol, respectively, for the pressure-populated transition (Fig. 1B).
of 3.9 ± 0.2 kcal/mol and −131 ± 7 mL/mol, respectively, for the pressure-populated transition (Fig. 1B).
At 0 bar, all holoMb EPR spectra at pH 4.1 are similar to their pH 6.0 counterparts, indicating that the backbone dynamics are largely unchanged, and no new spectral components are detected (compare black traces, columns 1 and 2, Fig. 4A). Specifically, residues 12R1, 42R1, 54R1, 66R1, 106R1, and 132R1 have EPR spectra that are nearly superimposable on the pH 6.0 spectra, whereas 35R1 contains an additional small population of a component corresponding to an immobilized state of the nitroxide. Both 35R1 and 87R1 exhibit slight sharpening of the mobile spectral components, indicating a small increase in the rate of motion at these sites. These results are consistent with the CD and Soret band data at 0 bar, which indicate the protein has a similar global secondary structure composition and local tertiary structure in the heme pocket, respectively.
Pressure application causes changes in the EPR spectra of most residues, unlike the case for the pH 6.0 state. For 12R1, 35R1, 42R1, 54R1, and 106R1, the spectra transition from single to two component at 2 kbar, consistent with the presence of alternative conformations involving helices A, B, C, D, and G. For 132R1, the small lineshape change with pressure results from changes in nitroxide mobility, and is similar to that observed for most residues at pH 6.0; such small changes indicate rigidity (incompressibility) of the protein at the corresponding sites. The most interesting responses are for 66R1 and 87R1, where pressure produces a well-resolved component with a sharp resonance line corresponding to a rapid isotropic motion of the nitroxide (arrows, column 2, Fig. 4A and shown in expanded view at the bottom of Fig. 4B). Such lineshapes are characteristic of dynamically disordered states (18, 60, 61), suggesting that the loss of helicity detected by CD under pressure may be attributed in part to regions involving helices E and F. This is in contrast to apoMb at pH 4.1, 2 kbar (see below), where sharp isotropic resonance lines are observed for residues 42R1 and 54R1 as well, indicating loss of helical content in the C and D helices (compare colored spectra in columns 2 and 4, Fig. 4A).
The High-Pressure “Unfolded” State of apoMb at pH 4.1 Retains Significant Secondary Structure.
At pH 4.1 and 0 bar, apoMb forms an MG state that is 48% helical according to the far-UV CD spectrum (Table 1). In relation to native apoMb and holoMb at pH 6.0, this is roughly a 10% and 20% reduction of helicity, respectively. A combination of 13Cα, 1H-15N HSQC, {1H}-15N heteronuclear NOE, and H/D exchange NMR experiments have shown that the “core” helices A, G, and H remain intact in the pH 4.1 MG state, whereas helices C, D, E, and F are partially or completely unfolded; the N-terminal half of helix B is largely disordered, whereas the C-terminal half is helical (42, 43, 52).
Pressurization of apoMb to 2 kbar at pH 4.1 reduces the helical content from 48% to 28%, at which point the drop in helicity levels off, for a final helical content of 26% at 2.4 kbar. Additionally, there is an increase in β-sheet population to ∼23% at 2.4 kbar, although the relative uncertainty in β-sheet estimation is higher than that for helical content. The ellipticity at 222 nm was used to obtain a ΔG0 of 1.1 ± 0.2 kcal/mol and Δ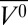 of −63 ± 9 mL/mol for the transition (Fig. 2B), in good agreement with earlier tryptophan fluorescence measurements (44). The pressure dependence of ellipticity at low pressures (0–0.75 kbar) is nonsigmoidal, indicating that a mixture of states is present at 0 bar, perhaps including a small population of a partially folded state distinct from the MG state known to be predominant at pH 4.1 and 0 bar (39, 42). However, the MG state is approximated to be fully populated at 0 bar for the purpose of fitting the data, as described in Methods. As a result, the estimates for ΔG0 and Δ
of −63 ± 9 mL/mol for the transition (Fig. 2B), in good agreement with earlier tryptophan fluorescence measurements (44). The pressure dependence of ellipticity at low pressures (0–0.75 kbar) is nonsigmoidal, indicating that a mixture of states is present at 0 bar, perhaps including a small population of a partially folded state distinct from the MG state known to be predominant at pH 4.1 and 0 bar (39, 42). However, the MG state is approximated to be fully populated at 0 bar for the purpose of fitting the data, as described in Methods. As a result, the estimates for ΔG0 and Δ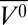 are a rough approximation and provide an upper limit for the transition.
are a rough approximation and provide an upper limit for the transition.
All spin-labeled residues in the pH 4.1 MG state of apoMb exhibit two-component EPR spectra at 0 bar (Fig. 4A), consistent with the widespread conformational exchange known to exist in this state. Detailed interpretations of the 0 bar spectra in terms of structure and dynamics recently were published (18). Briefly, the sharp spectral components of residues 54R1, 66R1, and 87R1 reflect essentially isotropic motion of the nitroxide, consistent with dynamically disordered states in the D, E, and F helices, respectively. Residues 12R1, 35R1, 42R1, 106R1, and 132R1 have two-component spectra consistent with conformational exchange in a partially folded state. Upon pressurization, the dominant spectral change observed is an increase in the relative population of sharp spectral components reflecting dynamically disordered conformational states (column 4, Fig. 4A). This is most prominent in residues 54R1, 66R1, and 87R1, which already exhibit isotropic mobile components at 0 bar; a small population of a component corresponding to high mobility appears at high pressure for 42R1. Residues 12R1, 35R1, 106R1, and 132R1 show only minor changes in mobility due to pressure. Based on these results, the loss of helicity detected by CD apparently has significant contributions from unfolding in the helical sequences around residues 54R1, 66R1, and 87R1 in helices D, E, and F.
Horse Heart Myoglobin Is Less Stable than Sperm Whale Myoglobin.
All the CD and EPR results presented above are for sperm whale myoglobin. Identical high-pressure CD experiments were performed on horse heart myoglobin in the same four states, and the results are qualitatively similar to those for sperm whale myoglobin (Table S1). In agreement with previous comparisons of these two variants (62, 63), horse heart myoglobin is slightly less stable. This is particularly apparent in horse heart apoMb at pH 6.0, in which there is a 17% drop in helicity upon pressurization to 2 kbar (from 60% to 43%; Table S1), compared with almost no change for sperm whale myoglobin under the same conditions (Table 1). SDSL-EPR studies were not done on the horse heart protein.
Discussion
One aim of this study was methodology development, i.e., to use myoglobin as a well-studied model system to provide an empirical foundation for interpretation of high-pressure SDSL-EPR and CD data in terms of structure and dynamics. To the extent they can be compared, the data obtained in this study are consistent with existing information from other established methods (44–48, 50, 64), an outcome that provides some level of validation for the approach. Additionally, because the time scale of the SDSL-EPR experiment provides an instantaneous sampling of an ensemble of structures fluctuating in the time scale of microseconds to milliseconds, the data offer a unique view of the conformationally flexible states populated by pressure, the highlights of which are summarized below.
The Pressure Response of holo- and apoMb at pH 6.0: The High-Pressure MG State of apoMb.
HoloMb at pH 6.0 was explored as a well-ordered reference state. At neutral pH and atmospheric pressure in solution, the protein is well ordered and devoid of high-amplitude conformational fluctuations (42, 65). The UV-Vis (Soret band), CD, and EPR data generally are consistent with this picture of a rigid, relatively incompressible structure at pH 6.0 in the pressure range investigated. However, the EPR spectrum of residue 87R1 in the F helix shows the appearance of an immobile component at 2 kbar, reflecting the onset of localized conformational exchange. This result is consistent with previous studies that identified rearrangements in the heme group upon pressurization that are transmitted to the F helix through the coordinating His93 (57, 58). In addition, the pressure sensitivity of 106R1 in helix G reveals a small and highly localized compressibility that may occur in an otherwise rigid structure.
In contrast to the relatively pressure-insensitive holoMb, apoMb at pH 6.0 undergoes a transition to a pressure-populated MG state around 1.5 kbar with a Δ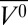 ∼−70 mL/mol detected by both tryptophan fluorescence (44) and NMR (48). In principle, NMR can provide atomic level information on the pressure-populated MG structure, but the loss of nearly all cross-peak intensity in the HSQC spectrum was observed at 2 kbar, with less than 20% of the atmospheric pressure signal intensity remaining at random coil positions. The significant loss of signal intensity due to line broadening allowed only the conclusion that the entire polypeptide chain had lost the native fold and was disordered to form a heterogeneous state exchanging just slowly enough to cause the observed broadening (48). The EPR spectra of apoMb at 2 kbar clearly reveal the heterogeneity via multicomponent EPR spectra, one component of which is very similar to apoMb at atmospheric pressure. Taken alone, the EPR data do not define the structures present, except that the heterogeneous population includes a conformation much like native apoMb. However, together with CD data that show no change in secondary structure in the MG relative to native apoMb, and the NMR data that reveal millisecond exchange between populations, a working model can be constructed. With the assumption that a defined thermodynamic state is present at 2 kbar, as concluded from the intrinsic fluorescence and NMR studies (44, 48), we propose that the high-pressure MG state consists of a native-like population with the helical segments fluctuating on the millisecond time scale. In some positions of the helices, the R1 side chain makes contact with neighboring structures, giving rise to the immobile component. The resolution of X-band EPR to discern different motions in the slow-motion regime is not high enough to conclude that the immobile state is a single dynamic mode of the nitroxide, so the immobile state itself might be heterogeneous and represent an ensemble. In future studies, this might be resolved by high-field EPR (66).
∼−70 mL/mol detected by both tryptophan fluorescence (44) and NMR (48). In principle, NMR can provide atomic level information on the pressure-populated MG structure, but the loss of nearly all cross-peak intensity in the HSQC spectrum was observed at 2 kbar, with less than 20% of the atmospheric pressure signal intensity remaining at random coil positions. The significant loss of signal intensity due to line broadening allowed only the conclusion that the entire polypeptide chain had lost the native fold and was disordered to form a heterogeneous state exchanging just slowly enough to cause the observed broadening (48). The EPR spectra of apoMb at 2 kbar clearly reveal the heterogeneity via multicomponent EPR spectra, one component of which is very similar to apoMb at atmospheric pressure. Taken alone, the EPR data do not define the structures present, except that the heterogeneous population includes a conformation much like native apoMb. However, together with CD data that show no change in secondary structure in the MG relative to native apoMb, and the NMR data that reveal millisecond exchange between populations, a working model can be constructed. With the assumption that a defined thermodynamic state is present at 2 kbar, as concluded from the intrinsic fluorescence and NMR studies (44, 48), we propose that the high-pressure MG state consists of a native-like population with the helical segments fluctuating on the millisecond time scale. In some positions of the helices, the R1 side chain makes contact with neighboring structures, giving rise to the immobile component. The resolution of X-band EPR to discern different motions in the slow-motion regime is not high enough to conclude that the immobile state is a single dynamic mode of the nitroxide, so the immobile state itself might be heterogeneous and represent an ensemble. In future studies, this might be resolved by high-field EPR (66).
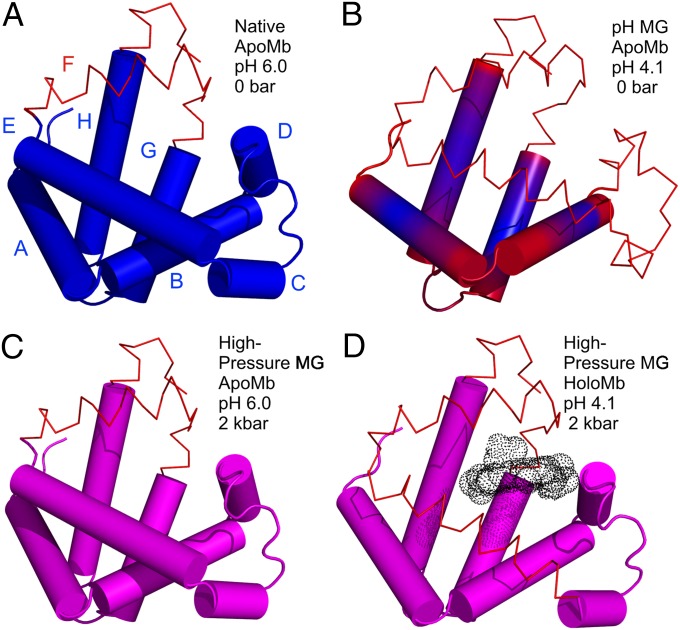
It is interesting to compare the high-pressure MG of apoMb with that produced at pH 4.1, atmospheric pressure. The HSQC spectrum of the pH-populated MG did not show the extensive cross-peak broadening observed at 2 kbar, pH 6.0; the difference was attributed to a presumed sensitivity of the MG state to details of experimental conditions, which included a difference in temperature (48). The results from the current work suggest that the discrepancies in the NMR spectra may instead be the result of structural and dynamical differences between the two states. In the present study, the MG states were compared with respect to helical content and EPR spectral signatures at the same temperature. The data reveal a difference in helical content of ∼10%, lower in the pH 4.1 MG, and the presence of sharp isotropic components in the spectra of R1 in helices D, E, and F (18) that are not observed in the pressure-populated MG state. Together, these results suggest local unfolding in these helices in the pH-populated MG but not in the pressure-populated MG. A working model summarizing the main features of the high-pressure MG in comparison with the pH 4.1 MG and native apoMb is shown in Fig. 5.
Fig. 5.
Models for the structure and dynamics of partially folded states of myoglobin. Each model is based on the crystal structure of holoMb; sequences that retain helical content are shown as cylinders; segments that have some fraction unfolded are shown in wire representation. (A and B) Models of native apoMb and the pH 4.1 MG state, respectively, based on solution NMR (42, 51). Regions undergoing conformational exchange are colored red, and more rigid regions are colored blue. The structure of native apoMb is similar to that of holoMb, but with localized unfolding in the F helix and portions of the G and H helices. In B, the conformational exchange implied by the gradient of color in the helices represents a gradient in the population of nonhelical states, increasing toward the helix termini, as inferred from 13Cα chemical shifts (42). (C and D) Models of the high-pressure MG of apoMb and holoMb, based on SDSL, CD, and NMR. Helices with a fluctuating tertiary structure are colored magenta. (C) The pressure-populated MG of apoMb at pH 6.0, 2 kbar, contains the full complement of native-state secondary structure according to CD, but line broadening in NMR and the spectral shifts in EPR indicate a fluctuating tertiary fold. (D) The pressure-populated MG of holoMb at pH 4.1 and 2 kbar experiences dynamic disorder in some fraction of the E and F helices, and a fluctuating tertiary fold in the remaining helices.
Specific details of the pressure-dependent changes in apoMb warrant comment. At first glance, the lack of change in 87R1 in helix F upon pressurization is perhaps unexpected. Numerous solution NMR studies have shown helix F to be conformationally disordered in the native state of apoMb (39, 42, 51); therefore, one might anticipate 87R1 to be mobile and malleable under pressure. However, the spectrum reveals a dominant immobile state in apoMb with only a minor change at 2 kbar. The difference may be attributed to the different time scales of EPR and NMR. For NMR, high flexibility would correspond to microsecond–millisecond motions, which are frozen on the EPR time scale. Earlier SDSL-EPR (18) and NMR (39) studies suggested that helix F sequence might be tucked into the heme pocket in native apoMb, where it could undergo motions on the NMR but not EPR time scales.
The Pressure Response of holoMb at pH 4.1: A holoMb MG State.
HoloMb was investigated at pH 4.1 at high pressure. Earlier crystallographic (53), UV-Vis absorbance (50), and CD (59) measurements all indicate that the secondary and tertiary structure of holoMb is invariant between pH 4.1 and 6.0 at atmospheric pressure, and the data presented here support this view (Fig. 4, Table 1). However, at pH 4.1, increasing pressure from 0 to 2 kbar results in a reversible loss of the Soret band and reversible loss of ∼29% helical content (Table 1) that accompany a structural transition with Δ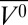 of −131 ± 7 mL/mol and ΔG0 of 3.9 ± 0.2 kcal/mol (Fig. 1B). This implies an atmospheric pressure population of ∼0.1% for the excited state, illustrating the ability of pressure to make extremely rare states accessible for study. Small-angle X-ray scattering studies at pH 4.5 identified a compact state at 2 kbar that was attributed to an MG state (64), and the large Δ
of −131 ± 7 mL/mol and ΔG0 of 3.9 ± 0.2 kcal/mol (Fig. 1B). This implies an atmospheric pressure population of ∼0.1% for the excited state, illustrating the ability of pressure to make extremely rare states accessible for study. Small-angle X-ray scattering studies at pH 4.5 identified a compact state at 2 kbar that was attributed to an MG state (64), and the large Δ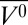 and pattern of EPR spectral changes (appearance of two-component spectra at most sites) generally is consistent with this idea (Fig. 4A). However, there are clear differences between the pressure-populated MGs of holoMb at pH 4.1 and apoMb at pH 6.0. In the holoMb MG, EPR spectral components corresponding to dynamically disordered states appear for R1 residues in the E and F helices; in the apoMb MG state, R1 residues in these helices become more immobilized. As suggested above, the F helix may occupy the empty heme pocket in the apoMb MG state. If so, this pocket is not available in the holo protein, and the response to pressure instead is unfolding, which apparently involves at least part of the contiguous E helix, contributing to the observed loss in helicity. In addition, an unfolding of helices E and F would move the heme ligands His64 and His93, resulting in the loss of the Soret band under pressure. A schematic representation comparing the proposed model for the holoMb MG state and other states of apoMb is given in Fig. 5.
and pattern of EPR spectral changes (appearance of two-component spectra at most sites) generally is consistent with this idea (Fig. 4A). However, there are clear differences between the pressure-populated MGs of holoMb at pH 4.1 and apoMb at pH 6.0. In the holoMb MG, EPR spectral components corresponding to dynamically disordered states appear for R1 residues in the E and F helices; in the apoMb MG state, R1 residues in these helices become more immobilized. As suggested above, the F helix may occupy the empty heme pocket in the apoMb MG state. If so, this pocket is not available in the holo protein, and the response to pressure instead is unfolding, which apparently involves at least part of the contiguous E helix, contributing to the observed loss in helicity. In addition, an unfolding of helices E and F would move the heme ligands His64 and His93, resulting in the loss of the Soret band under pressure. A schematic representation comparing the proposed model for the holoMb MG state and other states of apoMb is given in Fig. 5.
Recently, it was reported that MG states can retain the ability to bind native ligands, implying that close packing is not a requirement for this function (37). A comparison of the high-pressure EPR spectra of holoMb at pH 4.1 with that of apoMb under the same conditions reveals partial stabilization of the C and D helices by the heme group (Fig. 5). Furthermore, the rapid reversibility of all pressure-related changes in this state implies that the heme ligand remains bound to the protein. Thus, this system adds to existing examples of MG states that retain ligand-binding capability, albeit not in the native binding conformation.
The Pressure Response of apoMb at pH 4.1: The Pressure “Unfolded” State.
Earlier tryptophan fluorescence studies of the pH-populated MG state of apoMb at high pressure reported that the protein undergoes a transition to an unfolded state, with a Δ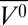 of −61 mL/mol (44). A similar value for Δ
of −61 mL/mol (44). A similar value for Δ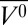 was found in this study, using CD as a monitor (Fig. 2B). However, the CD data also show that pressure causes only a partial loss of helical structure, resulting in a final value of 28% at 2 kbar with a concurrent appearance of ∼19% β-sheet. Although it cannot be ruled out that the reversible formation of β-sheet is the result of intermolecular interactions in small soluble aggregates, large-scale aggregation would cause scattering not detected in the CD data. Thus, the population existing at 2 kbar is not unfolded in the literal sense, and certainly retains a higher fraction of secondary structure than that produced by chemical perturbation techniques, including GdnHCl (∼5%), urea (∼7%), and pH (∼13%) (67, 68). Although EPR spectral changes at high pressure confirm the appearance of a dynamically disordered population in the noncore helices C, D, E, and F, the spectra at all sites retain two components, indicating the retention of a tertiary fold.
was found in this study, using CD as a monitor (Fig. 2B). However, the CD data also show that pressure causes only a partial loss of helical structure, resulting in a final value of 28% at 2 kbar with a concurrent appearance of ∼19% β-sheet. Although it cannot be ruled out that the reversible formation of β-sheet is the result of intermolecular interactions in small soluble aggregates, large-scale aggregation would cause scattering not detected in the CD data. Thus, the population existing at 2 kbar is not unfolded in the literal sense, and certainly retains a higher fraction of secondary structure than that produced by chemical perturbation techniques, including GdnHCl (∼5%), urea (∼7%), and pH (∼13%) (67, 68). Although EPR spectral changes at high pressure confirm the appearance of a dynamically disordered population in the noncore helices C, D, E, and F, the spectra at all sites retain two components, indicating the retention of a tertiary fold.
Kinetic studies of apoMb folding near neutral pH suggested the mechanism U ↔ Ia ↔ Ib ↔ N, where U is the unfolded state and Ia and Ib are transient intermediates (43, 69). The intermediate Ia is formed within 0.4 ms after initiation of folding from an acid denatured state, and relaxes to form Ib within 6 ms (43). In principle, these states are in equilibrium under all conditions, but at pH 4.1 and atmospheric pressure, at which the acid MG is formed, the dominant species is Ib (39, 69). Interestingly, the equilibrium CD and EPR data collected at pH 4.1 and 2 kbar are consistent with the Ia rather than the Ib intermediate. For example, the Ia intermediate has a stable core consisting of helices A, G, and H, giving a total helical content of about 30%, the same as that found by CD for the pH 4.1, 2-kbar state within experimental error. Moreover, the EPR results under these conditions reveal an increase in the population of disordered states for R1 residues in noncore helices, consistent with the lower protection factors for hydrogen exchange in these helices in Ia compared with Ib (43). Note that the Ib intermediate is distinct from Ia in having greater helical content (48%) due to partial folding of the noncore B, C, and E helices (42). Thus, pressure may cause a shift in the Ia ↔ Ib equilibrium toward the Ia state. The ability to populate a transiently formed kinetic intermediate under equilibrium conditions highlights the utility of the application of pressure for exploring the conformational ensemble of proteins.
Physical Origins of the Pressure Response.
Pressure reversibly shifts protein conformational equilibria in a direction to lower the total volume of the system (Le Chatelier’s principle), and excited states populated by pressure apparently have a smaller partial molar volume than the ground (native) state (9). The major contribution to the smaller molar volume for excited states is believed to be hydration of internal cavities (packing defects) enabled by conformational changes relative to the native state (70, 71). In the case of apoMb, the data presented here argue that at high pressure, hydration of internal cavities, either native or resulting from heme removal, occurs via population of an ensemble of conformations in which there is a global disruption of close packing without secondary structure change, perhaps involving solvent-separated hydrophobic interactions (72) that effectively lubricate transitions between members of the ensemble.
An alternative to the cavity hydration model is a structure-relaxation model in which excited states represent alternative packing modes of higher energy that reduce the size of internal cavities, with concomitant reduction in the overall protein molecular volume. Indeed, it is observed in crystal structures of cavity-creating mutants that the protein generally responds by shifts in atomic positions in a direction to reduce the cavity volume substantially (73, 74). Recent NMR and modeling studies of the T4 lysozyme L99A cavity-creating mutation showed that the protein responds to the mutation with a backbone structural rearrangement that allows Phe114 to move into and fill the cavity; this alternative structure is poorly populated because of strain (7). High pressure is expected to favor alternative packing modes of lower volume, and the fluctuating helix model for the pressure-populated MG state of apoMb can be understood in terms of an ensemble of alternative packing modes that reduce cavity volumes at the expense of strain. It remains to be elucidated whether one of these models dominates or whether both play a role.
Methods
Myoglobin Cloning, Expression, Purification, and Spin Labeling.
All single-cysteine mutants of sperm whale myoglobin and their spin-labeled derivatives in both the apo and holo states were reported previously and prepared following the procedures described (18). The purified protein was transferred to the appropriate buffer and concentrated to ∼500 μM for EPR experiments or ∼10 μM for CD experiments using an Amicon Ultra concentrator (10 kDa molecular weight cut-off; Millipore). ApoMb and holoMb samples were prepared in either 20 mM MES buffer at pH 6.0 or 10 mM sodium acetate at pH 4.1 for EPR experiments. For CD experiments, the same conditions were used, except the concentration of MES or sodium acetate was reduced to 5 mM. HoloMb samples prepared as described in ref. 18 were auto-oxidized to the aquomet form (Fe3+-H2O), confirmed by the ratio of the Soret band maxima at 409 nm to the absorption at 280 nm (ε280 = 31,000 L⋅mol−1⋅cm−1, ε409 = 157,000 L⋅mol−1⋅cm−1) (75). Protein concentration was measured via absorption spectroscopy at 280 nm for apoMb (ε280 = 15,400 L⋅mol−1⋅cm−1) and 409 nm for holoMb. Samples for EPR were diluted in a 1:1 ratio with 50% wt/wt Ficoll-70 (Sigma) in the appropriate buffer to reduce the effect of protein rotational diffusion on the EPR spectral lineshape (17, 26, 66); the final protein concentrations for EPR were ∼250 μM.
Horse heart holoMb (Sigma) was solubilized in buffer and used without further purification. ApoMb was prepared by the standard butanone extraction method (76). Briefly, 100 mg of holoMb was dissolved in water. The pH of the solution was adjusted to 1.5 with concentrated HCl over ice. The solution then was mixed with two volumes of 2-butanone, and the upper organic layer was decanted. The extraction procedure was repeated twice. The aqueous phase than was dialyzed at 4 °C for 2 d against 10 mM Tris buffer at pH 8, and then concentrated at 4 °C to ∼10 μM for CD experiments. Protein concentration was measured via absorption at 280 nm for apoMb (ε280 = 14,300 L⋅mol−1⋅cm−1) and 408 nm for holoMb (ε408 = 188,000 L⋅mol−1⋅cm−1) (77).
High-Pressure EPR Spectroscopy.
A computer-controlled pressure intensifier rated at 3.79 kbar and ceramic sample cells rated to 3 kbar were developed jointly with Pressure Biosciences, Inc. (PBI). The intensifier, now commercially available from PBI (model HUB440), is feedback regulated to maintain constant pressure, even in the presence of small leaks, and can be programmed to provide automated changes in pressure with various profiles in time. The ceramic cells, also now commercially available (HUB440-Cer), were fabricated for use in a five-loop four-gap resonator operating at X-band microwave frequency (78). The ceramic cell has a pressure-independent EPR signal that was subtracted from all spectra after data acquisition. The pressurization fluid was either water or buffer, and pressure was monitored with two separate transducers (Kistler model 6229A and Precise Sensors model 5550) connected inline with the sample cell. Pressure values given in this article are gauge pressure, i.e., 0 bar is equivalent to atmospheric pressure.
EPR spectra were acquired on either a Bruker ELEXSYS 580 or Varian E-109 spectrometer fitted with the abovementioned five-loop four-gap resonator operating with 5 mW incident power and at a temperature of 298 K. The sweep width for all spectra was 100 G. Atmospheric pressure spectra were acquired before and after pressurization to ensure reversibility; all reported pressure effects were reversible (Fig. S2).
High-Pressure CD and UV-Vis Spectroscopy.
Far-UV CD was performed on a Jasco 810 spectropolarimeter using a modified high-pressure optical cell (ISS model HP-200). Briefly, the standard windows were replaced with MgF2 windows (Karl Lambrecht Corporation), and the unsupported aperture was reduced to 3 mm, resulting in a cell capable of withstanding pressures up to 2.5 kbar. Details of the modifications to the high-pressure cell (Fig. S3) and evaluation of performance (Figs. S4 and S5) are provided in Supporting Information. Pressure was produced with a manually operated piston screw pump generator (High Pressure Equipment Company model 37–5.75–60) rated at 4.14 kbar and monitored with a pressure gauge (Enerpac Model T6010L) mounted inline with the sample cell. Each far-UV CD spectrum is the average of nine scans, acquired after a 3-min equilibration at each pressure. The pressure was increased in steps of 0.5 kbar from 0 to 2 kbar, and then in 0.2-kbar steps up to 2.4 kbar. For the thermodynamic analysis, the pressure was increased in steps of 0.2 kbar from 0 to 2.4 kbar. UV-Vis spectra were acquired using the same high-pressure cell with the absorption channel of the spectrometer. Reversibility was verified for each sample by collecting a spectrum at 0 bar before and after pressurization. For all experiments, the sample cell optical path length was 0.5 mm. The secondary structure parameters were estimated using the Contin algorithm (27, 28).
Thermodynamic Analysis of CD Spectra.
CD experiments detected a significant loss of helicity in the pH 4.1 states of both apoMb and holoMb due to pressure. In both cases presented here, the data were fit using a two-state model, 1↔2, where the equilibrium constant is given by Eq. 1 and the fi are the fractions of the respective states:
Structural changes were followed using CD ellipticity (θ) at 222 nm, which is related to the fraction of protein in state 2 (f2) by Eq. 2:
In Eq. 2, θ1 and θ2 are the ellipticity of states 1 and 2, respectively, and θp is the ellipticity at pressure p. The free energy difference (ΔG) in a two-state system at equilibrium is given by Eq. 3, and Eq. 4 is a first-order approximation of the pressure dependence of ΔG:
Combining Eqs. 2, 3, and 4 and solving for θp yields
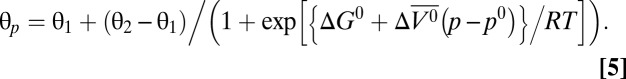 |
Plots of θp vs. p were fit using Eq. 5 to solve for free energy and partial molar volume differences at atmospheric pressure (ΔG0 and ∆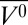 , respectively). In the pressure-populated transition of holoMb at pH 4.1, the fitted values for θ1 and θ2 were very similar to the θ at 0 and 2 kbar, respectively. This supports the two-state model for this transition and indicates that state 1 is fully populated at 0 bar and state 2 is fully populated at 2 kbar. The data for apoMb at pH 4.1 suggest that a mixture of states is populated at 0 bar, as reported previously (44), and as such, the two-state model for this transition is an approximation. The low-pH MG of apoMb is known to be populated at pH 4.1 and 0 bar; therefore, state 1 is taken to be the low-pH MG and θ1 was set to equal θ at 0 bar to generate a reasonable fit.
, respectively). In the pressure-populated transition of holoMb at pH 4.1, the fitted values for θ1 and θ2 were very similar to the θ at 0 and 2 kbar, respectively. This supports the two-state model for this transition and indicates that state 1 is fully populated at 0 bar and state 2 is fully populated at 2 kbar. The data for apoMb at pH 4.1 suggest that a mixture of states is populated at 0 bar, as reported previously (44), and as such, the two-state model for this transition is an approximation. The low-pH MG of apoMb is known to be populated at pH 4.1 and 0 bar; therefore, state 1 is taken to be the low-pH MG and θ1 was set to equal θ at 0 bar to generate a reasonable fit.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Peter Wright (The Scripps Institute) for calling our attention to the potential relationship between the high-pressure state of apoMb produced at pH 4.1 and a transient folding intermediate, as presented in Discussion. We thank Christian Altenbach, Margaux Kreitman, Carlos López, Ned Van Eps, and Zhongyu Yang for careful reading of the manuscript, and Lin Lin Ding for technical assistance with CD experiments. This work was supported by National Institutes of Health Grants EY05216, T32 EY07026, 5P41EB001980, and 2P30 EY00331 and by the Jules Stein Professorship endowment.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320124110/-/DCSupplemental.
References
- 1.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin AJ, Kay LE. NMR spectroscopy brings invisible protein states into focus. Nat Chem Biol. 2009;5(11):808–814. doi: 10.1038/nchembio.238. [DOI] [PubMed] [Google Scholar]
- 3.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: A kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89(18):8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: The energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. doi: 10.1146/annurev.physchem.48.1.545. [DOI] [PubMed] [Google Scholar]
- 5.Boehr DD, Nussinov R, Wright PE. The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol. 2009;5(11):789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma B, Shatsky M, Wolfson HJ, Nussinov R. Multiple diverse ligands binding at a single protein site: A matter of pre-existing populations. Protein Sci. 2002;11(2):184–197. doi: 10.1110/ps.21302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouvignies G, et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477(7362):111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trizac E, Levy Y, Wolynes PG. Capillarity theory for the fly-casting mechanism. Proc Natl Acad Sci USA. 2010;107(7):2746–2750. doi: 10.1073/pnas.0914727107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourme R, Girard E, Akasaka K. High-pressure macromolecular crystallography and NMR: Status, achievements and prospects. Curr Opin Struct Biol. 2012;22(5):636–642. doi: 10.1016/j.sbi.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Kitahara R, et al. High pressure NMR reveals active-site hinge motion of folate-bound Escherichia coli dihydrofolate reductase. Biochemistry. 2000;39(42):12789–12795. doi: 10.1021/bi0009993. [DOI] [PubMed] [Google Scholar]
- 11.Kitahara R, Yokoyama S, Akasaka K. NMR snapshots of a fluctuating protein structure: Ubiquitin at 30 bar-3 kbar. J Mol Biol. 2005;347(2):277–285. doi: 10.1016/j.jmb.2005.01.052. [DOI] [PubMed] [Google Scholar]
- 12.Girard E, et al. Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure. Biophys J. 2010;98(10):2365–2373. doi: 10.1016/j.bpj.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dellarole M, Roumestand C, Royer C, Lecomte JT. Volumetric properties underlying ligand binding in a monomeric hemoglobin: A high-pressure NMR study. Biochim Biophys Acta. 2013;1834(9):1910–1922. doi: 10.1016/j.bbapap.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 14.Kapoor S, et al. Revealing conformational substates of lipidated N-Ras protein by pressure modulation. Proc Natl Acad Sci USA. 2012;109(2):460–465. doi: 10.1073/pnas.1110553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akasaka K, Kitahara R, Kamatari YO. Exploring the folding energy landscape with pressure. Arch Biochem Biophys. 2013;531(1-2):110–115. doi: 10.1016/j.abb.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Bridges MD, Hideg K, Hubbell WL. Resolving conformational and rotameric exchange in spin-labeled proteins using saturation recovery EPR. Appl Magn Reson. 2010;37(1-4):363. doi: 10.1007/s00723-009-0079-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López CJ, Fleissner MR, Guo Z, Kusnetzow AK, Hubbell WL. Osmolyte perturbation reveals conformational equilibria in spin-labeled proteins. Protein Sci. 2009;18(8):1637–1652. doi: 10.1002/pro.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.López CJ, Oga S, Hubbell WL. Mapping molecular flexibility of proteins with site-directed spin labeling: A case study of myoglobin. Biochemistry. 2012;51(33):6568–6583. doi: 10.1021/bi3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altenbach C, Flitsch SL, Khorana HG, Hubbell WL. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989;28(19):7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- 20.Hubbell WL, López CJ, Altenbach C, Yang Z. Technological advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 2013;23(5):725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Cascio D, Hideg K, Kálái T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16(6):1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL. Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry. 1996;35(24):7692–7704. doi: 10.1021/bi960482k. [DOI] [PubMed] [Google Scholar]
- 23.Hubbell WL, Cafiso DS, Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat Struct Biol. 2000;7(9):735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 24.Hubbell WL, Gross A, Langen R, Lietzow MA. Recent advances in site-directed spin labeling of proteins. Curr Opin Struct Biol. 1998;8(5):649–656. doi: 10.1016/s0959-440x(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 25.Columbus L, Hubbell WL. Mapping backbone dynamics in solution with site-directed spin labeling: GCN4-58 bZip free and bound to DNA. Biochemistry. 2004;43(23):7273–7287. doi: 10.1021/bi0497906. [DOI] [PubMed] [Google Scholar]
- 26.McCoy J, Hubbell WL. High-pressure EPR reveals conformational equilibria and volumetric properties of spin-labeled proteins. Proc Natl Acad Sci USA. 2011;108(4):1331–1336. doi: 10.1073/pnas.1017877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287(2):252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 28.Fasman GD. Circular Dichroism and the Conformational Analysis of Biomolecules. New York: Plenum; 1996. p. 738. [Google Scholar]
- 29.Akasaka K. Probing conformational fluctuation of proteins by pressure perturbation. Chem Rev. 2006;106(5):1814–1835. doi: 10.1021/cr040440z. [DOI] [PubMed] [Google Scholar]
- 30.Harris RD, Jacobs M, Long MM, Urry DW. A high-pressure sample cell for circular dichroism studies. Anal Biochem. 1976;73(2):363–368. doi: 10.1016/0003-2697(76)90181-0. [DOI] [PubMed] [Google Scholar]
- 31.Di Venere A, et al. Characterization of monomeric substates of ascorbate oxidase. FEBS J. 2011;278(9):1585–1593. doi: 10.1111/j.1742-4658.2011.08084.x. [DOI] [PubMed] [Google Scholar]
- 32.Fraga TR, et al. Refolding of the recombinant protein OmpA70 from Leptospira interrogans from inclusion bodies using high hydrostatic pressure and partial characterization of its immunological properties. J Biotechnol. 2010;148(2-3):156–162. doi: 10.1016/j.jbiotec.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Gonçalves RB, Sanches D, Souza TL, Silva JL, Oliveira AC. The proapoptotic protein Smac/DIABLO dimer has the highest stability as measured by pressure and urea denaturation. Biochemistry. 2008;47(12):3832–3841. doi: 10.1021/bi702248n. [DOI] [PubMed] [Google Scholar]
- 34.Menéndez O, Rawel H, Schwarzenbolz U, Henle T. Structural changes of microbial transglutaminase during thermal and high-pressure treatment. J Agric Food Chem. 2006;54(5):1716–1721. doi: 10.1021/jf0522863. [DOI] [PubMed] [Google Scholar]
- 35.Ikeuchi Y, et al. Pressure-induced denaturation of monomer beta-lactoglobulin is partially irreversible: Comparison of monomer form (highly acidic pH) with dimer form (neutral pH) J Agric Food Chem. 2001;49(8):4052–4059. doi: 10.1021/jf001364d. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Dunker AK, Powers JR, Clark S, Swanson BG. Beta-lactoglobulin molten globule induced by high pressure. J Agric Food Chem. 2001;49(7):3236–3243. doi: 10.1021/jf001226o. [DOI] [PubMed] [Google Scholar]
- 37.Baldwin RL, Rose GD. Molten globules, entropy-driven conformational change and protein folding. Curr Opin Struct Biol. 2013;23(1):4–10. doi: 10.1016/j.sbi.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 38.Ptitsyn OB, Pain RH, Semisotnov GV, Zerovnik E, Razgulyaev OI. Evidence for a molten globule state as a general intermediate in protein folding. FEBS Lett. 1990;262(1):20–24. doi: 10.1016/0014-5793(90)80143-7. [DOI] [PubMed] [Google Scholar]
- 39.Meinhold DW, Wright PE. Measurement of protein unfolding/refolding kinetics and structural characterization of hidden intermediates by NMR relaxation dispersion. Proc Natl Acad Sci USA. 2011;108(22):9078–9083. doi: 10.1073/pnas.1105682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharyya S, Varadarajan R. Packing in molten globules and native states. Curr Opin Struct Biol. 2013;23(1):11–21. doi: 10.1016/j.sbi.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 41.Eliezer D, Jennings PA, Dyson HJ, Wright PE. Populating the equilibrium molten globule state of apomyoglobin under conditions suitable for structural characterization by NMR. FEBS Lett. 1997;417(1):92–96. doi: 10.1016/s0014-5793(97)01256-8. [DOI] [PubMed] [Google Scholar]
- 42.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat Struct Biol. 1998;5(2):148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 43.Uzawa T, et al. Hierarchical folding mechanism of apomyoglobin revealed by ultra-fast H/D exchange coupled with 2D NMR. Proc Natl Acad Sci USA. 2008;105(37):13859–13864. doi: 10.1073/pnas.0804033105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vidugiris GJ, Royer CA. Determination of the volume changes for pressure-induced transitions of apomyoglobin between the native, molten globule, and unfolded states. Biophys J. 1998;75(1):463–470. doi: 10.1016/S0006-3495(98)77534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bismuto E, Sirangelo I, Irace G, Gratton E. Pressure-induced perturbation of apomyoglobin structure: Fluorescence studies on native and acidic compact forms. Protein Sci. 1996;35(4):1173–1178. doi: 10.1021/bi951163g. [DOI] [PubMed] [Google Scholar]
- 46.Bismuto E, Sirangelo I, Irace G, Gratton E. Pressure-induced perturbation of apomyoglobin structure: Fluorescence studies on native and acidic compact forms. Biochemistry. 1996;35(4):1173–1178. doi: 10.1021/bi951163g. [DOI] [PubMed] [Google Scholar]
- 47.Bondos SE, Sligar S, Jonas J. High-pressure denaturation of apomyoglobin. Biochim Biophys Acta. 2000;1480(1-2):353–364. doi: 10.1016/s0167-4838(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 48.Kitahara R, Yamada H, Akasaka K, Wright PE. High pressure NMR reveals that apomyoglobin is an equilibrium mixture from the native to the unfolded. J Mol Biol. 2002;320(2):311–319. doi: 10.1016/S0022-2836(02)00449-7. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka N, et al. Pressure effect on the conformational fluctuation of apomyoglobin in the native state. Biochemistry. 2000;39(39):12063–12068. doi: 10.1021/bi001009g. [DOI] [PubMed] [Google Scholar]
- 50.Zipp A, Kauzmann W. Pressure denaturation of metmyoglobin. Biochemistry. 1973;12(21):4217–4228. doi: 10.1021/bi00745a028. [DOI] [PubMed] [Google Scholar]
- 51.Eliezer D, Wright PE. Is apomyoglobin a molten globule? Structural characterization by NMR. J Mol Biol. 1996;263(4):531–538. doi: 10.1006/jmbi.1996.0596. [DOI] [PubMed] [Google Scholar]
- 52.Hughson FM, Wright PE, Baldwin RL. Structural characterization of a partly folded apomyoglobin intermediate. Science. 1990;249(4976):1544–1548. doi: 10.1126/science.2218495. [DOI] [PubMed] [Google Scholar]
- 53.Yang F, Phillips GN., Jr Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J Mol Biol. 1996;256(4):762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 54.Nishii I, Kataoka M, Tokunaga F, Goto Y. Cold denaturation of the molten globule states of apomyoglobin and a profile for protein folding. Biochemistry. 1994;33(16):4903–4909. doi: 10.1021/bi00182a019. [DOI] [PubMed] [Google Scholar]
- 55.Columbus L, Kálai T, Jekö J, Hideg K, Hubbell WL. Molecular motion of spin labeled side chains in alpha-helices: Analysis by variation of side chain structure. Biochemistry. 2001;40(13):3828–3846. doi: 10.1021/bi002645h. [DOI] [PubMed] [Google Scholar]
- 56.Frauenfelder H, et al. Thermal expansion of a protein. Biochemistry. 1987;26(1):254–261. doi: 10.1021/bi00375a035. [DOI] [PubMed] [Google Scholar]
- 57.Galkin O, Buchter S, Tabirian A, Schulte A. Pressure effects on the proximal heme pocket in myoglobin probed by Raman and near-infrared absorption spectroscopy. Biophys J. 1997;73(5):2752–2763. doi: 10.1016/S0006-3495(97)78304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urayama P, Phillips GN, Jr, Gruner SM. Probing substates in sperm whale myoglobin using high-pressure crystallography. Structure. 2002;10(1):51–60. doi: 10.1016/s0969-2126(01)00699-2. [DOI] [PubMed] [Google Scholar]
- 59.Goto Y, Fink AL. Acid-induced folding of heme proteins. Methods Enzymol. 1994;232:3–15. doi: 10.1016/0076-6879(94)32039-x. [DOI] [PubMed] [Google Scholar]
- 60.Morin B, et al. Assessing induced folding of an intrinsically disordered protein by site-directed spin-labeling electron paramagnetic resonance spectroscopy. J Phys Chem B. 2006;110(41):20596–20608. doi: 10.1021/jp063708u. [DOI] [PubMed] [Google Scholar]
- 61.Belle V, et al. Mapping alpha-helical induced folding within the intrinsically disordered C-terminal domain of the measles virus nucleoprotein by site-directed spin-labeling EPR spectroscopy. Proteins. 2008;73(4):973–988. doi: 10.1002/prot.22125. [DOI] [PubMed] [Google Scholar]
- 62.Regis WC, Fattori J, Santoro MM, Jamin M, Ramos CH. On the difference in stability between horse and sperm whale myoglobins. Arch Biochem Biophys. 2005;436(1):168–177. doi: 10.1016/j.abb.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 63.Puett D. The equilibrium unfolding parameters of horse and sperm whale myoglobin. Effects of guanidine hydrochloride, urea, and acid. J Biol Chem. 1973;248(13):4623–4634. [PubMed] [Google Scholar]
- 64.Spinozzi F, et al. Met-myoglobin association in dilute solution during pressure-induced denaturation: An analysis at pH 4.5 by high-pressure small-angle X-ray scattering. J Phys Chem B. 2007;111(14):3822–3830. doi: 10.1021/jp063427m. [DOI] [PubMed] [Google Scholar]
- 65.Jennings PA, Stone MJ, Wright PE. Overexpression of myoglobin and assignment of its amide, C alpha and C beta resonances. J Biomol NMR. 1995;6(3):271–276. doi: 10.1007/BF00197808. [DOI] [PubMed] [Google Scholar]
- 66.Zhang Z, et al. Multifrequency electron spin resonance study of the dynamics of spin labeled T4 lysozyme. J Phys Chem B. 2010;114(16):5503–5521. doi: 10.1021/jp910606h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goto Y, Calciano LJ, Fink AL. Acid-induced folding of proteins. Proc Natl Acad Sci USA. 1990;87(2):573–577. doi: 10.1073/pnas.87.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barrick D, Baldwin RL. Three-state analysis of sperm whale apomyoglobin folding. Biochemistry. 1993;32(14):3790–3796. doi: 10.1021/bi00065a035. [DOI] [PubMed] [Google Scholar]
- 69.Jamin M, Baldwin RL. Two forms of the pH 4 folding intermediate of apomyoglobin. J Mol Biol. 1998;276(2):491–504. doi: 10.1006/jmbi.1997.1543. [DOI] [PubMed] [Google Scholar]
- 70.Roche J, et al. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci USA. 2012;109(18):6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamatari YO, Smith LJ, Dobson CM, Akasaka K. Cavity hydration as a gateway to unfolding: An NMR study of hen lysozyme at high pressure and low temperature. Biophys Chem. 2011;156(1):24–30. doi: 10.1016/j.bpc.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 72.Hummer G, Garde S, García AE, Paulaitis ME, Pratt LR. The pressure dependence of hydrophobic interactions is consistent with the observed pressure denaturation of proteins. Proc Natl Acad Sci USA. 1998;95(4):1552–1555. doi: 10.1073/pnas.95.4.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buckle AM, Cramer P, Fersht AR. Structural and energetic responses to cavity-creating mutations in hydrophobic cores: Observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities. Biochemistry. 1996;35(14):4298–4305. doi: 10.1021/bi9524676. [DOI] [PubMed] [Google Scholar]
- 74.Xu J, Baase WA, Baldwin E, Matthews BW. The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect. Protein Sci. 1998;7(1):158–177. doi: 10.1002/pro.5560070117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fasman GD. Practical Handbook of Biochemistry and Molecular Biology. Boca Raton, FL: CRC; 1992. p. 601. [Google Scholar]
- 76.Teale FW. Cleavage of the haem-protein link by acid methylethylketone. Biochim Biophys Acta. 1959;35:543. doi: 10.1016/0006-3002(59)90407-x. [DOI] [PubMed] [Google Scholar]
- 77.Crumpton MJ, Polson A. A comparison of the conformation of sperm whale metmyoglobin with that of apomyoglobin. J Mol Biol. 1965;11:722–729. doi: 10.1016/s0022-2836(65)80030-4. [DOI] [PubMed] [Google Scholar]
- 78.Rinard GA, Eaton GR. Biomedical EPR, part B: Methodology, instrumentation, and dynamics. In: Eaton SR, Eaton GR, Berliner LJ, editors. Biological Magnetic Resonance. New York: Springer; 2005. Vol 24, p 28. [Google Scholar]
- 79.Brucker EA, Olson JS, Phillips GN, Jr, Dou Y, Ikeda-Saito M. High resolution crystal structures of the deoxy, oxy, and aquomet forms of cobalt myoglobin. J Biol Chem. 1996;271(41):25419–25422. doi: 10.1074/jbc.271.41.25419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.