Significance
Bacteria stimulate host cells in part via secreted products, some of which are packaged in membrane vesicles (MV). MV released by the major human pathogen Mycobacterium tuberculosis (Mtb) carry lipoprotein LpqH, a major agonist for host Toll-like receptor 2 (TLR2). This study identifies a gene, rv0431, which appears to regulate mycobacterial MV formation, and therefore we suggest it be named “vesiculogenesis and immune response regulator” (virR). This gene encodes a protein that includes a unique fold, as determined by NMR spectroscopy, and a disordered domain suggestive of participation in a higher-order complex. By restraining the release of most of the material released by Mtb that activates host cells through TLR2, VirR reduces Mtb’s immunostimulatory potential and increases its virulence.
Keywords: TLR2, macrophages
Abstract
Mycobacterium tuberculosis (Mtb) restrains immune responses well enough to escape eradication but elicits enough immunopathology to ensure its transmission. Here we provide evidence that this host–pathogen relationship is regulated in part by a cytosolic, membrane-associated protein with a unique structural fold, encoded by the Mtb gene rv0431. The protein acts by regulating the quantity of Mtb-derived membrane vesicles bearing Toll-like receptor 2 ligands, including the lipoproteins LpqH and SodC. We propose that rv0431 be named “vesiculogenesis and immune response regulator.”
Mycobacterium tuberculosis (Mtb) has coevolved with its human hosts from the time of our hominid ancestors (1, 2). Millions of years of pathogenicity and an estimated cumulative toll of one billion human lives (3, 4) reflect Mtb’s ability to elicit a balanced immune response. The host response to Mtb is vigorous enough to facilitate aerosol transmission by liquefying infected lung in 5–10% of those infected but is insufficient to sterilize infection in the remainder of the one-third of the human population that is estimated to carry Mtb in a latent form (5, 6).
Mtb infection illustrates the complex balance between immunity and immunopathology that can dictate the host’s fate in infectious disease (7, 8). Unlike most bacterial pathogens, Mtb devotes the most conserved sequences in its genome to encode human T-cell epitopes (9). Mtb activates macrophages, its primary niche, to a heightened antimicrobial and proinflammatory state but not maximally, as evidenced by the fact that knocking out particular genes in the host (10–12) or the pathogen (13–15) can lead to a greater level of macrophage activation (16).
Recently, a forward genetic screen using a library of more than 10,000 loss-of-function transposon mutants of Mtb identified rv0431 as an endogenous regulator of Mtb’s immunostimulatory potential (17). Infection with Mtb rv0431:Tn led to markedly increased release of TNF-α, IL-12 p40, and IL-6 from primary mouse macrophages. Mtb rv0431:Tn grew normally in vitro but was attenuated in mice, suggesting that its proliferation may have been suppressed by a heightened host inflammatory or immune response. However, as with most other Mtb genes that have been found to restrain the host response, the mechanism by which rv0431 did so was unknown.
Mtb communicates with host cells by binding cellular receptors and by releasing products that do so. Toll-like receptor 2 (TLR2) is one prominent host-cell receptor through which Mtb and products such as the 19-kDa lipoprotein LpqH trigger macrophage responses (18). Mtb secretes proteins through four known pathways: general secretion SecA1, general secretion SecA2, twin arginine transport (Tat), and type VII (19, 20). Lipids released from Mtb can traffic out of infected macrophages in macrophage-derived exosomes (21). Recently, Mtb was discovered to be among the various bacteria that can release membrane vesicles (MV) (22). Mtb’s MV are spheres, 60–200 nm in diameter, that are rich in immunomodulatory factors (22), including LpqH, and another TLR2 agonist, SodC (23). Little is known about how bacteria form and release MV, although the formation of MV may involve the packaging of proteins and lipoproteins secreted by one or more of the canonical pathways. To our knowledge, no genes have yet been identified that regulate the quantity or content of mycobacterial MV.
Here, we identify Rv0431 as a regulator of Mtb’s MV. Through this means, Rv0431 exerts extensive control over the extent of TLR2-dependent stimulation of macrophages and over TLR2-dependent restriction of Mtb’s growth in the host. Based on evidence presented below, we propose that rv0431 be named “vesiculogenesis and immune response regulator” (virR). Characterization of VirR as a protein that controls Mtb’s MV and their inflammatory and immunostimulatory potential sets the stage for studies to explore the molecular machinery involved and the implications for immunization.
Results
VirR Deficiency Augments Responses of Mouse and Human Macrophages.
Earlier work demonstrated that the transposon disrupting virR had inserted between codons 121 and 122 (17). However, it remained to be established whether the VirR protein was absent, truncated, or modified with a neo-C terminus, whether the mutant Mtb had broad effects on the mouse macrophage transcriptome, and whether the hyperstimulatory phenotype extended to human macrophages.
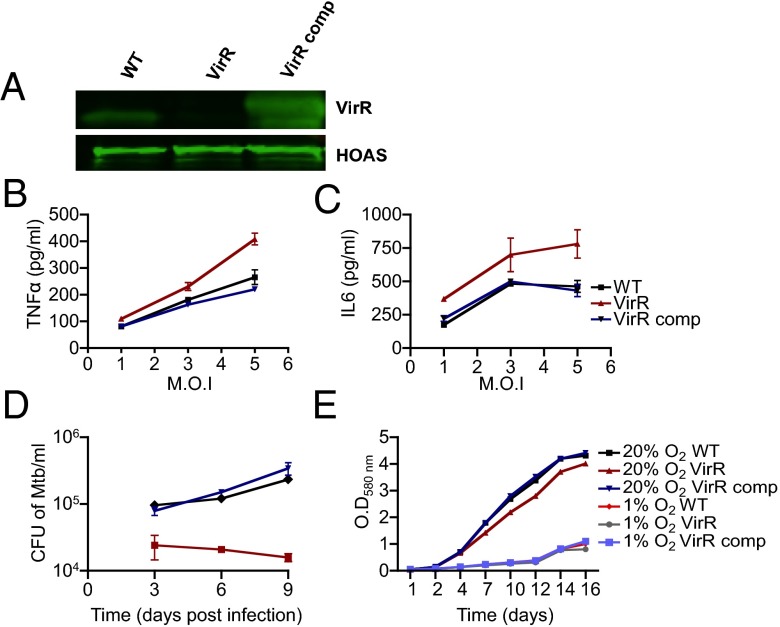
To address these questions, we began by raising polyclonal antisera to purified recombinant VirR. Immunoblot demonstrated that VirR migrated at ∼23 kDa, near its predicted molecular mass of 17 kDa, in lysates of WT Mtb but was undetectable in the lysates of the transposon mutant. Thus, the transposon insertion resulted in a null allele. Complementation with a WT allele of virR under the strong heat shock protein 60 (hsp60) promoter in an episomal plasmid led to overexpression (Fig. 1A), which proved useful in subsequent experiments to evaluate a protein dose effect. Complementation reverted the hyperinflammatory phenotype of VirR-deficient Mtb in primary mouse bone marrow-derived macrophages (BMM) and reverted its attenuation in C57BL/6 mice (SI Appendix, Fig. S1).
Fig. 1.
Mtb with transposon-disrupted Rv0431 lacks VirR protein and is attenuated and hyperinflammatory in primary human macrophages. (A) Whole-cell lysates of WT, VirR-deficient (Tn:Rv0431), and VirR-complemented (Tn:Rv0431::Rv0431) Mtb strains were immunoblotted with antiserum against VirR. The lysates also were blotted with antiserum against hydroxyoxoadipate synthase (HOAS) as a loading control. (B and C) Human macrophages were infected with WT, VirR-deficient, or VirR-complemented Mtb at an MOI of 1, 3, and 5, and induction of TNF-α (B) and IL-6 (C) was assessed 48 h postinfection. Data points represent the mean ± SD of samples in triplicate, and results are shown from one representative experiment of two. (D) Human macrophages cultured and maintained at 10% O2 were infected with WT, VirR-deficient, or VirR-complemented Mtb at MOI 0.1, and survival was assessed. Data points represent the mean ± SD of samples in triplicate, and results are shown from one representative experiment of five. (E) Growth of VirR-deficient Mtb was compared with WT and VirR-complemented Mtb in liquid 7H9 medium at either 1% O2 (hypoxic) or 20% O2 (hyperoxic for tissue).
We used global transcriptomic RNAseq profiles of uninfected BMM and BMM infected with WT or VirR-deficient Mtb to survey the extent of VirR’s restriction of Mtb’s immunostimulatory impact on macrophages. In three independent experiments at 8 h of infection at a multiplicity of infection (MOI) of 5, we observed increased levels of transcripts of 839 genes in response to WT Mtb infection. However, there was minimal differential impact on the macrophage transcriptome upon infection with VirR-deficient Mtb versus WT Mtb. Of the 839 genes, only 12 genes were up-regulated by ≥1.5-fold by infection with VirR-deficient Mtb relative to infection with WT Mtb (P ≤ 0.05) (SI Appendix, Table S1).These 12 genes did not include the genes encoding for cytokines and chemokines whose increased release from VirR-deficient infected macrophages was demonstrated by ELISA. The minimal differential impact on the macrophage transcriptome in response to infection with VirR-deficient Mtb might reflect that we analyzed the transcriptome at a single time point and/or suggest that posttranscriptional regulation played a role in increased expression of TNF-α and IL-12 p40.
Next, we tested the impact of VirR-deficient Mtb on primary human macrophages cultured at a physiologic tissue O2 tension of 10% (24). VirR-deficient Mtb hyperinduced TNF-α and IL-6 in the human cells in a dose-dependent manner (Fig. 1 B and C). The cytokine levels were elevated above those seen following infection with WT Mtb, even though the human macrophages sustained a far lower burden of VirR-deficient Mtb than WT Mtb (Fig. 1D), with the difference amounting to 1 log10 fewer cfu by day 9 postinfection. The macrophage monolayers were confluent after infection with either strain of Mtb, indicating that the reduced bacterial numbers did not result from fewer viable macrophages (SI Appendix, Fig. S2A). Because the intracellular O2 tensions may have been even lower than 10%, we tested the impact of O2 tension on Mtb growth rate in axenic cultures. VirR-deficient Mtb replicated at the same rate as WT Mtb at 1% oxygen and at 20% O2 (Fig. 1E). Thus, the inability of VirR-deficient Mtb to replicate in human macrophages did not result from a greater dependence on O2. We tested whether VirR-deficient Mtb might be more susceptible than WT Mtb to acidification or to oxidative or nitrosative stress, using reported methods (25), but we found no differences in its susceptibility compared with WT Mtb (SI Appendix, Fig. S2 B–D). Thus, VirR-deficient Mtb does not appear to be more sensitive than WT Mtb to several of the antimicrobial effectors of macrophages but does appear to activate human macrophages to a greater extent than WT Mtb.
VirR Regulates Release of Immunomodulatory Factors.
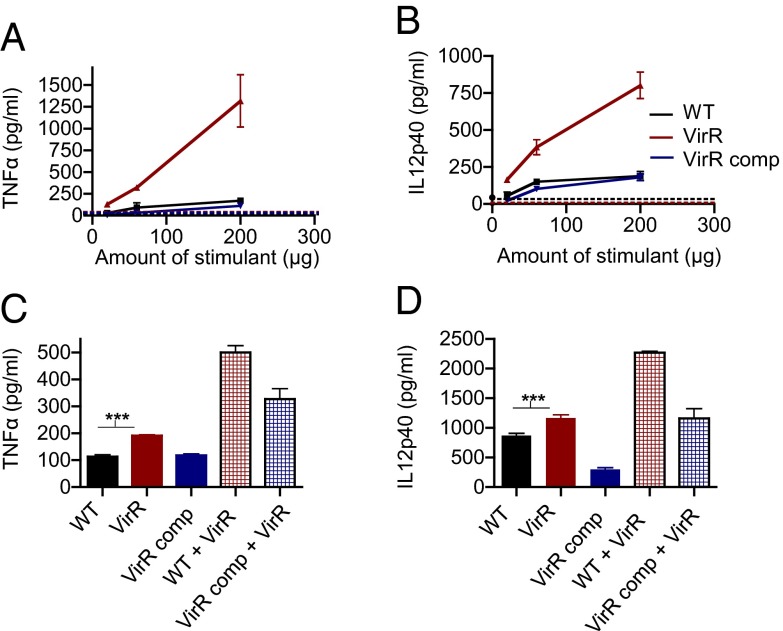
To analyze how VirR affects Mtb’s ability to stimulate macrophages, we tested if medium conditioned by VirR-deficient Mtb would stimulate macrophages more potently than medium conditioned by WT Mtb. WT, VirR-deficient, and VirR-complemented Mtb were grown in 7H9 medium in the absence of detergent. Cell-free culture filtrate was concentrated using an ultrafiltration system with a 3-kDa cutoff and was added to BMM in graded doses. Culture filtrate from VirR-deficient Mtb proved to be far more potent than WT culture filtrate in stimulating mouse macrophages to produce TNF-α and IL-12 p40 (Fig. 2A) and human macrophages to produce TNF-α and IL-6 (SI Appendix, Fig. S3).
Fig. 2.
Culture filtrate derived from VirR-deficient Mtb is hyperinflammatory in BMM. (A and B) WT BMM were treated with culture filtrate derived from WT Mtb, VirR-deficient Mtb, or VirR-complemented Mtb, in varying doses. The flow-through was used to assess background cytokine induction (dashed lines). TNF-α levels (A) and IL-12 p40 levels (B) were measured in the macrophage supernatants 24 h posttreatment. Data points represent the mean ± SD of triplicate wells from one representative experiment of three. (C and D) WT BMM were treated with culture filtrate derived from WT, VirR-deficient, or VirR-complemented Mtb. In addition, BMM were treated with a [1:1] mixture of culture filtrate derived from VirR-deficient Mtb and culture filtrate derived from either WT or VirR-complemented Mtb. TNF-α (C) and IL-12 p40 (D) levels in the macrophage supernatants were measured 24 h posttreatment. Data represent the mean ± SD of triplicate wells from one representative experiment of two. ***P < 0.001.
To explore whether the increased release of immunostimulatory factors might represent leakage across compromised cell walls, we assessed bacterial morphology and sensitivity to SDS and lipophilic drugs and found no differences between VirR-deficient and WT Mtb (SI Appendix, Fig. S4). Furthermore, the protein concentrations of the culture filtrates of VirR-deficient, WT, and complemented strains were comparable (7, 7, and 8 μg/mL, respectively). Thus, VirR-deficient Mtb had normal overall cell-wall integrity, and VirR deficiency did not lead to an overall increase in the amount of protein released by Mtb.
To distinguish increased release of immunostimulatory factors from decreased release of immunosuppressive factors—perhaps including VirR itself—we exposed BMM to mixtures of detergent-free culture filtrate from VirR-deficient and WT Mtb. Adding culture filtrate from VirR-deficient Mtb to filtrate from WT Mtb resulted in increased immunostimulation over an equal concentration of the latter filtrate alone (Fig. 2B). Thus, it appeared that VirR-deficient Mtb released a greater quantity of immunostimulatory factors than WT Mtb, not a lesser quantity of inhibitory factors.
Next, we investigated whether VirR might suppress the production of immunostimulatory factors by Mtb at the transcriptional level, so that the immunostimulatory factors might be released to a greater extent when VirR was lacking. Earlier, we performed whole-genome transcriptomic analysis of macrophages infected with VirR-deficient Mtb; now we performed the analysis on the Mtb itself, again using RNAseq. No significant differences were detected between the transcriptomes of WT and VirR-deficient Mtb (Gene Expression Omnibus database accession no. GSE50050).
We next investigated whether VirR might modulate specific secretory pathways. As noted, mycobacteria are known to secrete proteins by four defined mechanisms: SecA1, SecA2, Tat, and type VII. To investigate whether VirR might regulate these pathways, we used immunoblots to compare the quantities of a representative protein released by each pathway in the culture filtrate of VirR-deficient and WT Mtb. To ensure that the culture filtrate was not contaminated with cytosol from dying cells, we also assessed the levels of the cytosolic enzyme dihydrolipoamide acyltransferase (DlaT) (26, 27). DlaT was detectable in cytosol but not in the culture filtrate. The culture filtrates of the two strains contained comparable levels of antigen 85B (Ag85B) and early secreted antigenic target of 6 kDa (ESAT6), indicating that VirR deficiency did not globally alter the SecA1 and type VII pathways (SI Appendix, Fig. S5). We could detect HspX and KatG, both SecA2 substrates, and BlaC, a Tat substrate, in the cytosol of each strain, where their levels were comparable. Both proteins were below the level of detection in culture filtrates from both strains. Thus, we could find no evidence that VirR deficiency globally enhanced secretion through SecA2 or Tat pathways. It is possible that VirR might regulate the release of individual substrates of these pathways other than those we measured.
In none of these studies could we detect VirR itself in the culture filtrate (SI Appendix, Fig. S5). In considering how VirR might function within the cell to suppress release of immunostimulatory factors outside the cell, we sought clues from VirR’s relationship to the cell membrane and its structure.
Topological and Structural Features of VirR.
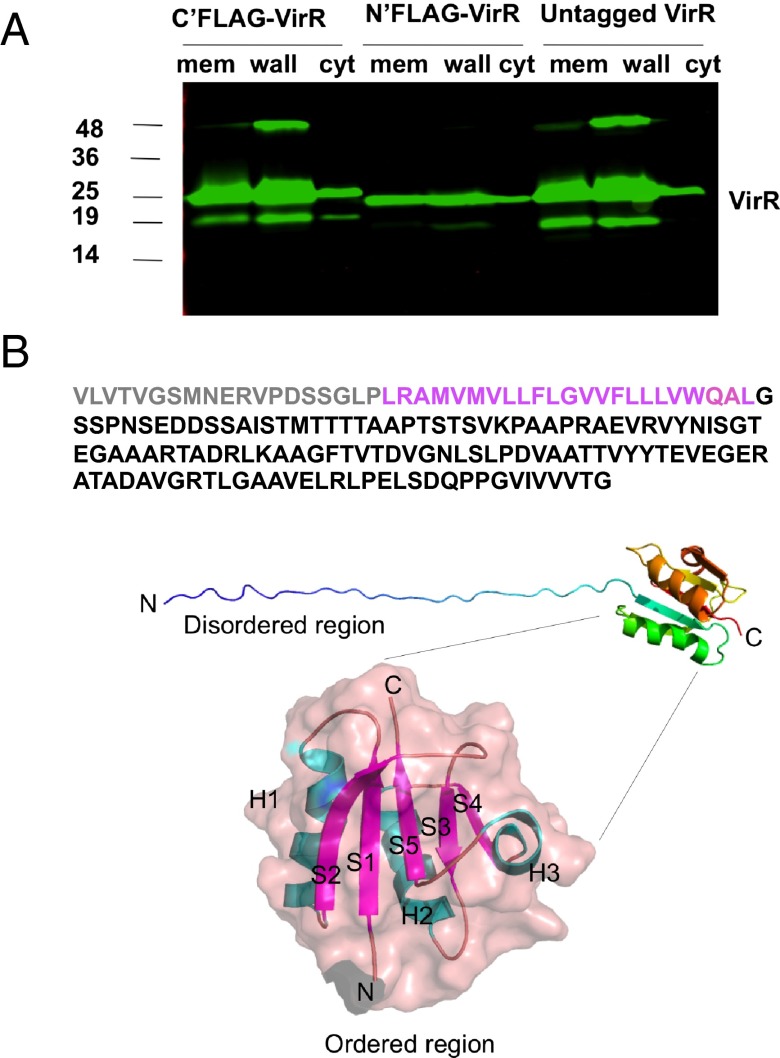
Bioinformatic analysis of VirR’s primary sequence identified no homologs outside of actinobacteria but predicted that VirR is a type I integral membrane protein, with its N terminus in the periplasm and C terminus in the cytoplasm. Immunoblotting subcellular fractions of WT Mtb with anti-VirR antibodies revealed a predominant association of VirR with the cell envelope fraction, which includes plasma membrane, periplasm, and outer membrane. However, we also detected VirR in the cytosolic fraction (Fig. 3A). VirR’s distribution was similar in WT Mtb and in VirR-deficient Mtb complemented with untagged VirR or with VirR bearing a FLAG tag at either terminus, although the expression of a FLAG-tag in the N terminus reduced expression of the protein (Fig. 3A). If VirR has a single predominant subcellular localization, then its distribution in both cell envelope and cytosol fractions would suggest that VirR may be reversibly associated with the inner surface of the plasma membrane.
Fig. 3.
Subcellular localization of VirR and partial structure of VirR. (A) Immunoblot of cellular fractions of the complemented strains expressing VirR with a C terminus FLAG-tag (C’FLAG-Rv0431), N terminus FLAG-tag (N’FLAG-Rv0431), or no tag (Untagged Rv0431) using antiserum against VirR. VirR appears predominantly as an ∼23-kDa protein. The band at 48 kDa was confirmed to be a homodimer of VirA by peptide mass fingerprinting. The band at 19 kDa is speculated to be a degradation product of VirR. (B) (Upper) The amino acid sequence of full-length ViR highlights the extensive disordered region shown in gray (truncated in the recombinant protein) and purple (disordered in the NMR structure). (Middle) The NMR structure of the truncated protein is shown in cartoon view and is colored in rainbow from N-terminal blue to C-terminal red. (Lower) The ordered region is shown as a transparent surface with the cartoon view superimposed with helices (H) in cyan and β sheets (S) in purple. C, C terminus; N, N terminus.
Because the experimental evidence challenged the bioinformatic prediction that VirR may be a single-pass transmembrane protein, we sought to define VirR’s relationship to the plasma membrane by topologic studies with fusion proteins. Terminal fusion constructs coupled varying lengths of VirR with alkaline phosphatase (PhoA) or β galactosidase (LacZ) in their C termini (SI Appendix, Fig. S6 A and B). Full-length VirR, a truncated form lacking the C-terminal 56 amino acids, and a more severely truncated form consisting only of residues 1–42 (that is, the first 20 amino acids and the predicted transmembrane domain) were each cloned as fusions with either of the reporters. The resulting six constructs were introduced individually into Mycobacterium smegmatis. All three strains transformed with PhoA constructs formed white colonies, suggesting that none of the constructs localized to the periplasm and that, instead, both termini of VirR were in the cytoplasm (SI Appendix, Fig. S6B). As a positive control, M. smegmatis expressing Ag85B C-terminally fused with PhoA yielded blue colonies. All the LacZ fusion constructs gave rise to blue colonies, again suggesting that both termini of the protein were cytosolic. Taken together, the subcellular fractionation and topologic analyses suggested that VirR is located in the cytosol, where it is associated with the plasma membrane without crossing it.
Efforts to crystallize recombinant VirR were confounded by its seemingly unlimited solubility. This feature suggested that VirR contains an intrinsically disordered region. Such regions may acquire structure in the presence of a binding partner (28, 29). Thus, we turned to NMR spectroscopy, using a truncated protein in which the highly hydrophobic stretch spanning amino acid residues 20–42 was deleted, along with the preceding 19 residues in the N terminus. The truncated protein, consisting of residues 42–164, was expressed in the pET28b vector system with a hexahistidine (His6) tag in its N terminus to aid in purification from recombinant Escherichia coli. First, we explored the biologic properties of truncated VirR by complementing VirR-deficient Mtb with the same construct, checking its distribution in cell fractions, infecting BMM, and measuring their release of cytokines. Deletion of residues 1–42 led to a purely cytosolic distribution (SI Appendix, Fig. S6C). The purely cytosolic form of VirR retained immunomodulatory capacity (SI Appendix, Fig. S6D).
With the reassurance that the purely cytosolic, truncated form of VirR was functional, we resolved its structure at 0.9-Å resolution (Fig. 3B). Consistent with its solution behavior, the first 35 amino acid residues in the N terminus of the truncated C-terminal two-thirds of the holoprotein were intrinsically disordered. The remainder of the protein, corresponding to residues 78–164 of the holoprotein, formed a globular structure with a unique fold. The fold consisted of a five-stranded β-sheet core flanked by two α-helices on one side and one α-helix on the other. Further details of the NMR analysis are shown in SI Appendix, Figs. S7–S11 and SI Appendix, Table S2.
Proteins Associated with VirR.
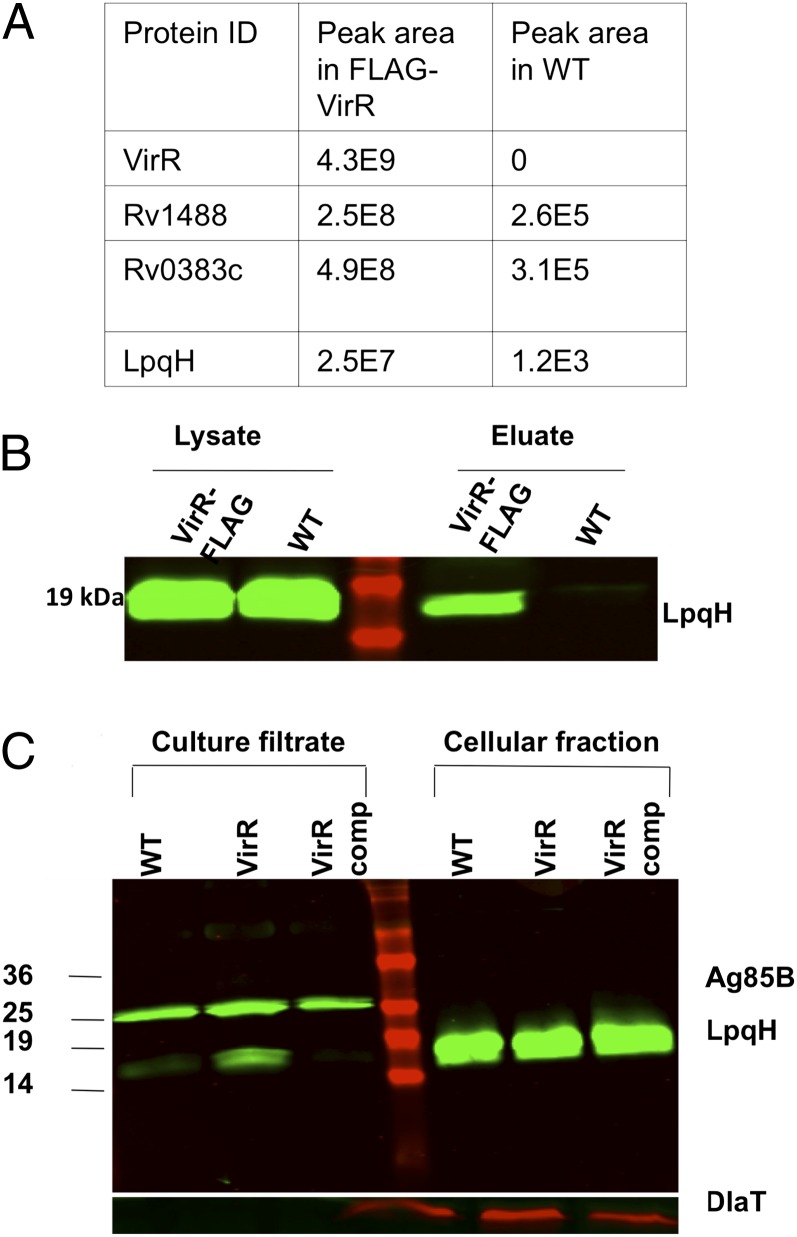
Given that VirR is predominantly cytosolic, albeit membrane associated, and contains a disordered domain suggestive of a binding partner, we considered that VirR might bind immunostimulatory factors within the cell and regulate their release. To investigate this possibility, we complemented the VirR-deficient strain with full-length FLAG-VirR with the C terminus tagged, confirmed that this complementation reverted the hyperstimulatory phenotype (SI Appendix, Fig. S6C), and used anti-FLAG antibody for coimmunoprecipitation. WT Mtb served as a negative control, because its VirR was untagged. Peptide mass fingerprinting of the immunoprecipitated proteins revealed VirR as the most abundant source of peptides. Three other proteins were relatively abundant in the anti-FLAG immunoprecipitate. Two are uncharacterized: Rv1488 and Rv0383c. The third was LpqH, the 19-kDa lipoprotein characterized as a major TLR2 agonist of Mtb (Fig. 4A) (18). We confirmed the association of VirR with LpqH using coimmunoprecipitation of native VirR from WT Mtb with anti-VirR antibody (Fig. 4B).
Fig. 4.
VirR is associated with LpqH, and culture filtrate derived from VirR-deficient Mtb is enriched in LpqH. (A) Lysates of FLAG-tagged VirR expressing Mtb and WT Mtb, grown to midlog phase, were immunoprecipitated with anti-FLAG antibody. The eluates were reduced, denatured, and subjected to SDS/PAGE, followed by mass spectrometry for identification of the binding partners. As indicated by the peak areas shown in the table, VirR was pulled down only in the FLAG-Rv0431 lysates, and LpqH was detected to a significantly greater extent in the FLAG-Rv0431 lysates, suggesting that the pulldown was anti-FLAG antibody specific. (B) To confirm binding of LpqH with VirR, native-coimmunoprecipitation was performed. WT and VirR-deficient Mtb lysates were immunoprecipitated with antiserum against VirR, and the eluates were immunoblotted with the same antiserum. The precleared lysates were loaded alongside eluates to confirm equal protein concentration between samples. (C) Culture filtrates and whole-cell lysates derived from WT, VirR-deficient, and VirR-complemented Mtb were immunoblotted with antiserum against LpqH. To ensure equal loading of culture filtrates among strains, samples were immunoblotted with antiserum against Ag85B. To ensure equal loading of lysates among strains and to rule out significant cell lysis, samples were immunoblotted with antisera against DlaT.
These experiments suggested that VirR plays an upstream regulatory role in modulating Mtb’s impact on the macrophage response; that it does so via a noncanonical secretion mechanism or a postsecretion process; and that it might do so by virtue of what it binds. LpqH can exit Mtb through the SecA1 pathway (30). Although the SecA1 substrate Ag85B was not overrepresented in the culture filtrate of VirR-deficient Mtb, we found that levels of LpqH were markedly higher in culture filtrate from VirR-deficient Mtb than in filtrate from WT Mtb (Fig. 4C). This observation suggested that VirR might regulate the release of LpqH to the medium by a mechanism other than regulation of the SecA1 pathway.
VirR Regulates Mtb’s Ability to Signal via TLR2.
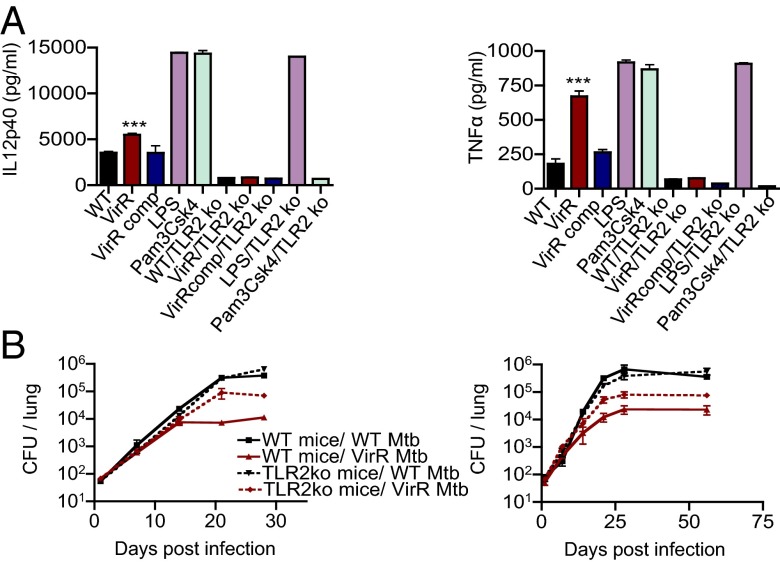
Because the culture filtrate of VirR-deficient Mtb was enriched in LpqH, a strong agonist of TLR2, we investigated the importance of TLR2 in mediating the hyperinflammatory properties of VirR-deficient culture filtrate and VirR-deficient Mtb itself. Compared with WT BMM, TLR2-knockout BMM treated with culture filtrate derived from VirR-deficient Mtb did not express enhanced levels of cytokines (Fig. 5A). In two independent experiments, when we infected WT mice with ∼100 bacilli of the WT or VirR-deficient strains, VirR-deficient Mtb already was attenuated between days 7 and 14 by 0.5 log10 compared with WT Mtb. By day 21, the attenuation amounted to 1 log10, and this deficit was maintained through the 2 mo of observation. In contrast to a recent report (31), the bacterial burden of WT Mtb was unaltered in TLR2-knockout mice compared with WT mice. However, the attenuation of VirR-deficient Mtb was substantially but not completely reversed in mice lacking TLR2 (Fig. 5B). Thus, the attenuation of VirR-deficient Mtb in WT mice was mediated in part by an enhanced host response to TLR2 stimulation.
Fig. 5.
The hyperinflammatory phenotype of VirR-deficient Mtb depends on TLR2 in vitro, and the virulence of VirR-deficient Mtb is partially restored in TLR2-KO mice. (A) BMM from WT and TLR2-KO were treated with culture filtrate derived from WT Mtb, VirR-deficient Mtb, or VirR-complemented Mtb. IL-12p40 and TNF-α levels were measured in the macrophage supernatants 24 h posttreatment. LPS and Pam3Csk4 were used as positive and negative controls, respectively, for cytokine induction via TLR2 stimulation. The left and right panels show IL12 p40 and TNFα levels, respectively. Data points represent the mean ± SD of triplicate wells from one representative experiment of two. ***P < 0.001. (B) WT C57BL/6 and TLR2-KO mice were infected with WT Mtb or VirR-deficient Mtb via aerosol. Lung lysates were plated on 7H11 agar at the indicated time points to enumerate bacterial burden. Each data point is the mean ± SD from four mice. The left and right panels represent data from two independent experiments. The virulence of VirR-deficient Mtb was partially restored in mice lacking TLR2.
MV from VirR-Deficient Mtb Are Hyperinflammatory.
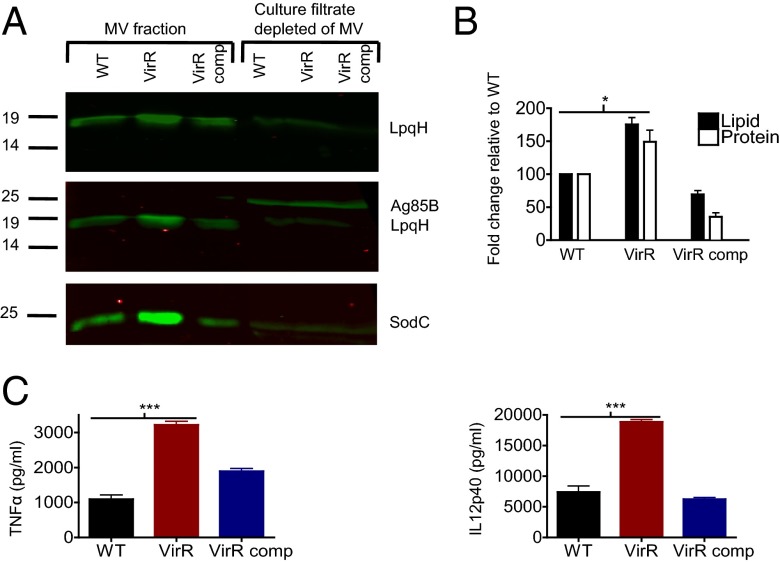
Because Mtb MV were found to contain LpqH (22), we compared the LpqH content of MV from VirR-deficient and WT Mtb. Mtb cultures were grown in minimal 7H9 medium without detergent, and their MV were purified by sequential centrifugation and ultracentrifugation as described (22). Compared with the MV fraction from WT Mtb, the MV fraction from VirR-deficient Mtb was indeed enriched in LpqH and also in SodC, another TLR2 agonist (Fig. 6A). In contrast, the remainder of the culture filtrate after depletion of MV by ultracentrifugation contained much less LpqH and SodC, and the levels of these proteins in the soluble fraction were comparable in the two strains. To detect MV released into the culture fluid, we used the lipophilic probe, 1,6-diphenyl-1,3,5-hexatriene (DPH), which fluoresces in hydrophobic environments but not in aqueous environments (32) and has been used to detect MV in Borrelia burgdorferi culture supernatants (33). By the DPH assay, VirR-deficient Mtb released 1.5-fold more lipid than WT Mtb, whereas the complemented strain released less than half as much as WT (Fig. 6B). The ratios of protein to lipid in the MV fraction from WT and VirR-deficient Mtb were similar, suggesting that physiologic levels of VirR have a greater impact on the number of MV released than on the average protein content per MV.
Fig. 6.
MV derived from VirR-deficient Mtb are enriched in LpqH and SodC and are hyperinflammatory in BMM. (A) MV isolated from the culture filtrates of WT, VirR-deficient, and VirR-complemented Mtb grown to the same biomass, alongside the remaining culture filtrates depleted of MV, were immunoblotted with antiserum against LpqH. (Top) The intensity of the LpqH band in the VirR-deficient MV fraction was 1.4 times higher than that in the WT fraction, as assessed by densitometric analysis. (Middle) To assess the purity of the MV fraction, samples were immunoblotted with antiserum against Ag85B. Ag85B was not detected in the MV fraction but only in total culture filtrates. (Bottom) Blotting with antiserum against SodC revealed that the intensity of the SodC band in the VirR-deficient MV fraction was 2.4 times higher than that in the WT fraction, whereas Ag85B levels were comparable in all three strains, as assessed by densitometric analysis. (B) Lipid content associated with MV was quantified using a lipid release assay. Protein concentration was quantified using a Bradford assay. Data points represent the mean ± SD of three independent experiments. *P < 0.05. (C) WT BMM were treated with MV isolated from WT, VirR-deficient, or VirR-complemented Mtb. TNF-α (Left) and IL-12 p40 (Right) levels were measured in the macrophage supernatants 24 h posttreatment. Data points represent the mean ± SD of triplicate wells from one representative experiment of two. ***P < 0.0005.
Consistent with their greater content of TLR2 agonists, MV isolated from VirR-deficient Mtb were more stimulatory to BMM than MV from WT Mtb at equivalent protein concentrations, as assessed by TNF-α and IL-12 p40 concentrations in the supernatants (Fig. 6C).
VirR Regulates Vesiculogenesis.
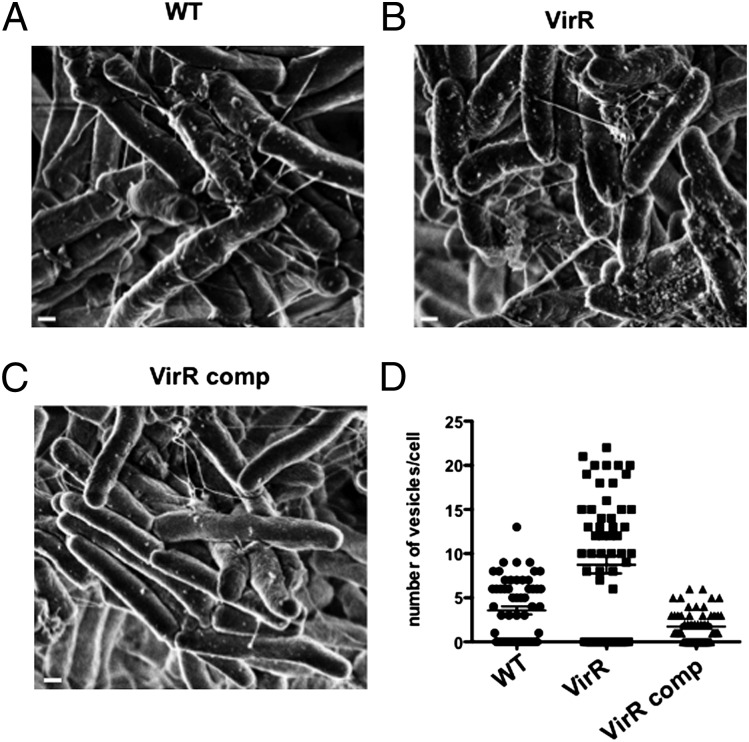
The foregoing evidence suggested that VirR regulates some aspect of the production of MV, such as their number, size, or composition. We turned to scanning electron microscopy of VirR-deficient and WT Mtb and their isolated MV. On average, markedly more MV were visible on the surface of VirR-deficient Mtb than on WT or complemented strains (Fig. 7 A–C), although the numbers on each strain were variable, and the visible portions of some bacteria appeared to be devoid of MV. A mean of 10, four, and two vesicles per cell were counted in VirR-deficient, WT, and complemented strains, respectively (Fig. 7D). The differences between VirR-deficient strain and the other two strains were statistically significant (P < 0.01). There also appeared to be more filamentous, pilus-like structures on VirR-deficient Mtb than on WT and complemented strains (Fig. 7 A–C).
Fig. 7.
Increased vesiculogenesis is observed in VirR-deficient Mtb. MV from WT, VirR-deficient, and VirR-complemented Mtb were visualized by SEM on the surface of (A) WT, (B) VirR-deficient, and (C) VirR-complemented bacilli. (Scale bars, 100 nm.) (D) MV were counted on 100–150 bacilli per strain in SEM images.
Discussion
The studies presented here provide evidence that vesiculogenesis in Mtb is subject to genetic regulation. VirR controls vesiculogenesis in such a way as to blunt the activation of macrophages and the host’s ability to restrict Mtb’s proliferation. VirR does so in large part, but not exclusively, by diminishing the release of TLR2 agonists carried in MV.
Bacteria and archaea release vesicles with diverse bioactivities (34). For example, commensal Bacterioides fragilis packages polysaccharide A capsular antigen (PSA) in MV. PSA helps mediates immune tolerance in the gastrointestinal tract, a process requiring communication among dendritic cells, regulatory T cells, and CD4 T cells (35). However, studies of bacterial vesiculogenesis (36–38) have only begun to suggest how the process is regulated. Thus, it is of considerable interest that VirR regulates vesiculogenesis in Mtb.
Studies in other bacteria have documented genes whose deletion results in either hyper- or hypovesiculation (39, 40). The lack of DegP, a periplasmic protease/chaperone in E. coli, resulted in an increased production of MV as a stress response, with an increased accumulation of DegP’s substrate, misfolded outer membrane porins. Lack of E. coli’s NlpA, an inner membrane-anchored lipoprotein, DsbA, a disulfide oxidoreductase, or BolA, a protein thought to regulate morphologic changes under stress conditions, resulted in hypovesiculation. Combining the deletion of degP with that of nlpA, dsbA, or bolA results in a growth defect in vitro (40). In contrast, deletion of peptidoglycan-associated outer membrane proteins OprF and OprI in Pseudomonas aeruginosa results in hypervesiculation (39). OprF has been shown to influence outer MV formation by modulating the Pseudomonas quinolone signal (39). Bioinformatic searches based on homology modeling do not show any structural similarity between VirR and any of these proteins.
The mechanism by which VirR regulates vesiculogenesis remains to be defined. The identification of an intrinsically unstructured region of VirR suggests that VirR binds another protein through interactions with those residues. VirR’s highly hydrophobic region is speculated to bind to the inner layer of the plasma membrane, but it may bind a protein with a hydrophobic surface. Coimmunoprecipitation led to the identification of three proteins associated with VirR: LpqH and two proteins about which almost nothing is known, Rv1488 and Rv0383c. We doubt that VirR restricts the content of LpqH in MV simply by binding LpqH and retaining it within the bacilli, because this process seems unlikely to account for VirR’s regulation of the SodC content of MV, the abundance of MV, and the prevalence of filamentous structures. Moreover, LpqH localized predominantly in membrane and cell-wall fractions in both the WT and VirR-deficient strain, with no detectable difference in relative abundance between WT and VirR strains (SI Appendix, Fig. S11). Therefore, binding of VirR to LpqH is unlikely to play a role in regulation of vesiculogenesis. Taken together, these considerations suggest that VirR may function as part of a higher-order complex. We have no evidence for a direct interaction of VirR with SodC, but it is intriguing that SodC is encoded by rv0432 in the same operon as virR (rv0431).
Also requiring further study is the relationship between VirR and filamentous structures on Mtb. Filamentous structures were described on mycobacteria more than 40 y ago (41). In recent work, such structures were characterized as laminin-binding pili comprised mainly of the protein Rv3312A, which is immunogenic in humans with tuberculosis and thus appears to be expressed in the host (42). VirR may regulate the release of Rv3312A or a factor that participates in its polymerization.
This possibility leads to the question of the relation of MV formation in general and of VirR in particular to the SecA1, SecA2, Tat, and type VII secretory pathways. VirR may control the extent to which certain substrates for those pathways are degraded before they are secreted. Alternatively, VirR may regulate whether some substrates of the SecA1, SecA2, or Tat pathways are packaged in MV for transit through, or budding from, the outer membrane rather than being retained in the periplasm.
Likewise, it is remains to be clarified how MV formation by Mtb relates to exosome formation by macrophages containing Mtb (21, 43). We speculate that extracellular Mtb use MV to communicate with the host, whereas MV released by intraphagosomal Mtb may become incorporated into macrophage exosomes. This two-step process of vesiculation would allow Mtb to influence not only the cell in which it resides but more distant host cells as well.
The involvement of TLR2 in mediating VirR-dependent immune modulation is consistent with the reported enrichment of TLR2 agonists in MV (22) and with the presence of TLR2 agonists in mycobacterial products recovered from exosomes shed by infected macrophages (21). The partial restoration of virulence of VirR-deficient Mtb in mice lacking TLR2 suggested both that TLR2 is important in the host response to VirR-regulated processes and also that Mtb engages additional host signaling pathways in a manner influenced by VirR.
VirR regulates Mtb’s stimulation of TLR2 in mouse macrophages and in mice, but we have not established to what extent the marked impact of VirR on the response of human macrophages to Mtb involves agonism of TLR2. A role for human TLR2 seems likely, given that engagement of TLR2 is important in inducing mycobactericidal factors, such as β-defensins, cathelicidin LL-37, granulysin, and ubiquitin-derived peptides (44), in human macrophages (18). It remains to be determined if these or other effectors contribute to the marked attenuation of VirR-deficient Mtb in human macrophages.
In a classic experiment by Jung and North (45), mice were immunized by infection with WT Mtb and then were cured with chemotherapy. When the mice were challenged again with WT Mtb, their protective immune response was manifest only 3 d earlier than in mice undergoing the challenge infection alone. In contrast, VirR-deficient Mtb activated the host in such a way that mice began to exert control over the infection as early as 7–14 d after inhaling ∼100 bacilli, rather than at 21–28 d, as seen with WT Mtb. This experiment raises the possibility that Mtb lacking VirR and additionally attenuated for safety (46) might be a more effective vaccine than Mtb bearing the additional attenuating mutations alone. Alternatively, cell-free, nonreplicative MV from VirR-deficient Mtb might be useful as an adjuvant or immunogen.
Materials and Methods
Assays for VirR topology and stress resistance of VirR-deficient Mtb, methods for transcriptomic analysis, and details of the NMR studies are described in SI Appendix.
Mice.
Young adult female C57BL/6 WT and TLR2-knockout mice from Jackson Laboratories were housed in specific pathogen-free facilities. Under protocols approved by the Weill Cornell Medical College or Rockefeller University Institutional Animal Care and Use Committees, mice were infected by aerosol with ∼100 bacilli. Tissues were collected at time points and processed to enumerate cfu as described (17).
Macrophages.
Mouse BMM and human monocyte-derived macrophages collected under a Weill Cornell Medical College Institutional Review Board-approved protocol were differentiated and infected as described (17, 24). Cytokines were measured by ELISA. For transcriptomic studies, 107 BMM were infected with Mtb at an MOI of 5 for 8 h or were left uninfected. Cells were lysed with TRIzol, and RNA was extracted with a kit from Qiagen. cDNA libraries were synthesized following the protocol of Illumina Inc. and were sequenced on an Illumina HiSeq.
Mtb.
Mtb H37Rv from American Type Culture Collection and derivative strains (SI Appendix, Table S3) were grown at 37 °C in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol (Sigma), 0.5% BSA fraction V (Roche), 0.2% dextrose (Sigma-Aldrich), and 0.085% sodium chloride (Sigma-Aldrich) (7H9-ADN). The cfu in serial dilutions were enumerated on Middlebrook 7H11 agar (Difco) plates containing 0.5% glycerol and 10% (vol/vol) oleic acid-albumin-dextrose-catalase (Becton Dickinson) after 2–3 wk at 37 °C. Strains containing antibiotic-resistance cassettes were cultured in or plated on medium containing hygromycin (50 µg/mL), streptomycin (30 µg/mL), or kanamycin (15 µg/mL). Complementation plasmids were cloned using Gateway Cloning Technology (Invitrogen) as described (47). Target sequences from Mtb H37Rv genomic DNA were amplified by PCR using primers designed so that the amplicons would be flanked with Gateway cloning-specific attachment (att) sites. PCR products were recombined into Gateway donor vectors using bacteriophage-derived integrase and integration host factor. The Gateway entry vectors generated were recombined with another entry vector containing the hsp60 promoter, an empty entry vector, or a complementation vector (episomal) to give rise to the final destination vectors containing a hygromycin-resistance cassette. The final strain overexpressed Rv0431 from an episomal plasmid (details in SI Appendix, Methods). Truncated VirR cloned in the pET28 vector system containing a His6 tag in the C′ terminus was purified by FPLC using a Ni-NTA column and used by Covance to raise antibodies in rabbits.
Mtb Culture Filtrates, Lysates, and Fractionations.
Mtb strains were grown for 10 d in minimal 7H9 or minimal Sauton’s medium without detergent. Cultures were filtered (0.22-µm pore size) to yield early culture filtrates that were concentrated (3-kDa cutoff; Amicon Ultrafiltration System; Millipore) in the presence of 1 mM PMSF. Protein concentrations were measured using a Bio-Rad DC kit. Immunoblots after SDS/PAGE were performed with anti-ESAT 6 (1:5,000; Abcam), anti-Ag85B (1:5,000; Abcam), anti-HspX (1:5,000; Abcam), anti-LpqH (1:1,000; BEI Resources), anti-BlaC [1:1,000; courtesy of Miriam Braunstein (University of North Carolina, Chapel Hill, NC)], anti-SodC [1:1,000; courtesy of John Belisle (Colorado State University, Fort Collins, CO)], and anti-HOAS (48). The secondary antibodies were anti-rabbit IgG (1:5,000) or anti-mouse IgG (1:5,000) labeled with infrared dyes. Blots were visualized with an Infrared Odyssey reader (LI-COR). For whole-cell lysates, Mtb cultures grown to midlog phase (∼OD 0.6) in 7H9 medium were pelleted, washed in PBS, and resuspended in 25 mM potassium phosphate buffer containing 1 mM EDTA and 1 mM PMSF at 4 °C for lysis by bead-beating. Lysates were clarified at 20,000 × g for 15 min. The protein concentration of the supernatant was measured, and the supernatant was subjected to SDS/PAGE followed by immunoblotting with anti-VirR (1:10,000), anti-DlaT (1:10,000) (26), or anti-FLAG (1:5,000; Sigma). For subcellular fractionation, lysates were centrifuged at 20,000 × g for 2 h at 4 °C to pellet the cell-wall fraction. The supernatants were centrifuged at 100,000 × g for 1 h at 4 °C to pellet the cell-membrane fraction. The pellets were washed and resuspended in SDS sample buffer, and the supernatants were reserved as the cytosolic fraction. For SDS/PAGE, 10 µg of cytosolic fraction and an equal volume of cell envelope fraction were loaded, based on total volume of the fractions.
MV.
Mtb cultures (100 mL) were grown in roller bottles in minimal 7H9 medium without detergent for 2 wk at 37 °C and were centrifuged (3,450 × g). Supernatants were filtered (0.22-μm pore size), concentrated to ∼1 mL (100-kDa cutoff; Amicon), and centrifuged at 4,000 × g for 15 min and then at 15,000 × g for 15 min. MV were pelleted at 100,000 × g for 1 h at 4 °C. The pellet was resuspended in 500 μL of 10 mM Hepes, 0.15 M NaCl, mixed with 1 mL Optiprep (Sigma-Aldrich), yielding a 40% (wt/vol) concentration, and were overlaid with a series of Optiprep solutions ranging from 35–30% (wt/vol). The step gradients were ultracentrifuged at 100,000 × g for 16 h at 4 °C. Then 1 mL was collected from the top, dialyzed in PBS overnight, ultracentrifuged at 100,000 × g for 1 h, and resuspended in sterile PBS. For quantification of released membrane material, MV were labeled with 1 µg/mL of the probe DPH (Invitrogen) for 30 min at 33 °C. After 20 min, the supernatants were analyzed for fluorescence in a SpectraMax M2 plate reader using an excitation of 360 nm and emission of 430 nm.
Electron Microscopy.
For transmission electron microscopy, bacilli were fixed in 2% (vol/vol) glutaraldehyde in 0.1 M cacodylate at room temperature for 2 h and then were incubated overnight in 4% (vol/vol) formaldehyde, 1% glutaraldehyde, and 0.1% PBS. Fixed samples were stained for 90 min in 2% (vol/vol) osmium and then were serially dehydrated in ethanol and embedded in Spurr’s epoxy resin. Thin sections were obtained on an Ultracut UCT (Reichert) and were stained with 0.5% uranyl acetate and 0.5% lead citrate (Reichart). Samples were observed in a JEOL 1200EX transmission electron microscope operating at 80 kV. For scanning electron microscopy (SEM), cells were fixed with 2.5% glutaraldehyde, 0.1 M sodium cacodylate, 0.2 M sucrose, 5 mM MgCl2 (pH 7.4) and were dehydrated through a graded series of ethanol solutions before critical-point drying using liquid carbon dioxide in a Toumisis Samdri 795 device and sputter-coating with gold-palladium in a Denton Vacuum Desk-2 device. Samples were examined in a Zeiss Supra Field Emission Scanning Electron Microscope (Carl Zeiss Microscopy, LLC North America), using an accelerating voltage of 5 KV. Quantification of MV associated with bacterial cells was performed by counting the MV associated with 100–150 bacilli per strain in SEM images.
Statistical Analysis.
The statistical significance of the difference between experimental groups was determined by the two-tailed Student’s t-test using PRISM software. P values less than 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank X. Jiang for help with mouse experiments, K. Burns-Huang and A. Ding for discussions, R. Bryk, M. Braunstein and J. Belisle for antibodies, E. Pamer for TLR2-knockout mice, D. Schnappinger and S. Ehrt for gateway vectors, and J. Schneider and M. Glickman for backbone plasmids pMVB and pJSMG. This work was supported by the Milstein Program in Chemical Biology of Infectious Disease (C.N.) and National Institutes of Health Grant R01 AI70285 (to H.L.). A.C. and R.P.-R. were supported by a grant from the Bill and Melinda Gates Foundation. The Weill Cornell Medical College Department of Microbiology and Immunology is supported by the William Randolph Hearst Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The RNAseq data (raw reads and reads per kilobase per million) are available at the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE50050). The NMR chemical shift assignments for backbone 1H, 13C, and 15N of VirR (Rv0431) have been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession no. 19087). The solution structure has been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2M5Y).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320118110/-/DCSupplemental.
References
- 1.Gutierrez MC, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1(1):e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel TM. The history of tuberculosis. Respir Med. 2006;100(11):1862–1870. doi: 10.1016/j.rmed.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous WHO global tuberculosis control report 2010. Summary. Cent Eur J Public Health. 2010;18(4):237. [PubMed] [Google Scholar]
- 4.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328(5980):856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann SH. Envisioning future strategies for vaccination against tuberculosis. Nat Rev Immunol. 2006;6(9):699–704. doi: 10.1038/nri1920. [DOI] [PubMed] [Google Scholar]
- 6.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12(8):581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 7.Jamieson AM, et al. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340(6137):1230–1234. doi: 10.1126/science.1233632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335(6071):936–941. doi: 10.1126/science.1214935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42(6):498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu K, et al. Improved control of tuberculosis and activation of macrophages in mice lacking protein kinase R. PLoS ONE. 2012;7(2):e30512. doi: 10.1371/journal.pone.0030512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tobin DM, et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148(3):434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar D, et al. Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell. 2010;140(5):731–743. doi: 10.1016/j.cell.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Dao DN, et al. Mycolic acid modification by the mmaA4 gene of M. tuberculosis modulates IL-12 production. PLoS Pathog. 2008;4(6):e1000081. doi: 10.1371/journal.ppat.1000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rengarajan J, et al. Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc Natl Acad Sci USA. 2008;105(1):264–269. doi: 10.1073/pnas.0710601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madan-Lala R, Peixoto KV, Re F, Rengarajan J. Mycobacterium tuberculosis Hip1 dampens macrophage proinflammatory responses by limiting toll-like receptor 2 activation. Infect Immun. 2011;79(12):4828–4838. doi: 10.1128/IAI.05574-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton SM, et al. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc Natl Acad Sci USA. 2006;103(42):15594–15598. doi: 10.1073/pnas.0604283103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beaulieu AM, et al. Genome-wide screen for Mycobacterium tuberculosis genes that regulate host immunity. PLoS ONE. 2010;5(12):e15120. doi: 10.1371/journal.pone.0015120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thoma-Uszynski S, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291(5508):1544–1547. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]
- 19.Feltcher ME, Braunstein M. Emerging themes in SecA2-mediated protein export. Nat Rev Microbiol. 2012;10(11):779–789. doi: 10.1038/nrmicro2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feltcher ME, Sullivan JT, Braunstein M. Protein export systems of Mycobacterium tuberculosis: Novel targets for drug development? Future Microbiol. 2010;5(10):1581–1597. doi: 10.2217/fmb.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beatty WL, et al. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1(3):235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 22.Prados-Rosales R, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121(4):1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piddington DL, et al. Cu,Zn superoxide dismutase of Mycobacterium tuberculosis contributes to survival in activated macrophages that are generating an oxidative burst. Infect Immun. 2001;69(8):4980–4987. doi: 10.1128/IAI.69.8.4980-4987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt G, Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J Clin Invest. 2011;121(10):3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venugopal A, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9(1):21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295(5557):1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 27.Bryk R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3(3):137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Darwin KH, Li H. Binding-induced folding of prokaryotic ubiquitin-like protein on the Mycobacterium proteasomal ATPase targets substrates for degradation. Nat Struct Mol Biol. 2010;17(11):1352–1357. doi: 10.1038/nsmb.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grupi A, Haas E. Segmental conformational disorder and dynamics in the intrinsically disordered protein α-synuclein and its chain length dependence. J Mol Biol. 2011;405(5):1267–1283. doi: 10.1016/j.jmb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Feltcher ME, Gibbons HS, Ligon LS, Braunstein M. Protein export by the mycobacterial SecA2 system is determined by the preprotein mature domain. J Bacteriol. 2013;195(4):672–681. doi: 10.1128/JB.02032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McBride A, Konowich J, Salgame P. Host defense and recruitment of Foxp3⁺ T regulatory cells to the lungs in chronic Mycobacterium tuberculosis infection requires toll-like receptor 2. PLoS Pathog. 2013;9(6):e1003397. doi: 10.1371/journal.ppat.1003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.London E, Feligenson GW. A convenient and sensitive fluorescence assay for phospholipid vesicles using diphenylhexatriene. Anal Biochem. 1978;88(1):203–211. doi: 10.1016/0003-2697(78)90412-8. [DOI] [PubMed] [Google Scholar]
- 33.Crowley JT, et al. Lipid exchange between Borrelia burgdorferi and host cells. PLoS Pathog. 2013;9(1):e1003109. doi: 10.1371/journal.ppat.1003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacDonald IA, Kuehn MJ. Offense and defense: Microbial membrane vesicles play both ways. Res Microbiol. 2012;163(9-10):607–618. doi: 10.1016/j.resmic.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen Y, et al. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12(4):509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect Immun. 2012;80(6):1948–1957. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi DS, et al. Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa. Proteomics. 2011;11(16):3424–3429. doi: 10.1002/pmic.201000212. [DOI] [PubMed] [Google Scholar]
- 38.Pierson T, et al. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J Proteome Res. 2011;10(3):954–967. doi: 10.1021/pr1009756. [DOI] [PubMed] [Google Scholar]
- 39.Wessel AK, Liew J, Kwon T, Marcotte EM, Whiteley M. Role of Pseudomonas aeruginosa peptidoglycan-associated outer membrane proteins in vesicle formation. J Bacteriol. 2013;195(2):213–219. doi: 10.1128/JB.01253-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwechheimer C, Kuehn MJ. Synthetic effect between envelope stress and lack of outer membrane vesicle production in Escherichia coli. J Bacteriol. 2013;195(18):4161–4173. doi: 10.1128/JB.02192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gordon J, White RG. Surface peptido-glycolipid filaments on Mycobacterium leprae. Clin Exp Immunol. 1971;9(4):539–547. [PMC free article] [PubMed] [Google Scholar]
- 42.Alteri CJ, et al. Mycobacterium tuberculosis produces pili during human infection. Proc Natl Acad Sci USA. 2007;104(12):5145–5150. doi: 10.1073/pnas.0602304104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh PP, Smith VL, Karakousis PC, Schorey JS. Exosomes isolated from mycobacteria-infected mice or cultured macrophages can recruit and activate immune cells in vitro and in vivo. J Immunol. 2012;189(2):777–785. doi: 10.4049/jimmunol.1103638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu PT, et al. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS ONE. 2009;4(6):e5810. doi: 10.1371/journal.pone.0005810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jung YJ, Ryan L, LaCourse R, North RJ. Properties and protective value of the secondary versus primary T helper type 1 response to airborne Mycobacterium tuberculosis infection in mice. J Exp Med. 2005;201(12):1915–1924. doi: 10.1084/jem.20050265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambandamurthy VK, et al. Long-term protection against tuberculosis following vaccination with a severely attenuated double lysine and pantothenate auxotroph of Mycobacterium tuberculosis. Infect Immun. 2005;73(2):1196–1203. doi: 10.1128/IAI.73.2.1196-1203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blumenthal A, Trujillo C, Ehrt S, Schnappinger D. Simultaneous analysis of multiple Mycobacterium tuberculosis knockdown mutants in vitro and in vivo. PLoS ONE. 2010;5(12):e15667. doi: 10.1371/journal.pone.0015667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian J, Bryk R, Itoh M, Suematsu M, Nathan C. Variant tricarboxylic acid cycle in Mycobacterium tuberculosis: Identification of alpha-ketoglutarate decarboxylase. Proc Natl Acad Sci USA. 2005;102(30):10670–10675. doi: 10.1073/pnas.0501605102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.