Significance
Infection by a given pathogen results in stimulation of multiple classes of innate receptors in a cell, leading to activation of distinct signaling pathways. However, these pathways are not always beneficial to immune response against the pathogen. This study shows that, on infection by pathogenic bacterium Listeria monocytogenes, Toll-like receptor (TLR) pathways suppress type I IFN gene induction that is mediated by stimulator of IFN genes. Type I IFNs are critical for antiviral immunity but detrimental to macrophage bactericidal activity. The TLR pathways selectively suppress IFN regulatory factor 3, an essential transcription factor for type I IFN gene induction, through induction/activation of mitogen-activated protein kinase phosphatases, revealing a unique mechanism of beneficial innate signaling interference against bacterial infection.
Keywords: signaling cross-talk, host defense, innate immune response
Abstract
A major function of innate immune receptors is to recognize pathogen-associated molecular patterns and then evoke immune responses appropriate to the nature of the invading pathogen(s). Because innate immune cells express various types of these receptors, distinct combinations of signaling pathways are activated in response to a given pathogen. Although the conventional wisdom is that these signaling pathways cooperate with one another to ensure an effective host response, a more nuanced view recognizes antagonism between the individual pathways, where the attenuation of a signaling pathway(s) by others may shape the immune response. In this study, we show that, on Listeria monocytogenes infection, Toll-like receptor-triggered MyD88 signaling pathways suppress type I IFN gene induction, which is detrimental to macrophage bactericidal activity. These pathways target and suppress the IFN regulatory factor 3 (IRF3) transcription factor that is activated by the stimulator of IFN genes–TANK-binding kinase-1 kinase pathway. We also provide evidence for the involvement of the MAPK phosphatase family members, which renders IRF3 hypophosphorylated on Toll-like receptor signaling by enhancing the formation of an MAPK phosphatase–IRF3–TANK-binding kinase-1 ternary complex. This study, therefore, reveals a hitherto unrecognized and important contribution of a beneficial innate signaling interference against bacterial infections.
One of the most critical challenges for the innate immune system is how it responds to a wide range of rapidly evolving pathogens with a limited repertoire of germ line-encoded, pathogen-sensing innate receptors (1–5). Two features of the innate receptors help accomplish this task; one feature is that the innate receptors generally recognize structures on pathogens that are conserved because of their vital nature for their survival, and the other feature is that the limited but diverse innate signaling pathways triggered by more than one receptor to a given pathogen cooperate with each other to ensure best fit and robust immune responses (1–5).
Indeed, the cardinal feature of signal-transducing innate receptors, now known to consist of several families, is their ability to recognize conserved pathogen-associated molecular patterns to activate innate antimicrobial responses (1–5). Given the vast evolutionary pressures on pattern recognition receptors (PRRs), most pathogens are detected by more than one class of receptor (1–5). On recognition of their microbial ligands, these receptors activate several distinct combinations of signal transduction pathways that converge on several transcription factors, such as NF-κBs and IFN regulatory factors (IRFs) (1–5). These transcription factors often function combined with each other to induce the expression of several classes of genes, including antimicrobial effectors such as cytokines and chemokines (1–5).
Bacterial pathogens can be recognized by several innate receptors, including Toll-like receptors (TLRs), nucleotide binding domain and leucine-rich repeat-containing protein receptors, and intracellular DNA sensors in innate immune cells, typically antigen-presenting cells (APCs) (1–5). Robust immune responses by APCs are thought to be driven by the cooperation of signaling pathways that emanate from each of the innate receptors (1–5). However, a given innate signaling pathway, essential against some pathogens, may be counterproductive for responses against others (4, 6–11). Thus, although the main tenet still holds true that innate signaling pathways can engage by cooperation, complementation, and compensation to ensure effective immune responses (12), one may also envisage an additional engagement, namely interference. To date, whether and how such a beneficial signaling interference operates in antibacterial responses remains unknown.
In this study, we show one such example of beneficial signaling interference in the context of bacterial infection. Macrophage infection by Listeria monocytogenes results in the activation of the DNA-sensing pathway mediated by stimulator of IFN genes (STING) to evoke a type I IFN response, which is harmful to the host because of the IFN-mediated induction of apoptosis of macrophages (13). Our current study stems from the observation that, in the absence of MyD88 (the critical adaptor protein for TLR signaling), type I IFN gene induction becomes elevated on L. monocytogenes infection in macrophages. We show that antibacterial TLR activation causes a selective suppression of STING-mediated type I IFN gene expression by targeting the IRF3 transcription factor without affecting TANK-binding kinase-1 (TBK1), the critical serine/threonine kinase for IRF3 activation. We provide evidence for TLR-induced suppression of a dissociation of the IRF3–TBK1 complex, thereby delaying the IRF3 activation cycle. We also provide evidence that this suppression is mediated by the mitogen-activated protein kinase phosphatases (MKPs), the expressions of some of which are induced on TLR signaling (14, 15) by interacting with TBK1-bound IRF3, consequently causing hypophosphorylation of IRF3.
These findings, therefore, reveal a unique facet of innate immune receptor signaling, namely the beneficial innate signaling interference, which we show is vital during bacterial infection. We will discuss the significance of these findings in terms of evolution of the innate receptor signaling pathways for infections.
Results
Suppression of Type I IFN Gene Expression on Bacterial Infection.
Infection by L. monocytogenes, a Gram-positive facultative intracellular bacterium, involves the activation of several classes of PRRs, such as TLRs, inflammasomes, and STING (13). As an approach to examine signaling interference among these innate pathways, we first examined cytokine gene expression profiles in L. monocytogenes-infected macrophages from WT or mutant mice that lack a critical protein in the pathway of these PRRs, namely MyD88 or Toll/IL-1R domain-containing adaptor inducing IFNb (TRIF) for TLRs, apoptosis-associated speck-like protein containing a carboxy-terminal CARD (ASC) for inflammasomes, or STING (1, 4, 5, 16). Interestingly, type I IFN mRNA induction, which is almost abrogated in STING-deficient macrophages, was markedly enhanced in MyD88-deficient cells, whereas IL12b gene induction was abrogated (Fig. 1A and Fig. S1 A and B). Expectedly, a notable elevation of type I IFN gene induction was also observed in L. monocytogenes-infected cells lacking TLR4, although an elevation of mRNA levels was not as pronounced as mRNA levels found in MyD88-deficient cells (Fig. S1C); this finding indicates the additional contribution of other TLR–MyD88 pathways, such as TLR2 and TLR5, which are also activated by L. monocytogenes infection (13). However, such an enhancement was not seen in cells deficient in TRIF or ASC, the common adaptor for inflammasome-activating pathways (16) (Fig. S1 A and D).
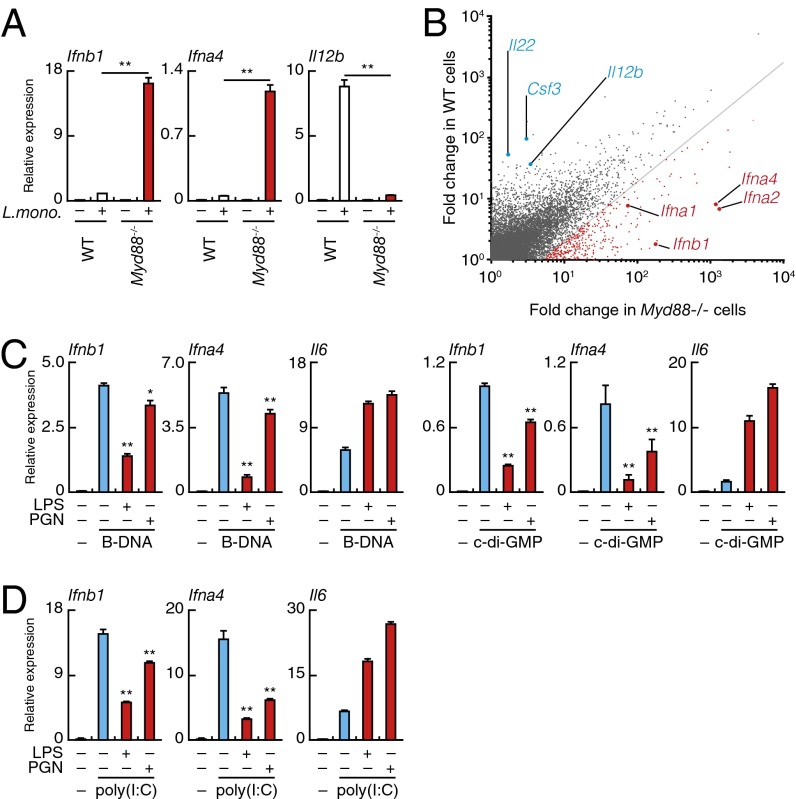
Fig. 1.
TLR-induced suppression of STING/RLR-mediated type I IFN gene expression. (A) Quantitative RT-PCR analysis of Ifnb1, Ifna4, and Il12b mRNAs in WT and MyD88-deficient (Myd88−/−) peritoneal macrophages infected for 6 h with L. monocytogenes. **P < 0.01. (B) Microarray analysis of mRNA induction in WT (vertical axis) and Myd88−/− (horizontal axis) peritoneal macrophages on infection with L. monocytogenes. Red, mRNAs with induction in Myd88−/− cells that is more than fivefold compared with WT cells. (C) Quantitative RT-PCR analysis of Ifb1, Ifna4, and Il6 mRNAs in peritoneal macrophages stimulated for 6 h with indicated combinations of B-DNA, cyclic di-GMP (c-di-GMP), LPS, or peptidoglycan (PGN). **P < 0.01; *P < 0.05 compared with cells stimulated with B-DNA or c-di-GMP. (D) Quantitative RT-PCR analysis of Ifnb1, Ifna4, and Il6 mRNAs in the peritoneal macrophages stimulated with indicated combinations of poly(I:C), LPS, or PGN for 6 h. **P < 0.01 compared with cells stimulated with poly(I:C).
These observations indicate that TLR–MyD88 signaling selectively interferes with signaling through STING, thereby suppressing type I IFN gene induction. Consistent with this notion, microarray analysis revealed a marked enhancement of type I IFN signature genes in the MyD88-deficient macrophages infected by L. monocytogenes (Fig. 1B). Furthermore, similar results were obtained on infection of these cells by a Gram-negative bacterial pathogen, Salmonella typhimurium, known to activate TLR4 and TLR2 (17) (Fig. S1E). TLRs and inflammasomes are both critical for the protective immune responses against these bacteria, whereas the STING–type I IFN axis, mediated by IRF3, is harmful to the host because of type I IFN-mediated apoptosis of macrophages (13, 18). Therefore, the suppression of the STING-mediated type I IFN gene induction by TLR–MyD88 signaling may be considered beneficial signal interference.
Suppression of Type I IFN Gene Expression by TLR Agonists.
To gain additional insight into the TLR-mediated suppression of type I IFN responses after bacterial infection, we next examined whether the above signaling interference can also be seen with mimetic ligands for TLRs and cytosolic nucleic acid-sensing receptors. As shown in Fig. 1C, the induction of type I IFN mRNAs in macrophages stimulated by poly(dA-dT)⋅poly(dT-dA) (B-DNA) or cyclic di-GMP, each activating the STING–IRF3–type I IFN pathway (19, 20), was suppressed by simultaneous stimulation by the TLR4 ligand LPS (4, 5). Of note, this suppressive effect of LPS is partially relieved not only in MyD88- but also, TRIF-deficient cells, indicating the involvement of both pathways (Fig. S1F).
In light of the above result showing that L. monocytogenes-induced type I IFN gene expression was not affected in TRIF-deficient cells (Fig. S1D), it is perhaps perplexing prima facie that LPS-mediated type I IFN gene suppression is also partially dependent on the TRIF pathway that is known to activate rather than suppress the IRF3-type IFN pathway (4, 5) (Fig. S1F). Because infection by L. monocytogenes triggers other TLRs that also activate MyD88 but not TRIF (13), we infer that the suppressive effect of TLR4-linked TRIF is overwhelmed by the strength of the MyD88 pathways activated by these TLRs. This situation would obviate a role for the TLR4–TRIF pathway in the outcome of the type I IFN gene induction (that is, elevated type I IFN gene induction in MyD88-deficient cells and no overt difference in TRIF-deficient cells on L. monocytogenes infection) (Fig. 1A and Fig. S1D).
Similarly, LPS-mediated suppression occurred in cells stimulated by poly(I:C), a synthetic dsRNA that activates retinoic acid-inducible gene I-like receptors (RLRs) for the IRF3-type I IFN pathway (2, 4) (Fig. 1D). Furthermore, suppression was also observed in cells stimulated with mimetics that activate other TLRs (Fig. 1 C and D and Fig. S1G). In contrast, the induction of IL-6 mRNA is further enhanced by these costimulation regimens, perhaps owing to the signaling cooperation between the TLR and STING pathways for this gene activation pathway (Fig. 1 C and D and Fig. S1G). These results further lend support the notion that STING/RLR-mediated type I IFN gene induction is specifically inhibited by the TLR signaling.
Selective Suppression of IRF3 Phosphorylation by TLR Signaling.
How does TLR signal function in the suppression of type I gene induction? We first examined the activation status of IRF3 because of its essential role for type I IFN response (2, 4, 5). Cytosolic stimulation of macrophages by B-DNA caused IRF3 phosphorylation at Ser-396, an essential event for IRF3 activation by TBK1 or inducible IκB kinase-ε (2, 4, 5), that was markedly inhibited by LPS stimulation (Fig. 2A). Similar observation was made when the cells were stimulated by poly(I:C) (Fig. S2A), and expectedly, dimerization of IRF3, the hallmark of IRF3 activation that is contingent on Ser-396 phosphorylation, is also suppressed by TLR4 signaling (Fig. S2B).
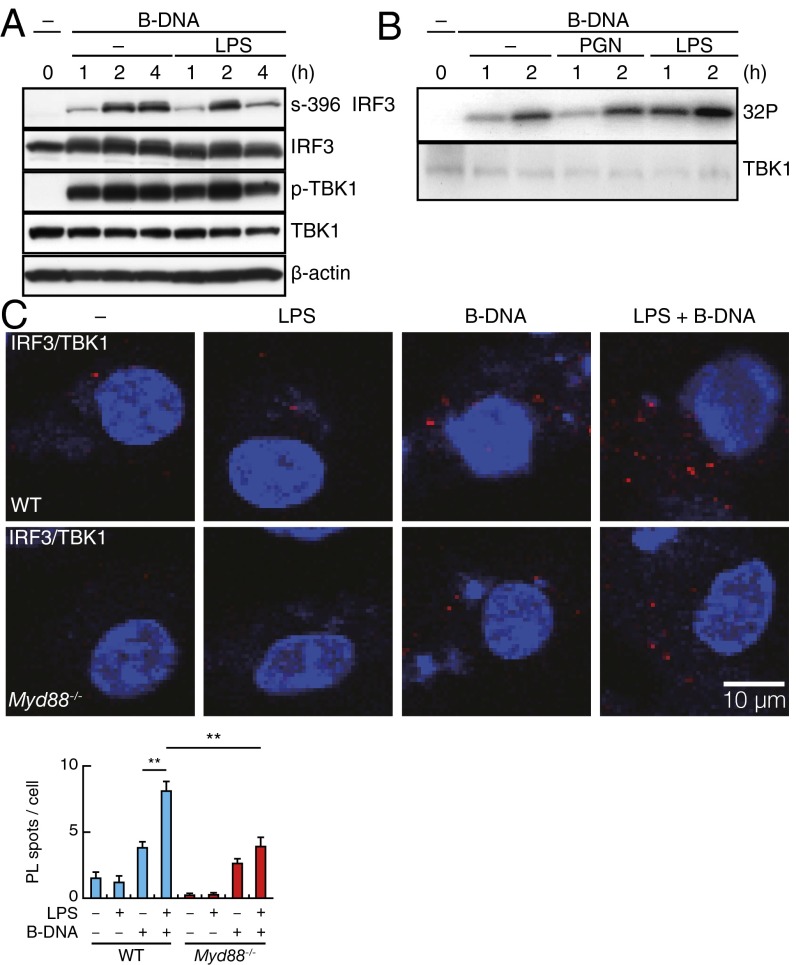
Fig. 2.
TLR-induced suppression of STING/RLR-mediated IRF3 activation. (A) Immunoblot analysis of IRF3, TBK1, and β-actin in peritoneal macrophages stimulated for indicated times with B-DNA or B-DNA plus LPS and assessed by SDS/PAGE. (B) In vitro kinase assay for TBK1 from peritoneal macrophages stimulated for indicated times with B-DNA, B-DNA plus peptidoglycan (PGN), or B-DNA plus LPS. The amount of TBK1 was examined by immunoblot analysis. (C, Upper) PLA for IRF3–TBK1 complex in WT and Myd88−/− peritoneal macrophages stimulated for 5 h with LPS, B-DNA, or B-DNA plus LPS. Red spots indicate the IRF3–TBK1 complex, whereas nuclei are stained in blue (DAPI staining). In C, Lower, data for quantitative analysis for the PL spots in a cell are shown (SI Text). **P < 0.01.
We next examined the effect of LPS on TBK1, because this kinase, but not IκB kinase-ε, is shown to be responsible for IRF3 activation in macrophages (21). As shown in Fig. 2B, the TBK1 kinase activity induced by B-DNA stimulation was not suppressed by LPS or peptidoglycan stimulation as measured by an in vitro kinase assay. The activation of other protein kinases, some of which are implicated for IRF3 activation (22, 23), as well as the phosphorylation of IκBα that leads to NF-κB activation were also not inhibited or even enhanced by the additional LPS stimulation when examined (Fig. S2C). Furthermore, the activation of IRF7, which also participates in the type I IFN gene (2, 4, 5), was not inhibited by LPS stimulation as monitored by its nuclear translocation (Fig. S2D).
Consistent with the above data, we also observed a marked enhancement of IRF3 phosphorylation levels on infection of MyD88-deficient macrophages with L. monocytogenes compared with MyD88-sufficient cells, whereas TBK1 phosphorylation levels remained essentially unaffected (Fig. S2E). As expected, enhanced type I IFN gene expression in MyD88-deficient cells was quasiabrogated in IRF3–MyD88 doubly deficient cells (Fig. S2F). These results are congruent with the above results obtained using the mimetics and indicate that the TLR–MyD88 pathway specifically interferes with IRF3 activation during bacterial infection.
Enhancement of IRF3–TBK1 Association by TLR–MyD88 Signaling.
Because the TLR activation of the MyD88 pathway rather than the TRIF pathway mainly contributes to the suppression of IRF3 activation by STING during bacterial infection (Fig. 1A and Fig. S1D), we next examined the intermolecular association between IRF3 and TBK1 with or without MyD88 signaling. For this purpose, we used in situ proximity ligation assays (PLAs) that enable the visualization of endogenous protein–protein interactions in vivo (24). As shown in Fig. 2C, the signals (referred to as PL spots) were scarce in unstimulated macrophages but notably increased on stimulation by B-DNA, indicating the STING signal-induced association of IRF3 with TBK1. Interestingly, a considerable increase of about fivefold in the number of PL spots was observed when B-DNA–stimulated cells were costimulated by LPS (Fig. 2C), whereas LPS stimulation alone did not give rise to such PL spots to a detectable level (Fig. 2C). These observations indicate an enhancement of the STING-induced IRF3–TBK1 association by TLR4 stimulation.
Notably, such an enhancement of the PL spot number was barely detectable when MyD88-deficient macrophages were subjected to the same assay, indicating that the LPS–TLR4–MyD88 pathway is mainly, if not entirely, responsible for the enhancement of the IRF3–TBK1 association (Fig. 2C). As expected, a similar observation was made when the cells were stimulated by poly(I:C) in lieu of B-DNA and together with LPS (Fig. S3). These results can be interpreted as follows. The activation of the STING (or RLR) pathway enhances the association of IRF3 and TBK1, a process essential to IRF3 activation by TBK1. However, phosphorylated IRF3 must promptly dissociate from TBK1 to undergo the conformational changes required for its dimerization (5, 25). Hence, if IRF3 remains associated with TBK1 by additionally interacting molecules induced/activated by TLR4–MyD88 signaling, the IRF3 activation processes would be inhibited. The fact that phosphorylation levels of IRF3, induced by the STING or RLR pathway, diminish on LPS stimulation suggests that such interacting molecules may be phosphatases that render the TBK1-bound IRF3 hypophosphorylated.
Association of MKP Family Phosphatases with TBK1-Associated IRF3.
It has been shown that dissociation of kinase and its substrate protein is triggered by a conformational change in the phosphorylated substrate (26). In the case of IRF3, its TBK1-dependent phosphorylation leads to a conformational change in IRF3, resulting in IRF3 dimerization and nuclear translocation (2, 4, 5, 25); hence, dephosphorylation of TBK1-bound IRF3 may result in a sustained association between TBK1–IRF3 and suppression of IRF3 activation, which we observed in B-DNA–LPS-costimulated cells (Fig. 2C).
Because the activation of TBK1 and other kinases is not inhibited by TLR4 activation (Fig. 2B and Fig. S2C), the most likely mechanism causing the suppression of the IRF3 dissociation from TBK1 may be the dephosphorylation of IRF3 by phosphatases. Along this line, treatment with sodium fluoride, which broadly inhibits Ser/Thr phosphatase activity, inhibited TLR-induced type I IFN gene suppression (Fig. S4A). In this context, it is interesting that expression of some of the MKP family is induced on TLR signaling (14, 15).
We, therefore, examined the association of IRF3 with MKP1 or MKP5, both of which are induced by TLR signaling (14, 15), by PLA. Although B-DNA stimulation slightly enhanced the numbers of PL spots that are diagnostic for an interaction of IRF3 with these MKPs, B-DNA–LPS costimulation resulted in a notable enhancement of the PL spot numbers (Fig. 3A). Because LPS stimulation alone gives only a small enhancement of PL spot (Fig. 3A), these MKPs likely target the IRF3–TBK1 complex induced by the B-DNA–STING pathway. Interestingly, PL spots for the TBK1–MKP interaction were also observed in B-DNA–stimulated WT macrophages cells, but the PL spots, detectable only at low levels in unstimulated cells, remained unaffected when IRF3-deficient macrophages were stimulated by B-DNA with or without LPS costimulation (Fig. 3B).
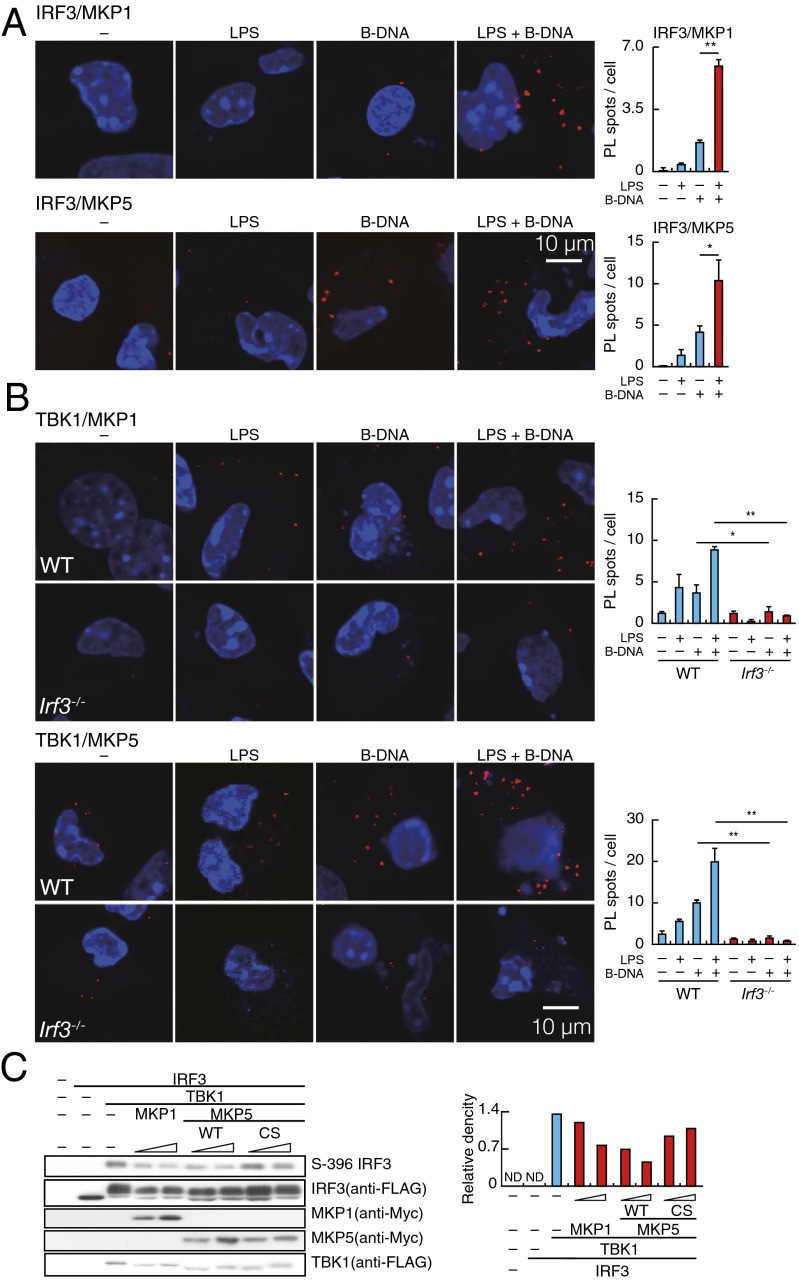
Fig. 3.
TLR-induced suppression of IRF3 by MKP proteins. (A, Left) PLA for IRF3–MKP1 or IRF3–MKP5 complex in peritoneal macrophages stimulated for 4 h with LPS, B-DNA, or B-DNA plus LPS. Red spots indicate IRF3–MKP, whereas nuclei are stained in blue (DAPI staining). In A, Right, data for quantitative analysis for the PL spots in a cell are shown. **P < 0.01; *P < 0.05. (B, Left) PLA for TBK1–MKP1 or TBK1–MKP5 complex in WT and IRF3-deficient (Irf3−/−) peritoneal macrophages stimulated with LPS, B-DNA, or B-DNA plus LPS. Red spots indicate the TBK1–MKP complex, whereas nuclei are stained in blue (DAPI staining). In B, Right, data for quantitative analysis for the PL spots in a cell are shown. **P < 0.01; *P < 0.05. (C, Left) Immunoblot analysis of IRF3, TBK1, and MKPs in HEK293T cells that overexpress FLAG-tagged IRF3, FLAG-tagged TBK1, and myelocytomatosis oncogene (Myc)-tagged MKPs, respectively, as assessed by SDS/PAGE. Relative band intensity (phospho-IRF3/IRF3) is shown in C, Right.
These results indicate that these MKPs each interacts with TBK1-bound IRF3 and suggest a scenario that the MKP–IRF3–TBK1 complex induced to form by the B-DNA–STING pathway is further enhanced by the LPS–TLR4 pathway, thereby shifting the overall equilibrium of IRF3 to a hypophosphorylated state. As discussed later, we also observed an increase in the number of PL spots for the IRF3 interaction with MKP3 but not MKP2 or MKP7 on B-DNA–LPS costimulation (Fig. S4B); therefore, MKP3 may also be involved in the IRF3 hypophosphorylation.
Regulation of IRF3 Activity by MKPs.
As another approach to examine the intermolecular association of IRF3 and MKPs, we expressed these molecules in HEK293 cells by cotransfecting expression vectors for these molecules and found that all MKPs tested interacted with IRF3 as revealed by coimmunoprecipitation assay (Fig. S4C). To gain additional mechanistic insight into the MKP-mediated regulation of IRF3, we performed a similar transient assay, where expression vectors for TBK1, IRF3, and either of the MKPs were cotransfected in HEK293 cells (27, 28). As shown in Fig. 3C, the IRF3 phosphorylation, which occurs on expression of TBK1, was inhibited by expressing MKP1 or MKP5 in a dose-dependent manner. Of note, expression of a phosphatase-defective MKP5 mutant also showed an inhibitory effect, albeit considerably weaker than WT MKP5, on IRF3 phosphorylation (Fig. 3C). Considering previous reports that show that MKP2 and MKP3 can inhibit the target protein without phosphatase activity (29, 30), we infer that the binding of MKP5 itself also has an inhibitory effect on the TBK1-mediated IRF3 phosphorylation, possibly by physically interfering with the conformation of the kinase/substrate complex that is required for phosphorylation. These results, therefore, suggest that MKPs exert a suppressive function by interaction with and dephosphorylation of the TBK1-bound IRF3.
We also examined the role of MKP1 and MKP5 by examining macrophages deficient in either one of these phosphatases. However, no dramatic difference was observed in any of these cells compared with WT cells on L. monocytogenes infection (Fig. S4D). These results may reflect functional redundancy among MKP1, MKP5 and other MKPs (Discussion).
Significance of TLR-Induced Suppression of IRF3 Against L. monocytogenes Infection.
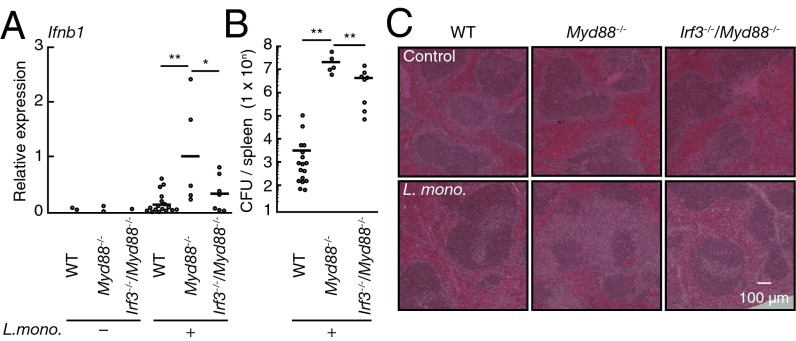
Finally, we examined the significance of TLR-induced IRF3 suppression. Consistent with our in vitro results, IRF3-dependent type I IFN gene induction significantly increased in the spleen from MyD88-deficient mice infected with L. monocytogenes (Fig. 4A). As expected, MyD88-deficient mice showed a significant increase in bacterial load that was accompanied with more severe tissue injury. However, both of these pathogenic features were attenuated by introducing an additional genetic deficiency in the IRF3 gene (Fig. 4 B and C). Thus, these observations lend support for the critical role of TLR-induced suppression of IRF3 activation described above.
Fig. 4.
Biological significance of TLR-induced suppression of IRF3. (A) Quantitative RT-PCR analysis of IFNb1 mRNA in the spleen from WT (control, n = 2; infected, n = 17), Myd88−/− (control, n = 2; infected, n = 5), and IRF3–MyD88 doubly deficient (Irf3−/−Myd88−/−; control, n = 1; infected, n = 7) mice infected with L. monocytogenes for 2.5 d. **P < 0.01; *P < 0.05. (B) Colony formation assay of L. monocytogenes titers in the spleen of WT (n = 16), Myd88−/− (n = 7), and Irf3−/−Myd88−/− (n = 5) mice infected as in A. Each symbol represents an individual mouse; small horizontal lines indicate the mean. **P < 0.01. (C) Histological analysis of the spleens from WT, Myd88−/−, and Irf3−/−Myd88−/− mice infected as in A and assessed by microscopy of sections stained with H&E. (Magnification: 40×.)
Discussion
Innate immune cells, such as APCs, must evoke immune response against a huge variety of pathogens while using only a limited repertoire of innate immune receptors, termed PRRs (1–5). As such, unlike antigen receptors of the adaptive immune system, a given PRR recognizes a variety of pathogens, each of which expresses numerous pathogen-associated molecular patterns, and infection of a pathogen triggers activation of multiple PRRs in a cell (1–5). The activation of multiple PRRs has the effect of enhancing the magnitude of innate response by cooperation of the signals emanating from each PRR, thereby providing advantages for a robust innate response against the invading pathogen (12). However, the activation of a multiple innate signaling pathway by a given pathogen may also evoke harmful responses and therefore, needs to be regulated. Our present study offers one such example, in which TLR signal-mediated signaling suppresses an STING/RLR-mediated type I IFN response that is critical to innate responses against viral infection but detrimental to antibacterial responses (6, 13, 31). Indeed, in the context of L. monocytogenes infection, type I IFNs also negatively affect adaptive immunity by sensitizing lymphocytes to undergo apoptosis during infection (32).
The TLR-mediated signaling interference is a unique system of fine tuning by PRRs, where the STING–IRF3-mediated type I IFN response is selectively suppressed during bacterial infection, which activates both TLR and STING pathways. Indeed, as shown in Fig. S5, several antibacterial cytokines were synergistically activated by simultaneous stimulation of STING and TLR pathways, indicating that these pathways cooperate with each other to induce IRF3-independent genes that may include protective genes for the antibacterial response. Indeed, although mice deficient in either IRF3 or IFN a/b receptor 1 (IFNAR1), a component of the type I IFN receptor, are both highly resistant to L. monocytogenes infection (18), STING-deficient mice show no resistance (33). This fact supports the notion that the STING-dependent pathway activates not only the harmful type I IFN response but also, the protective immune responses to this pathogen. Based on our results, we conclude that the TLR–MyD88 pathway provides a beneficial signaling interference, which inhibits only the harmful IRF3-mediated type I IFN response and does not affect other responses that would ensure protective cooperation between the TLR and STING pathways for the antibacterial innate responses.
Although we used intracellular bacteria to analyze the physiological role of MyD88-dependent cross-interference in this study, recent reports support the possible involvement of this protective system in the host defense against a variety of bacteria. First, bacteria increase the pathogenicity of bacteriophage infection, which has the potential to activate STING/RLR-mediated type I IFN gene induction (34). This fact suggests the possibility that MyD88-dependent cross-interference may protect the host from pathogenic bacteria infected with bacteriophages. Second, because various types of bacteria express the STING ligands cyclic di-GMP/AMP (35, 36), it is plausible that the STING-type I IFN pathway is activated by these bacteria-derived ligands. Thus, the MyD88-dependent signaling interference, described here, may protect the host from extracellular bacteria as well. Given the well-documented fact that the TLR–TRIF pathway leads to IRF3 activation and type I IFN gene induction (4, 5), it is somewhat enigmatic that type I IFN gene induction is also elevated on LPS stimulation. It is our conjecture that, although the TRIF pathway mediates the type I IFN induction pathway during TLR4 activation, this pathway also simultaneously activates a suppressive pathway to minimize type I IFN gene induction for the benefit of the antibacterial responses of the host.
Our results strongly implicate the involvement of the MKP family members, which on TLR stimulation, are induced, activated, or both by an unknown mechanism. MKP proteins then target IRF3 to form the TBK1–IRF3–MKP ternary complex. This complex formation may allow MKP to dephosphorylate the TBK1-bound IRF3 and/or physically interfere with the kinase action of the IRF3-bound TBK1, resulting in suppression of IRF3 phosphorylation at Ser-396 and probably, other serine residues that are critical for the activation of IRF3 and type I IFN responses. We tried to examine where such a complex is formed in a specified cell organelle by staining the PLA samples with antibodies for mitochondria, endoplasmic reticulum, and endosome, but there was no correlation in the staining pattern between PL spots and these organelles (Fig. S6), suggesting that the formation occurs perhaps in other cellular compartments. In a strict sense, our data do not impeccably show the role of these MKPs, because type I IFN mRNA induction remained largely unaffected in cells deficient in either MKP1 or MKP5; however, a likely possibility is that MKPs function redundantly for the negative regulation of IRF3 activation. Indeed, we also found by PLA an enhancement of B-DNA–induced MKP3–IRF3 association by TLR stimulation (Fig. S4B). As such, additional work will be required to clarify this issue.
In conclusion, our study offers insight into the regulation of the PRR-mediated antipathological immune response and highlights the importance of the TLR–MyD88–MKP axis in the beneficial signaling interference during bacterial infections. We infer that the signaling interference mechanism described here could have evolved during the acquisition of multiple innate receptors and their signaling pathways to optimize the outcome of the immune responses triggered by these pathways against pathogen infection.
Materials and Methods
Mice.
All mice were maintained on a C57BL/6 (B6) genetic background. About 6- to 12-wk-old mice were used for each experiment. The generation of mutant mice is described in SI Text. All animal experiments were done in accordance with the animal use guidelines of the University of Tokyo.
Cell Culture.
Cells were cultured or prepared by commonly used methods with reagents as described in SI Text.
Reagents and RNA Analysis.
Total RNA was extracted by using NucleoSpin RNA (from cells; TaKaRa) or RNAiso Plus (from spleens; TaKaRa). Reverse transcription was performed using PrimeScript RT Master Mix (TaKaRa). Quantitative RT-PCR was performed with specific primer sets as described in SI Text. Other reagents are described in SI Text.
Immunoblot Analysis.
Immunoblot analysis was performed by commonly used methods. More detailed experimental settings and antibodies are in SI Text.
PLA.
PLA was performed with a Duolink In Situ PLA Kit (Olink Bioscience) according to the manufacturer’s protocol. More detailed PLA experimental settings and antibodies are in SI Text.
Infection.
Mice were i.p. infected with L. monocytogenes (105 cfu) and analyzed 2.5 d after infection as described in SI Text.
Supplementary Material
Acknowledgments
We thank G. N. Berber and S. Saijo for Sting−/− mice; S. Akira for Myd88−/− mice; C. Dong for Mkp5−/− mice; T. Tamura, S. Hida, H. Tani, and T. Negishi for helpful discussions; and R. Koshiba, K. Ogami, T. Osawa, M. Taniguchi, H. Tanabe, R. Fujii, K. Adachi, and M. Shishido for technical assistance. This work was supported, in part, by the Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Agency (JST), The Naito Foundation, The Nakajima Foundation, a Grant-in-Aid for Exploratory Research of Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a Grant-in-Aid for Scientific Research on Innovative Areas of MEXT. H.S., S.M., A.M., and N.T.-A. are research fellows of the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320145110/-/DCSupplemental.
References
- 1.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32(3):305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Loo YM, Gale M., Jr Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–692. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 6.Kearney S, Delgado C, Lenz LL. Differential effects of type I and II interferons on myeloid cells and resistance to intracellular bacterial infections. Immunol Res. 2013;55(1–3):187–200. doi: 10.1007/s12026-012-8362-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker D, Prince A. Type I interferon response to extracellular bacteria in the airway epithelium. Trends Immunol. 2011;32(12):582–588. doi: 10.1016/j.it.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball EA, et al. IFNAR1 controls progression to cerebral malaria in children and CD8+ T cell brain pathology in Plasmodium berghei-infected mice. J Immunol. 2013;190(10):5118–5127. doi: 10.4049/jimmunol.1300114. [DOI] [PubMed] [Google Scholar]
- 9.Xin L, et al. Type I IFN receptor regulates neutrophil functions and innate immunity to Leishmania parasites. J Immunol. 2010;184(12):7047–7056. doi: 10.4049/jimmunol.0903273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34(2):213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Inglis DO, Berkes CA, Hocking Murray DR, Sil A. Conidia but not yeast cells of the fungal pathogen Histoplasma capsulatum trigger a type I interferon innate immune response in murine macrophages. Infect Immun. 2010;78(9):3871–3882. doi: 10.1128/IAI.00204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nish S, Medzhitov R. Host defense pathways: Role of redundancy and compensation in infectious disease phenotypes. Immunity. 2011;34(5):629–636. doi: 10.1016/j.immuni.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stavru F, Archambaud C, Cossart P. Cell biology and immunology of Listeria monocytogenes infections: Novel insights. Immunol Rev. 2011;240(1):160–184. doi: 10.1111/j.1600-065X.2010.00993.x. [DOI] [PubMed] [Google Scholar]
- 14.Chi H, et al. Dynamic regulation of pro- and anti-inflammatory cytokines by MAPK phosphatase 1 (MKP-1) in innate immune responses. Proc Natl Acad Sci USA. 2006;103(7):2274–2279. doi: 10.1073/pnas.0510965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondoh K, Nishida E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim Biophys Acta. 2007;1773(8):1227–1237. doi: 10.1016/j.bbamcr.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi O, et al. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11(4):443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 18.O’Connell RM, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200(4):437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdette DL, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med. 2004;199(12):1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsushima K, et al. IRF3 regulates cardiac fibrosis but not hypertrophy in mice during angiotensin II-induced hypertension. FASEB J. 2011;25(5):1531–1543. doi: 10.1096/fj.10-174615. [DOI] [PubMed] [Google Scholar]
- 23.Joung SM, et al. Akt contributes to activation of the TRIF-dependent signaling pathways of TLRs by interacting with TANK-binding kinase 1. J Immunol. 2011;186(1):499–507. doi: 10.4049/jimmunol.0903534. [DOI] [PubMed] [Google Scholar]
- 24.Söderberg O, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3(12):995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 25.Chen W, et al. Insights into interferon regulatory factor activation from the crystal structure of dimeric IRF5. Nat Struct Mol Biol. 2008;15(11):1213–1220. doi: 10.1038/nsmb.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai-Tamai Y, Mizuno K, Hirose T, Suzuki A, Ohno S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells. 2002;7(11):1161–1171. doi: 10.1046/j.1365-2443.2002.00590.x. [DOI] [PubMed] [Google Scholar]
- 27.Tanoue T, Moriguchi T, Nishida E. Molecular cloning and characterization of a novel dual specificity phosphatase, MKP-5. J Biol Chem. 1999;274(28):19949–19956. doi: 10.1074/jbc.274.28.19949. [DOI] [PubMed] [Google Scholar]
- 28.Tanoue T, Yamamoto T, Maeda R, Nishida E. A Novel MAPK phosphatase MKP-7 acts preferentially on JNK/SAPK and p38 alpha and beta MAPKs. J Biol Chem. 2001;276(28):26629–26639. doi: 10.1074/jbc.M101981200. [DOI] [PubMed] [Google Scholar]
- 29.Brunet A, et al. Nuclear translocation of p42/p44 mitogen-activated protein kinase is required for growth factor-induced gene expression and cell cycle entry. EMBO J. 1999;18(3):664–674. doi: 10.1093/emboj/18.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong MW, Kang TH, Kim W, Choi YH, Kim KT. Mitogen-activated protein kinase phosphatase 2 regulates histone H3 phosphorylation via interaction with vaccinia-related kinase 1. Mol Biol Cell. 2013;24(3):373–384. doi: 10.1091/mbc.E12-06-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rayamajhi M, Humann J, Kearney S, Hill KK, Lenz LL. Antagonistic crosstalk between type I and II interferons and increased host susceptibility to bacterial infections. Virulence. 2010;1(5):418–422. doi: 10.4161/viru.1.5.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200(4):535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin L, et al. STING/MPYS mediates host defense against Listeria monocytogenes infection by regulating Ly6C(hi) monocyte migration. J Immunol. 2013;190(6):2835–2843. doi: 10.4049/jimmunol.1201788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nat Immunol. 2013;14(7):654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamayo R, Pratt JT, Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu Rev Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrigan RM, Gründling A. Cyclic di-AMP: Another second messenger enters the fray. Nat Rev Microbiol. 2013;11(8):513–524. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.