Significance
The human immune system fights disease by eradicating sick cells after first recognizing that they are infected or cancerous. This is achieved by specialized cells that detect on the surface of other cells small molecules called antigenic peptides. Pathogens and cancer can evade the immune system by stopping the generation of antigenic peptides. We designed, synthesized and evaluated artificial small molecules that can effectively block a group of enzymes that are key for the production or destruction of antigenic peptides. We show that these compounds can enhance the generation of antigenic peptides in cells and enhance the immune system reaction toward cancer. Inhibitors of this kind may provide a new approach to coax the immune system into recognizing and eliminating cancer cells.
Keywords: molecular structure, adaptive immunity, major histocompatibility molecules, specificity, kinetics
Abstract
Intracellular aminopeptidases endoplasmic reticulum aminopeptidases 1 and 2 (ERAP1 and ERAP2), and as well as insulin-regulated aminopeptidase (IRAP) process antigenic epitope precursors for loading onto MHC class I molecules and regulate the adaptive immune response. Their activity greatly affects the antigenic peptide repertoire presented to cytotoxic T lymphocytes and as a result can regulate cytotoxic cellular responses contributing to autoimmunity or immune evasion by viruses and cancer cells. Therefore, pharmacological regulation of their activity is a promising avenue for modulating the adaptive immune response with possible applications in controlling autoimmunity, in boosting immune responses to pathogens, and in cancer immunotherapy. In this study we exploited recent structural and biochemical analysis of ERAP1 and ERAP2 to design and develop phosphinic pseudopeptide transition state analogs that can inhibit this family of enzymes with nM affinity. X-ray crystallographic analysis of one such inhibitor in complex with ERAP2 validated our design, revealing a canonical mode of binding in the active site of the enzyme, and highlighted the importance of the S2’ pocket for achieving inhibitor potency. Antigen processing and presentation assays in HeLa and murine colon carcinoma (CT26) cells showed that these inhibitors induce increased cell-surface antigen presentation of transfected and endogenous antigens and enhance cytotoxic T-cell responses, indicating that these enzymes primarily destroy epitopes in those systems. This class of inhibitors constitutes a promising tool for controlling the cellular adaptive immune response in humans by modulating the antigen processing and presentation pathway.
The human adaptive cellular immune response relies on cell-surface presentation of small peptides, 8–10 amino acids long, bound on specialized receptors of the major histocompatibility complex (MHC). Such peptides are derived from the proteolytic degradation of intracellular proteins and constitute a representative sample of the protein content of the cell (1). Infected or malignantly transformed cells express additional protein molecules that upon degradation give rise to distinct antigenic peptides that are presented on the cell surface complexed with MHC class I molecules (MHCI). Cytotoxic T cells can recognize these complexes and induce apoptotic cell death. Aberrant generation of antigenic peptides can lead to immune system evasion or to autoimmune reactions (2–6).
Most antigenic peptides are initially produced by the proteasome, but many of them are larger than the final antigenic epitope, containing one or more additional amino acids at their N termini (7). These antigenic peptide precursors are transported into the endoplasmic reticulum (ER), where they are further trimmed by at least two different aminopeptidases, endoplasmic reticulum aminopeptidase 1 and 2 (ERAP1 and ERAP2), to generate the mature antigenic peptides of the optimal length for loading onto MHCI molecules (8). During recent years, the importance of these two aminopeptidases has been established in several in vitro and in vivo systems, including mouse disease models (reviewed in refs. 9 and 10). Furthermore, these two aminopeptidases actively regulate the presentation of antigenic peptides, not only by generating the correct epitopes but also by destroying many of them by trimming them to lengths too short to bind onto MHCI (11). In the absence of these aminopeptidases, specific immunodominant epitopes are no longer generated and previously unrepresented epitopes can be detected on the cell surface. This can lead to either suppression or activation of existing cytotoxic responses or the generation of novel responses by both T cells and NK cells (2, 5, 12, 13). In this context, the activity of ERAP1 and ERAP2 directly affects the presented antigenic peptide repertoire altering the adaptive immune response both qualitatively and quantitatively. Single coding nucleotide polymorphisms in these enzymes have been recently associated with predisposition to a large array of infectious and autoimmune diseases (14–17). Changes in the enzymes’ activity and specificity have been proposed to be the molecular basis behind these associations (14, 18, 19).
In the cellular pathway of cross-presentation, ERAP1 and ERAP2 can also trim antigenic peptide precursors in endosomal compartments of professional antigen-presenting cells such as dendritic cells. A homologous aminopeptidase named insulin-regulated aminopeptidase (IRAP) has also been recently implicated to operate in a newly discovered cross-presentation pathway (20, 21). All three aminopeptidases are highly homologous (∼50% sequence identity) and use identical catalytic mechanisms but have differences in substrate specificity (22–24).
The important role played by these three aminopeptidases in modulating the adaptive immune response has spurred interest toward finding ways to either inhibit or enhance their action. Genetic down-regulation of ERAP1 in mice has been shown to lead to generation of some unstable MHCI molecules on the cell surface altering cytotoxic T-lymphocytes (CTL) responses and to also elicit nonclassical MHCIb-restricted CTL responses in vivo (2, 12). In murine tumor models, ERAP1 down-regulation by siRNA was sufficient to induce protective NK or cytotoxic T-cell responses and lead to tumor rejection (5, 13). These findings suggest that the pharmacological regulation of ERAP1 and possibly ERAP2 and IRAP may have important therapeutic applications in a large array of diseases ranging from viral infections, autoimmunity, and cancer.
Despite these possible applications, to our knowledge, no potent inhibitors have been described for ERAP1 and ERAP2. The broad-spectrum metallopeptidase inhibitor leucinethiol is a moderate inhibitor of ERAP1 with an affinity of ∼5–10 μM and has been used successfully to reproduce some genetic down-regulation effects (2, 12, 25). A novel class of inhibitors for aminopeptidases has been recently described, but with only moderate affinity for ERAP1 (26). Potent inhibitors for IRAP have been described but displayed low efficacy for ERAP1 and ERAP2, and their role in antigen processing has not been evaluated (27).
The recently solved crystal structures of ERAP1 and ERAP2 as well as the accumulation of a wide array of biochemical and functional data about these enzymes provide an opportunity for the rational design of potent, mechanism-based inhibitors (reviewed in ref. 28). Using this knowledge, we designed, synthesized, and evaluated two pseudopeptidic compounds carrying a phosphinic group that were expected to act as transition-state analogs for these enzymes. One of the compounds inhibited all three enzymes with high potency, having affinity in the nM range. Our compounds were able to affect antigen processing in cultured cells and elicit cytotoxic T-cell responses in a dose-dependent and affinity-dependent manner. Analysis of ERAP2 cocrystallized with one of the compounds validated our rational design strategy and provided insight on the mechanism of inhibition. We propose that these or similar compounds provide a basis by which to regulate the adaptive immune response for the treatment of autoimmunity and for enhancing cancer immunotherapy regimens.
Results
Design Rationale.
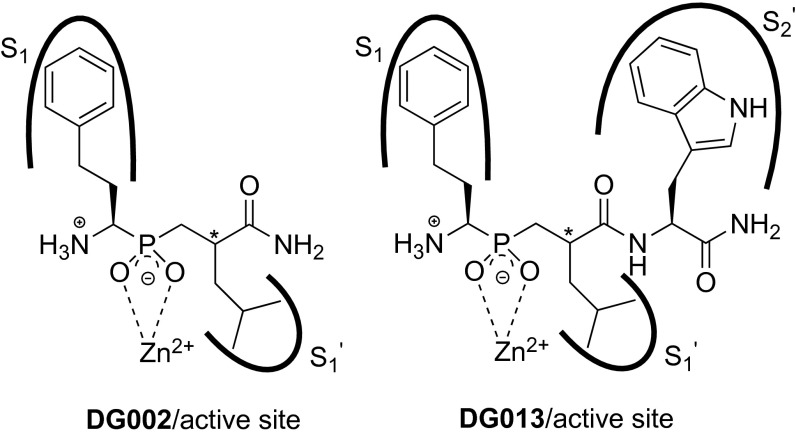
Phosphinic pseudopeptides have been developed as both potent and selective mechanism-based inhibitors of metalloproteinases (29, 30). One advantage of the phosphinic functional group is that it is a relatively weak zinc ligand, and as a result the binding affinity attained is mainly attributed to specific interactions between the side chains of the inhibitor and the active site specificity pockets of the enzyme. After optimization, this specific binding can lead to potent and selective inhibitors. Because ERAP1, ERAP2, and IRAP have the same fundamental catalytic mechanism as other Zn(II) metalloproteinases, we hypothesized that using a phosphinic backbone would be a valid approach to developing highly potent inhibitors for these aminopeptidases. Recent structural and biochemical analyses of ERAP1 and ERAP2 have revealed a significant amount of information regarding their specificity pockets that could be exploited for rational inhibitor design. Specifically, all three enzymes share key common characteristics in their S1 specificity pocket, a pocket that is structurally rigid and contains a hydrophobic base (24, 28). Substrate library screening revealed several side chains optimal for recognition by all three enzymes (24). Based on those studies, we selected phenylethyl (the side chain of the nonnatural amino acid homophenylalanine) for the S1 pocket, as it can be accommodated in that pocket in a near optimal fashion. Additionally, a previous study on the specificity of ERAP1 has revealed a preference for small hydrophobic residues in the S1’ specificity pocket, and we therefore selected a leucine residue for this position (31). Lastly, the recently solved structure of ERAP2 [Protein Data Bank (PDB) ID code 3SE6] showed a molecule of 2-(N-morpholino)ethanesulfonic acid tightly bound near the active site, with its morpholino ring stabilized by stacking interactions between Tyr455 and Tyr892, two residues that form a hydrophobic pocket that may act as the S2’ specificity pocket of the enzyme (32).
We therefore designed two pseudopeptides: both compounds contain a homophenylalanine and a leucine residue on each side of a phosphinic group. The second compound additionally contains a tryptophan residue as the C-terminal residue so that we could best exploit the hydrophobic and π-stacking properties of the postulated S2’ specificity pocket. The chemical structures of these two compounds are shown in Fig. 1. Each compound was synthesized with a single chiral center in residue 2 (indicated by an asterisk in Fig. 1).
Fig. 1.
Chemical structures of synthesized compounds. Black lines indicate the relative locations of the enzyme specificity pockets targeted by side chains in the compound. An asterisk indicates the chiral center in each compound.
Inhibitor Synthesis and Purification.
For the synthesis of inhibitors DG002 and DG013, a solid phase peptide synthesis -based approach was used (Fig. S1). A suitably protected phosphinic building block was synthesized to deliver the final structures after standard TFA cleavage from a Rink amide resin (33). Assembly of the phosphinodipeptidic scaffold was performed by a P-Michael addition of the enantiomerically resolved aminophosphinic analog and acrylate, which leads to an unresolved stereogenic center at P1′ position. Therefore, inhibitors DG002 and DG013 were finally obtained as mixtures of two diastereoisomers ([R,S], [R,R] for DG002 and [R,S,S], [R,R,S] for DG013).
Compounds were purified by reversed-phase HPLC using a linear acetonitrile gradient. This way it was possible to separate the two stereoisomers (Fig. S1 B and C). For all characterizations each stereoisomer was treated as a separate compound. Based on previous work that characterized the stereochemistry of structurally related pseudophosphinic peptides, the first eluted peak from the HPLC (compounds DG002A and DG013A) was expected to correspond to the [R,S] and [R,S,S] stereoisomers, respectively, and the second (compounds DG002B and DG013B) to the [R,R] and [R,R,S] stereochemistry (34). This was also found to be consistent with the crystallographic analysis (see Structure of ERAP2 with DG013).
In Vitro Potency.
We used a previously established fluorigenic assay to characterize the ability of each compound to inhibit the hydrolysis of model substrates L-Leucine 7-amido-4-methyl coumarin hydrochloride (L-AMC) and L-arginine 7-amido-4-methylcoumarin hydrochloride (R-AMC) by ERAP1, ERAP2, and IRAP (24). Representative titrations are shown in Fig. S2, and calculated IC 50 values are shown in Table 1. DG002 was a moderately potent inhibitor of all three enzymes regardless of its stereochemistry. In contrast, addition of a tryptophan residue at the C terminus resulted in very potent inhibition of all three enzymes that was more pronounced for ERAP2, with calculated IC50 values in the low nM range (Fig. S2C and Table 1). The stereochemical composition of the DG013 compound was important for inhibition, as evidenced from the much lower IC50 values calculated for the second isomer. Overall, the DG013A compound was found to be a very potent inhibitor of ERAP1, ERAP2, and IRAP and, to our knowledge, has the highest affinity of any inhibitor described for ERAP1 and ERAP2. Furthermore, it presented some selectivity for ERAP2, although it was equally effective versus ERAP1 and IRAP.
Table 1.
In vitro and bio-efficacy of synthesized inhibitors
| Compound name | ERAP1 | ERAP1 | Mouse ERAAP | ERAP2 | IRAP | HeLa cells | CT26 cells |
| IC50, nM* | IC50, nM† | IC50, nM* | IC50, nM* | IC50, nM* | ED50, μM | ED50, μM | |
| DG002A [R,S] | 520 ± 75 | 403 ± 71 | 650 ± 148 | 547 ± 110 | 218 ± 37 | 1.9 ± 1.1 | 0.626 ± 0.053 |
| DG002B [R,R] | 513 ± 51 | 481 ± 98 | 872 ± 137 | 571 ± 95 | 344 ± 68 | ND‡ | 1.2 ± 0.2 |
| DG013A [R,S,S] | 33 ± 5 | 55.7 ± 5.8 | 69 ± 13 | 11 ± 2 | 30 ± 4 | 0.44 ± 0.16 | 0.033 ± 0.015 |
| DG013B [R,R,S] | 3,600 ± 500 | 1,574 ± 754 | 1,333 ± 451 | 1,700 ± 200 | 2,200 ± 300 | >100 | 32 ± 19 |
X-AMC substrate.
10-mer substrate.
Not determined.
Because the small fluorigenic substrates used in the in vitro characterization of the inhibitors are not perfect models of the natural substrates of these enzymes (that usually are longer peptides consisting of 9–15 amino acids), we also tested the ability of the compounds to inhibit hydrolysis of a 10-mer fluorigenic peptide that has been designed to be a good ERAP1 substrate (35). The IC50 values for inhibition of the hydrolysis of this substrate were found to be largely similar to the IC50 values calculated for the L-AMC substrate (Fig. S2 and Table 1).
Mechanism of Inhibition.
To gain insight on the mechanism of inhibition by this group of compounds, we performed Michaelis–Menten (MM) analysis in the presence or absence of the compounds (Figs. S3–S5). This analysis suggested that DG013A as well as both stereoisomers of DG002 are competitive inhibitors of ERAP1 and ERAP2, affecting only the KM value of the substrate, as expected based on their design as mechanism-based inhibitors (Fig. S4 A–C and Fig. S5 A–C). In contrast, MM analysis for DG013B indicated that this weaker inhibitor acted noncompetitively for both ERAP1 and ERAP2, affecting the Vmax of the catalysis and not the substrate KM (Fig. S4D and Fig. S5D). This suggests that inhibition by DG013B may involve a different binding mode near the active site or even a distinct binding site. Because the presence of a secondary regulatory site has been proposed for ERAP1, we further investigated this phenomenon by performing MM analysis using the larger 10-mer fluorigenic peptide described above (Fig. S5). Interestingly, DG013B acted as competitive inhibitor for this larger substrate, suggesting that it may indeed be binding to a distinct site within the extended binding cavity of the enzyme (36).
Structure of ERAP2 with DG013.
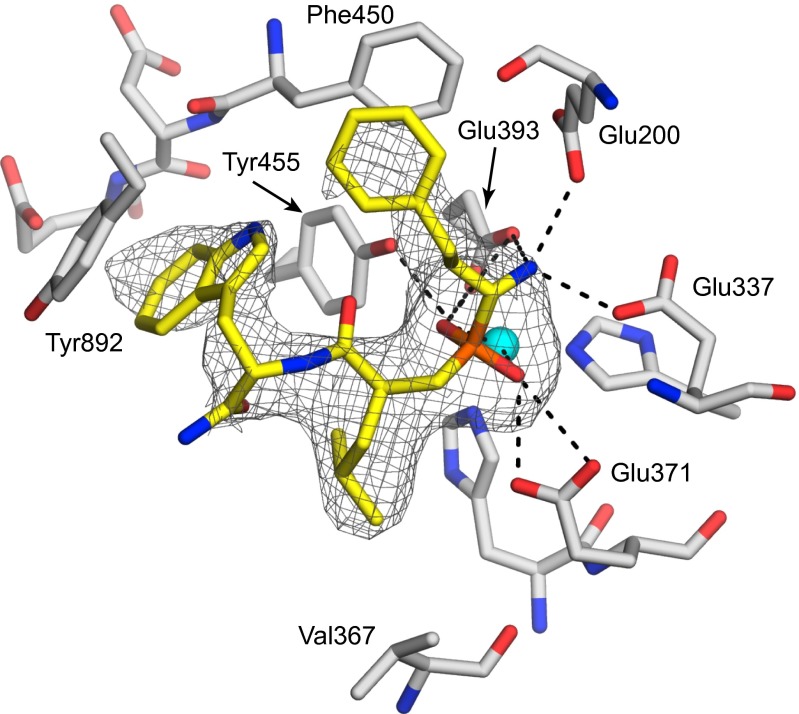
To better understand the mode of binding of the best inhibitor, DG013A, and to help guide further development of such inhibitors, we cocrystallized the compound with the ERAP2 N392K allele for which DG013A is also a potent inhibitor (18) (Fig. S6). The crystals diffracted to 2.8 Å, and the structure was solved by molecular replacement based on the recently determined crystallographic structure of ERAP2 (Table S1). The difference density found in the catalytic center of the enzyme was directly attributable to the presence of DG013A in the structure (Fig. 2). The electron density map was consistent with DG013A being the [R,S,S] stereoisomer in accord with the HPLC analysis. Analysis of the refined structure revealed a canonical mode of binding for the inhibitor, according to which the phosphinic group coordinates the active site Zn(II) atom and its two oxygen atoms are further stabilized by hydrogen bonding interactions with Glu371 and the hydroxyl group of Tyr455. In this context, the pseudopeptide is bound in a conformation resembling the intermediate of the cleavage reaction (tetrahedral carbanion intermediate) acting as a true transition-state analog inhibitor. The first residue of the inhibitor that bears the phosphinic group is further stabilized by (i) hydrogen bonds between the free N terminus and the carboxylic groups of Glu337 (2.78 Ǻ), Glu200 (2.81 Ǻ), and Glu393 (3.25 Ǻ) and (ii) hydrophobic interactions between the homophenylalanine side chain and the conserved Phe450 that lines the base of the S1 specificity pocket (the two phenyl rings are almost parallel, with a shortest distance of 3.68 Ǻ). The leucine residue of the inhibitor is stabilized by hydrophobic interactions with Val367 that defines the bottom of a shallow hydrophobic S1’ pocket. Finally, the tryptophan residue is found stacked between Tyr455 (closest distance 3.97 Ǻ) and Tyr892 (closest distance 3.31 Ǻ and almost parallel to it) (Fig. 2). Tyr892 is unique to ERAP2 and this additional interaction is probably sufficient to explain the higher affinity of the inhibitor for ERAP2 compared with ERAP1 or IRAP (Table 1). Overall, the inhibitor configuration helps define the first three specificity pockets of ERAP2. The main residues responsible for the specificity of binding were Phe450 (stabilizes the N-terminal homophenylalanine), Tyr455 (stabilizes the phosphinic group and the C-terminal tryptophan), as well as Glu200, Glu337, and Glu393 (stabilize the N terminus of the inhibitor). All these residues are conserved between ERAP1, ERAP2, and IRAP, something that explains how this inhibitor can effectively target all three enzymes (Fig. S7).
Fig. 2.
Schematic representation of the crystal structure of DG013A bound inside the ERAP2 catalytic site. The mesh indicates the |Fo–Fc| unbiased electron density at 2.5 σ calculated before ligand addition to the structure. DG013A is indicated as yellow sticks. ERAP2 residues within 4 Å of the inhibitor are indicated as gray sticks. Oxygen atoms are shown in red, nitrogen atoms in blue, phosphorus in orange, and Zn(II) in cyan. Hydrogen bonding interactions that stabilize the bound inhibitor are shown as dashed lines.
Inhibition of ERAP1 and ERAP2 in HeLa Cells Leads to Increased Presentation of an Antigenic Epitope.
To evaluate the ability of our compounds to inhibit ERAP1 or ERAP2 and therefore influence antigen presentation in live cells, we used a previously established antigen presentation rescue assay (19). In this assay, delivery of an antigenic peptide precursor into the ER is achieved after transfection by a suitable vector coding for a miniprotein that carries an ER-targeting signal sequence. Upon ER translocation of the miniprotein, the signal sequence is removed by the enzyme Signal Peptidase, releasing an antigenic peptide precursor. Processing of this antigenic peptide precursor by ERAP1 and/or ERAP2 controls mature antigenic epitope generation and loading onto nascent MHCI molecules (HLA-B27 subtype) that subsequently translocate to the cell surface, where they can be detected by a specialized antibody. Overprocessing of the precursor to peptides too small to bind onto MHCI can lead to reduced surface presentation. Using similar assays, researchers have demonstrated that ERAP1-mediated trimming can be absolutely necessary for the generation of many epitopes but can also lead to the destruction of many epitopes by trimming them to lengths that are too small for MHCI binding (11). We incubated HeLa cells transfected with a plasmid vector expressing an ER-targeted minigene with increasing amounts of compounds DG002A, DG013A, and DG013B. After 48 h, cell-surface antigen presentation was measured by flow cytometry and compared with a control in which no inhibitor was present (Fig. 3A and Table 1). The best inhibitor of the three, DG013A, resulted in a 2.5-fold enhancement of antigen presentation, whereas the medium potency inhibitor DG002A had a similar but weaker effect and the poor inhibitor DG013B showed a marginal effect only at the highest concentration tested. Overall, the in vitro potency of the inhibitors correlates well with their effect in the cell-based assay, indicating that their biological effects are indeed mediated by inhibition of ERAP1 and/or ERAP2.
Fig. 3.
Effects of inhibitors on antigen presentation and cytotoxic responses. (A) Enhancement of cell-surface antigen presentation by addition of inhibitors to HeLa cells expressing infected cell protein 47 (ICP47) and HLA-B27. Indicated inhibitors were added on cultured cells immediately after transfection with an ER-targeted miniprotein that after signal sequence cleavage gives rise to an HLA-B27–specific peptide precursor with the sequence ASRHHAFSFR. Cells were incubated for 48 h, and cell-surface translocation of peptide loaded HLA-B27 was followed by flow cytometry using an MHC-specific antibody. Signal was normalized to cells not treated with an inhibitor. (B) Inhibitor dose-dependent enhancement of cell-surface presentation of GSW11 epitope by CT26. CT26 cells were incubated for 48 h with indicated concentrations of inhibitors DG002A, DG013A, or DG013B and assessed for generation and presentation of the GSW11 epitope. GSW11 presentation was detected by the GSW11-specific lacZ-inducible T-cell hybridoma CCD2Z. Error bars indicate SD for each data point calculated from three separate measurements.
Inhibition of Endoplasmic Reticulum Aminopeptidase Associated with Antigen Processing Enhances Presentation of a Tumor Antigen in Mouse Cells.
Given that these inhibitors can enhance antigen presentation of a transfected miniprotein, we next investigated whether they can affect presentation of endogenous antigens and thus whether ERAP1 inhibition can influence antitumor immunity. To this end we used the murine colorectal carcinoma tumor model CT26. Mice challenged with CT26 succumb to the tumor after 20–25 d and generate CD8+ T-cell responses to an immunodominant antigen AH1. Depletion of regulatory T lymphocytes before challenge leads to rejection of the tumor in >90% of mice, with codominant CD8+ T-cell responses directed to both AH1 and a cryptic antigen GSW11. The GSW11-specific CD8+ T cells are responsible for CT26 tumor clearance and cross-protective antitumor immunity (13). Murine cells express endoplasmic reticulum aminopeptidase associated with antigen processing (ERAAP), the mouse homolog of human ERAP1, that shares the same active site and specificity (37). Therefore, we first tested whether our compounds can inhibit murine ERAAP. Indeed, ERAAP was inhibited by our compounds in vitro following a pattern similar to that of human ERAP1 (Table 1 and Fig. S8). We next incubated increasing amounts of each compound with CT26 for 48 h before assessing the generation and cell surface presentation of GSW11 by stimulation of the GSW11-specific T-cell hybridoma, CCD2Z. Titrations of inhibitor amount sufficient to inhibit ERAAP in vitro led to enhanced generation of GSW11, suggesting that ERAAP actively destroys this epitope (Fig. 3B). Furthermore, the T-cell hybridoma response was dose dependent and followed the same trend as the in vitro potency of the inhibitors (DG013A > DG002A ≅ DG002B > DG013B; Table 1). Strikingly, CTL stimulation by the best inhibitor, DG013A, was effective even at very low concentrations (in the nM range), indicating that this cellular system is particularly sensitive to changes in antigen processing. Furthermore, the highest concentration of DG013A resulted in reduced CCD2Z stimulation (and a bell-shaped curve), suggesting that the generation of GSW11 requires some ERAAP trimming function. This finding suggests that it may be possible to pharmacologically fine-tune antigen processing to selectively enhance cytotoxic responses toward cancer cells.
Discussion
Phosphinic Tripeptide Transition-State Analogs as a Promising Route for Controlling Antigen Presentation.
The important biological functions of ERAP1, ERAP2, and IRAP in modulating the adaptive immune response have generated interest in the development of pharmacological tools that can regulate their activity. In an effort to address this, we describe here a very potent inhibitor for these enzymes that is highly active in cellular proof-of-principle assays. This inhibitor (DG013A) was designed using a rational, structure-based approach and can inhibit all three enzymes at the nM level. Cell-based assays suggest that this inhibitor can quantitatively influence antigen presentation in cultured cells, indicating that it is at least partially cell-permeable and can target ERAP1 inside the ER. Additionally, it can enhance cytotoxic T-cell responses to a tumor antigen. Overall, our results validate our rational design approach for these enzymes and suggest that this class of inhibitors may hold promise for the pharmacological manipulation of antigen presentation for applications in the treatment of several diseases ranging from autoimmunity to cancer.
Targeting All Three Antigen-Processing Enzymes.
Although DG013A is about 10-fold more effective for ERAP2 compared with ERAP1 and IRAP, it is still a potent inhibitor for all three enzymes. This phenomenon is probably a direct result of our design strategy, which focuses on structural features in the catalytic and substrate binding site that are conserved between these three highly homologous enzymes (Fig. S7). Given the existence of functional redundancy between these three enzymes, parallel inhibition of ERAP1, ERAP2, as well as IRAP may be a powerful approach for enhancing the inhibitor’s overall biological effect. However, selective inhibition may be desirable for fine-tuning biological effects induced by this type of inhibitor, and this may become possible in the future by exploiting key residue differences between the active sites of these enzymes (see ref. 24 and Fig. S7).
Although the compartmentalized antigen processing in the ER or endosomal compartments by ERAP1/ERAP2 and IRAP has been shown to play a dominant role in antigen presentation, cytosolic aminopeptidases with primarily metabolic functions have, in some cases, been shown to play roles in antigenic peptide generation. Several of the effects described, however, were only evident after the coordinated down-regulation of more than one enzyme and often led to down-regulation of antigen presentation, an effect opposite to the one described in Fig. 3 (38–40). Additionally, other cytosolic aminopeptidases have been found to display either none or very limited effects on antigen presentation and CTL responses (41–43). It is, however, possible that pharmacological inhibition of cytosolic aminopeptidases can also influence antigen processing. This is probably not the main driving factor behind the cellular effects described in this study, as the role of ERAP1 is well established and dominant in these systems and there is a good correlation between in vitro affinity and ED50 values for the two independent cellular assays (13, 19). Regardless of this, and although not addressed in this proof-of-principle study, the selectivity of inhibitors such as the ones described here versus other aminopeptidases in the cell is an important issue to be addressed to minimize undesired off-target effects relating either to antigen processing or other cellular functions especially in view of potential clinical applications.
Antigenic Epitope Destruction Versus Epitope Generation.
The effect of the inhibitors on antigen presentation by HeLa and CT26 cells provides important clues regarding the role of ERAP1 in these two systems. It has been previously demonstrated that ERAP1 plays an active role in destroying antigenic epitopes, although this has not been studied extensively. The effect of the inhibitors on HeLa cells presenting the SRHHAFSFR epitope bound on the HLA-B27 MHCI allele indicate that the role of ERAP1 in this system is primarily destructive. This has been recently validated in this system by a separate study that indicated that an ERAP1 allele with higher enzymatic activity leads to lower presentation of virtually all HLA-B27 epitopes tested (44). Furthermore, enhancement of the GSW11-specific T-cell response in the CT26 tumor cells also corroborates the destructive role of ERAAP. Interestingly the bell-shaped curve seen for DG013A in Fig. 3 can be explained by ERAAP’s dual role in this system: some ERAAP activity is important for the generation of GSW11 epitope, whereas additional activity leads to epitope destruction. This dual function of ERAP1 and its mouse homolog ERAAP generates additional opportunities for the fine-tuning of antigen presentation by careful pharmacological inhibition. Overall, the effects of these inhibitors on antigen presentation are expected to be epitope-dependent; epitopes normally destroyed by the enzyme will have increased presentation in the presence of inhibitors, whereas epitopes requiring enzymatic processing for their generation will have decreased presentation in the presence of inhibitors. This may lead to changes in the repertoire of antigenic peptides presented by a cell, affecting immune responses. In this context, these inhibitors may be very useful in seemingly contradictory pharmaceutical approaches, although care should be taken to first study their specific effects in each biological system.
Potential Applications in Cancer Immunotherapy—Possible Shortcomings.
The inhibitors described in this study and in particular DG013A are of sufficient potency to warrant examination in in vivo systems for possible therapeutic effects. Possible applications may vary depending on how the epitope of interest is normally processed (generated or destroyed) by the targeted enzymes. Furthermore, dose-dependent effects such as the ones seen for the GSW11 epitope may be especially useful when subtle regulation of antigen presentation is desirable. Such therapeutic opportunities have recently been demonstrated using the moderate inhibitor leucinethiol which, upon administration, caused modest tumor growth attenuation in some established tumors (13). The high potency of DG013A may therefore allow it to induce highly effective cytotoxic responses useful in cancer immunotherapy either by enhancing existing or by inducing novel CTL or NK responses similar to ones found by the genetic down-regulation of ERAP1 (2, 5, 13). Additionally, ERAP1 down-regulation by inhibitors may initiate alternative, nonclassical MHCI responses (12). Finally, ERAP1 down-regulation may reduce autoimmune reactions to self-mimic peptides that initiate or sustain autoimmunity. Significant care should be taken, however, because large alterations of the antigen presentation pathway can in theory elicit unwanted side effects, either by assisting the evasion of pathogens or by itself creating autoimmunogenic CTL responses. Careful pharmacokinetic control, as well as the generation of highly selective inhibitors that preferentially target only one of the three enzymes, may help regulate this pathway more selectively and manipulate antigen presentation with the necessary precision.
Experimental Procedures
Inhibitor Synthesis.
For the synthesis of DG002 and DG013, the common building block precursor phosphinic block Boc-(R)-hPhe[PO(OAd)-CH2]-(R,S)-LeuOH 7 was synthesized based on a previously described protocol (45) (Fig. S1). More details about the synthesis can be found in the SI Experimental Procedures.
Enzymatic and Cell-Based Assays.
In vitro enzymatic assays using model fluorigenic substrates and peptides have been described before (24, 35). The HLA-B27 rescue assay has been described (18, 19). Furthermore, the method for assaying GSW11 epitope surface presentation by CT26 cells using T-cell hybridoma CCD2Z has been described (13). Both cell-based assays were performed in the presence of inhibitors for 48 h before epitope surface presentation quantification.
X-Ray Crystallography.
For cocrystallization experiments, the ERAP2(N392K) naturally occurring variant was expressed, purified, and crystallized as previously described (18). Crystallization conditions were identical as the ones used for free ERAP2 (18, 32), with the difference that no free amino acids were present in the mixture, but instead fourfold excess of the inhibitor was present. The crystal was practically isomorphous with the ERAP2 crystal described before (32) and belongs to the space group P21, a = 75.1 Å, b = 134.8 Å, c = 128.7 Å, and β = 90.3°. The structure was determined by molecular replacement using the ERAP2 coordinates (PDB ID code 4E36). Atomic coordinates and structure factors for the ERAP2-DG013A crystal structure have been deposited in PDB (www.pdb.org) (PDB ID code 4JBS).
Additional Methods.
Additional methods and details can be found in SI Experimental Procedures.
Supplementary Material
Acknowledgments
This research was financed by the European Union (European Social Fund) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework: Research Funding Program of the General Secretariat for Research & Technology (Grant ERC-14 to E. Stratikos). Funding was also provided by the FP7–Marie Curie–Industry-Academia Partnerships and Pathways (IAPP)–Novel Tools for Crystallization of Macromolecules project (European Commission Grant 217979 to E. Saridakis), and the Cancer Research United Kingdom (to E.J.). Partial funding for crystallographic data collection from European Molecular Biology Laboratory–Hamburg at the Doppel-Ring Speicher storage ring, Deutsches Elektronen Synchrotron, Germany, and the Swiss Light Source, Paul Scherrer Institut, Villigen, Switzerland (Grants 226716 and 20111173) is acknowledged.
Footnotes
Conflict of interest statement: D.G. and E. Stratikos declare that they have filed a patent application that includes compounds described in this article.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4JBS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1309781110/-/DCSupplemental.
References
- 1.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 2.Hammer GE, Gonzalez F, James E, Nolla H, Shastri N. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2007;8(1):101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 3.Blanchard N, et al. Immunodominant, protective response to the parasite Toxoplasma gondii requires antigen processing in the endoplasmic reticulum. Nat Immunol. 2008;9(8):937–944. doi: 10.1038/ni.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci USA. 2006;103(24):9202–9207. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cifaldi L, et al. Natural killer cells efficiently reject lymphoma silenced for the endoplasmic reticulum aminopeptidase associated with antigen processing. Cancer Res. 2011;71(5):1597–1606. doi: 10.1158/0008-5472.CAN-10-3326. [DOI] [PubMed] [Google Scholar]
- 6.Tenzer S, et al. Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat Immunol. 2009;10(6):636–646. doi: 10.1038/ni.1728. [DOI] [PubMed] [Google Scholar]
- 7.Cascio P, Hilton C, Kisselev AF, Rock KL, Goldberg AL. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 2001;20(10):2357–2366. doi: 10.1093/emboj/20.10.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evnouchidou I, Papakyriakou A, Stratikos E. A new role for Zn(II) aminopeptidases: Antigenic peptide generation and destruction. Curr Pharm Des. 2009;15(31):3656–3670. doi: 10.2174/138161209789271816. [DOI] [PubMed] [Google Scholar]
- 9.Blanchard N, Shastri N. Coping with loss of perfection in the MHC class I peptide repertoire. Curr Opin Immunol. 2008;20(1):82–88. doi: 10.1016/j.coi.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haroon N, Inman RD. Endoplasmic reticulum aminopeptidases: Biology and pathogenic potential. Nat Rev Rheumatol. 2010;6(8):461–467. doi: 10.1038/nrrheum.2010.85. [DOI] [PubMed] [Google Scholar]
- 11.York IA, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat Immunol. 2002;3(12):1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 12.Nagarajan NA, Gonzalez F, Shastri N. Nonclassical MHC class Ib-restricted cytotoxic T cells monitor antigen processing in the endoplasmic reticulum. Nat Immunol. 2012;13(6):579–586. doi: 10.1038/ni.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James E, Bailey I, Sugiyarto G, Elliott T. Induction of protective antitumor immunity through attenuation of ERAAP function. J Immunol. 2013;190(11):5839–5846. doi: 10.4049/jimmunol.1300220. [DOI] [PubMed] [Google Scholar]
- 14.Evans DM, et al. Spondyloarthritis Research Consortium of Canada (SPARCC) Australo-Anglo-American Spondyloarthritis Consortium (TASC) Wellcome Trust Case Control Consortium 2 (WTCCC2) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 2011;43(8):761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cagliani R, et al. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet. 2010;19(23):4705–4714. doi: 10.1093/hmg/ddq401. [DOI] [PubMed] [Google Scholar]
- 16.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 2010;42(11):1005–1009. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta AM, Jordanova ES, Kenter GG, Ferrone S, Fleuren GJ. Association of antigen processing machinery and HLA class I defects with clinicopathological outcome in cervical carcinoma. Cancer Immunol Immunother. 2008;57(2):197–206. doi: 10.1007/s00262-007-0362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evnouchidou I, et al. A common single nucleotide polymorphism in endoplasmic reticulum aminopeptidase 2 induces a specificity switch that leads to altered antigen processing. J Immunol. 2012;189(5):2383–2392. doi: 10.4049/jimmunol.1200918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evnouchidou I, et al. Cutting Edge: Coding single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 can affect antigenic peptide generation in vitro by influencing basic enzymatic properties of the enzyme. J Immunol. 2011;186(4):1909–1913. doi: 10.4049/jimmunol.1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saveanu L, et al. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325(5937):213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- 21.Segura E, Albiston AL, Wicks IP, Chai SY, Villadangos JA. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc Natl Acad Sci USA. 2009;106(48):20377–20381. doi: 10.1073/pnas.0910295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Georgiadou D, et al. Placental leucine aminopeptidase efficiently generates mature antigenic peptides in vitro but in patterns distinct from endoplasmic reticulum aminopeptidase 1. J Immunol. 2010;185(3):1584–1592. doi: 10.4049/jimmunol.0902502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim Biophys Acta. 2005;1751(1):9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 24.Zervoudi E, et al. Probing the S1 specificity pocket of the aminopeptidases that generate antigenic peptides. Biochem J. 2011;435(2):411–420. doi: 10.1042/BJ20102049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanaseki T, Blanchard N, Hammer GE, Gonzalez F, Shastri N. ERAAP synergizes with MHC class I molecules to make the final cut in the antigenic peptide precursors in the endoplasmic reticulum. Immunity. 2006;25(5):795–806. doi: 10.1016/j.immuni.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papakyriakou A, et al. Novel selective inhibitors of aminopeptidases that generate antigenic peptides. Bioorg Med Chem Lett. 2013;23(17):4832–4836. doi: 10.1016/j.bmcl.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 27.Albiston AL, et al. Identification and characterization of a new cognitive enhancer based on inhibition of insulin-regulated aminopeptidase. FASEB J. 2008;22(12):4209–4217. doi: 10.1096/fj.08-112227. [DOI] [PubMed] [Google Scholar]
- 28.Stratikos E, Stern LJ. Antigenic peptide trimming by ER aminopeptidases—Insights from structural studies. Mol Immunol. 2013;55(3-4):212–219. doi: 10.1016/j.molimm.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yiotakis A, Georgiadis D, Matziari M, Makaritis A, Dive V. Phosphinic peptides: Synthetic approaches and biochemical evaluation as Zn-metalloprotease inhibitors. Curr Org Chem. 2004;8(12):1135–1158. [Google Scholar]
- 30.Dive V, et al. Phosphinic peptides as zinc metalloproteinase inhibitors. Cell Mol Life Sci. 2004;61(16):2010–2019. doi: 10.1007/s00018-004-4050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evnouchidou I, et al. The internal sequence of the peptide-substrate determines its N-terminus trimming by ERAP1. PLoS ONE. 2008;3(11):e3658. doi: 10.1371/journal.pone.0003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birtley JR, Saridakis E, Stratikos E, Mavridis IM. The crystal structure of human endoplasmic reticulum aminopeptidase 2 reveals the atomic basis for distinct roles in antigen processing. Biochemistry. 2012;51(1):286–295. doi: 10.1021/bi201230p. [DOI] [PubMed] [Google Scholar]
- 33.Yiotakis A, Vassiliou S, Jirácek J, Dive V. Protection of the hydroxyphosphinyl function of phosphinic dipeptides by adamantyl. Application to the solid-phase synthesis of phosphinic peptides. J Org Chem. 1996;61(19):6601–6605. doi: 10.1021/jo9603439. [DOI] [PubMed] [Google Scholar]
- 34.Chen H, et al. Phosphinic derivatives as new dual enkephalin-degrading enzyme inhibitors: Synthesis, biological properties, and antinociceptive activities. J Med Chem. 2000;43(7):1398–1408. doi: 10.1021/jm990483l. [DOI] [PubMed] [Google Scholar]
- 35.Evnouchidou I, Berardi MJ, Stratikos E. A continuous fluorigenic assay for the measurement of the activity of endoplasmic reticulum aminopeptidase 1: Competition kinetics as a tool for enzyme specificity investigation. Anal Biochem. 2009;395(1):33–40. doi: 10.1016/j.ab.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen TT, et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18(5):604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serwold T, Gonzalez F, Kim J, Jacob R, Shastri N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419(6906):480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 38.Lévy F, et al. The final N-terminal trimming of a subaminoterminal proline-containing HLA class I-restricted antigenic peptide in the cytosol is mediated by two peptidases. J Immunol. 2002;169(8):4161–4171. doi: 10.4049/jimmunol.169.8.4161. [DOI] [PubMed] [Google Scholar]
- 39.Kim E, Kwak H, Ahn K. Cytosolic aminopeptidases influence MHC class I-mediated antigen presentation in an allele-dependent manner. J Immunol. 2009;183(11):7379–7387. doi: 10.4049/jimmunol.0901489. [DOI] [PubMed] [Google Scholar]
- 40.Altrich-VanLith ML, et al. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J Immunol. 2006;177(8):5440–5450. doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 41.Towne CF, et al. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J Immunol. 2005;175(10):6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 42.Towne CF, York IA, Watkin LB, Lazo JS, Rock KL. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J Immunol. 2007;178(11):6923–6930. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 43.Towne CF, et al. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J Immunol. 2008;180(3):1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 44.Seregin SS, et al. Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity. 2013 doi: 10.3109/08916934.2013.819855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Georgiadis D, Vazeux G, Llorens-Cortes C, Yiotakis A, Dive V. Potent and selective inhibition of zinc aminopeptidase A (EC 3.4.11.7, APA) by glutamyl aminophosphinic peptides: Importance of glutamyl aminophosphinic residue in the P1 position. Biochemistry. 2000;39(5):1152–1155. doi: 10.1021/bi9922345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.