Significance
When wild-derived laboratory mice are reintroduced to socially competitive populations, they quickly adapt by producing attractive sons that otherwise have no fitness advantages, consistent with the sexy sons model of sexual selection. These attractive sons inherit up-regulated expression of several pheromones belonging to the major urinary protein (MUP) family. Up-regulation is controlled by maternal social experience, and is associated with epigenetic modifications of MUP promoters that could enhance transcription. Inheritance of up-regulated MUPs is likely adaptive because females have odor preferences for male scent marks with higher MUP concentration. These results represent one of only a few cases where parental social experience adaptively modifies progeny phenotype.
Keywords: social selection, sexy sons, epigenetics
Abstract
When brought into captivity, wild animals can adapt to domestication within 10 generations. Such adaptations may decrease fitness in natural conditions. Many selective pressures are disrupted in captivity, including social behavioral networks. Although lack of sociality in captivity appears to mediate domestication, the underlying mechanisms are not well understood. Additionally, determining the contribution of genetic inheritance vs. transgenerational effects during relaxed selection may provide insight into the flexibility of adaptation. When wild-derived mice kept under laboratory conditions for eight generations were reintroduced to sociality and promiscuity (free mate choice), they adapted within two generations. Fitness assessments between this promiscuous lineage and a monogamous laboratory lineage revealed male-specific effects. Promiscuous-line males had deficits in viability, but a striking advantage in attracting mates, and their scent marks were also more attractive to females. Here, we investigate mechanistic details underlying this olfactory signal and identify a role of major urinary protein (MUP) pheromones. Promiscuous-line males inherit higher MUP expression than monogamous-line males through transgenerational inheritance. Sociality-driven maternal and paternal effects reveal intriguing conflicts among parents and offspring over pheromone expression. MUP up-regulation is not driven by hormone-driven transduction pathways, but rather is associated with reduction in DNA methylation of a CpG dinucleotide in the promoter. This reduction in methylation could enhance transcription by promoting the binding of transcription factor USF1 (upstream stimulatory factor 1). Finally, we experimentally demonstrate that increased MUP expression is a female attractant. These results identify molecular mechanisms guiding domestication and adaptive responses to fluctuating sociality.
Wild animals bred in captivity have been observed to adapt to domestication within 10 generations (1–4). As a result, captive bred animals often have reduced fitness when reintroduced to natural conditions (5). Selective pressures disrupted by captivity include inbreeding avoidance, effective population size, disease exposure, predation, and sexual selection. For the latter example, evidence suggests that lack of social context for mate choice mediates adaptation to captivity (6). This context is especially relevant for social animals because the opportunity for mating success is regulated by hierarchical behavioral networks (i.e., social selection) (7), and this information is missing in captivity. The molecular mechanisms underlying phenotypes affected by lack of social selection are poorly understood. Furthermore, rapid adaptation to captivity raises questions about the roles of genetic inheritance vs. transgenerational effects (i.e., inheritance independent of genetic variation) (8). Resolving the mechanistic and hereditary basis of these transitions can provide insight into the flexibility of adaptation (9) and to the management of captive breeding programs (1, 2, 6).
To address the effects of domestication on social selection, wild-derived mice that had been kept under laboratory conditions for eight generations were reintroduced to sociality and a promiscuous breeding system [where males and females have multiple partners (10)] for three generations (see SI Materials and Methods and Figs. S1 and S2 for detailed description of mouse stocks). The “promiscuous line” competed for social dominance, territorial breeding sites, and mating success in seminatural enclosures designed to capture mouse social ecology (11, 12), and the “monogamous line” bred under enforced mate assignment in cages where social selection is eliminated (13). To minimize environmental effects on offspring, pregnant promiscuous-line females were removed from enclosures and singly housed in cages. Thus, before testing, neither promiscuous-line nor monogamous-line individuals experienced seminatural conditions. A direct competition experiment in enclosures revealed fitness effects on males, but not females: compared with monogamous-line males, promiscuous-line males sired significantly more offspring and were favored by females during extraterritorial mating (14). Strikingly, promiscuous-line males had reduced survivorship and equivalent success in territory defense. A mate-choice experiment subsequently revealed female mating and odor preferences for promiscuous-line males (14). These results suggested that returning mice to sociality favored an attractive male-specific phenotype characterized by enhanced pheromone signaling, but conspicuously lacking in vigor and viability, a result consistent with the sexy sons hypothesis of sexual selection (14).
Here, we address the molecular and hereditary basis of this adaptive male phenotype. Using a candidate-gene approach, we examine the major urinary proteins (MUPs), a primary component of urinary pheromones in mice. MUPs belong to a multigene family that conveys information about genetic identity and kin relatedness in wild mice (15, 16). The quantitative role of MUP expression level also appears to have behavioral relevance: MUPs are expressed in high quantities in a sexually dimorphic fashion, with males expressing roughly four times that of females (17). Several researchers have proposed that such expression is a result of sexual selection. This “honest-signal” model predicts that females favor males who can afford high expression despite potential costs (e.g., protein loss or attracting predators and competitors) (18–22). Although this hypothesis has not been explicitly tested, the observation that MUP concentration declines as mice adapt to captivity (17, 23) suggests a possible role of relaxed social and sexual selection. Intriguingly, Mups were the first mammalian locus found to exhibit transgenerational epigenetic inheritance (i.e., resulting from epigenetic modifications passed via the gametes that escape reprogramming) (8, 24), but the biological significance of such regulation has not been identified.
We show that social competition induces heritable up-regulation of MUPs in sons, and this up-regulation is due to a maternal effect. Unexpectedly, a relatively weaker paternal effect down-regulates MUPs in sons. MUP up-regulation is not due to higher levels of circulating testosterone or activity of the growth hormone/STAT5b transduction pathway. Rather, it is associated with reduction in methylation of a CpG dinucleotide proximal to the transcription start site that is a likely binding site for the transcription factor USF1. We support the honest-signal model by experimentally demonstrating that MUP up-regulation serves as a female attractant.
Results
Testosterone.
Testosterone contributes to male sexual characteristics such as social status (25), attractiveness to females (26), and pheromones, including MUPs (27). Testosterone is also involved in tradeoffs with life-history characteristics, including decreased survival (25). We predicted that elevated testosterone would be associated with the increased mating success, but decreased survival found in promiscuous-line males. We measured circulating testosterone in F4 males before and after exposure to estrus females (28). Repeated measures analysis found no treatment effects on circulating testosterone (Fig. S3) (F = 0.36, P = 0.56), nor were there differences at either time point [before, t (28) = 0.84, P = 0.41; after, t (28) = −0.080, P = 0.94].
MUPs Are Up-Regulated in Promiscuous-Line Males.
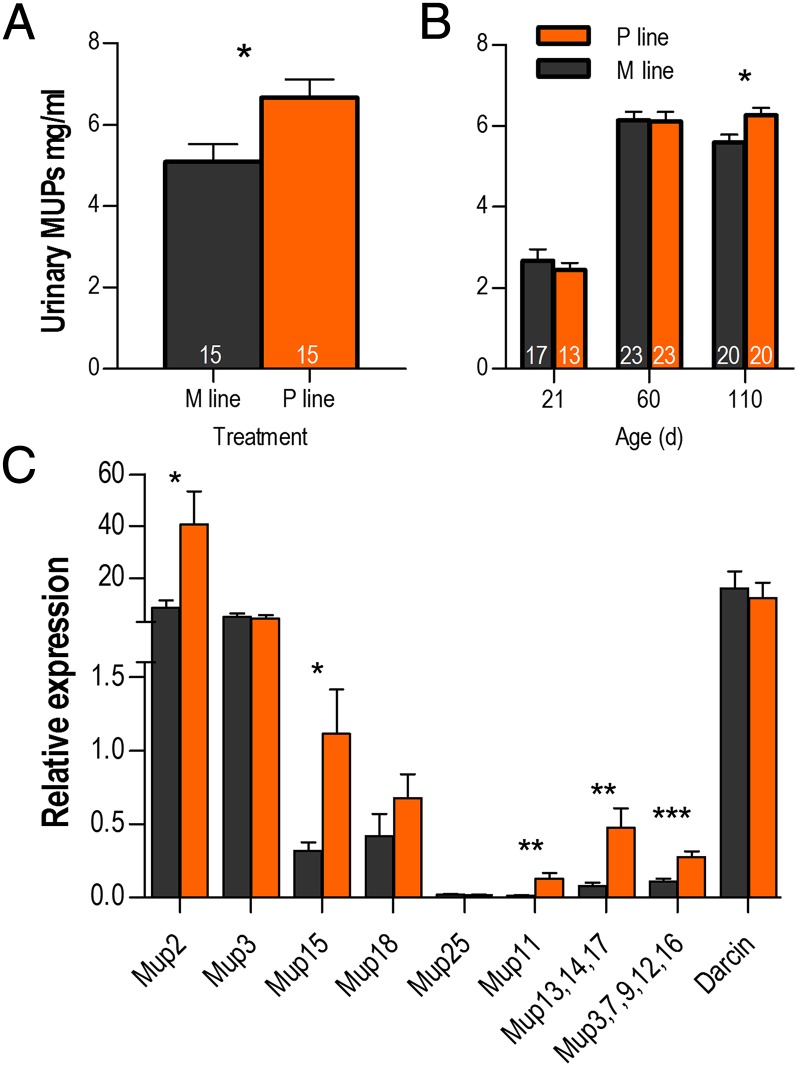
Because MUPs represent substantial investment in excreted protein, we hypothesized that increased mating success and decreased survival in promiscuous-line males could be associated with up-regulated MUP expression. Urine collected from fourth-generation, individually housed adult males following the testosterone assay revealed that promiscuous-line males had higher MUP expression than monogamous-line males (Fig. 1A) [t (26) = 2.56, P = 0.017]. To understand the developmental regulation of this phenotype, we compared MUP expression at three time points under varying conditions in naïve fifth-generation males (Fig. 1B). Results showed no effects on MUP expression in prepubertal males in home cages with brother siblings [t (27) = −0.59, P = 0.56], nor in subadult males in home cages with brother siblings [t (44) = −0.13, P = 0.90]. However, when males were adults and housed individually, promiscuous-line males had significantly higher MUP expression than monogamous-line males [t (45) = 2.42, P = 0.019].
Fig. 1.
Heritable up-regulation of MUPs. (A) F4 promiscuous-line (P line) males had higher urinary MUP expression than monogamous line (M line) males at age 180 d. (B) F5 P line males had higher MUP expression than M line males at 110 d, but not at 21 or 60 d. Means ± SEM. (C) F4 P line males (n = 8) had higher hepatic expression of five Mup targets than M line males (n = 10). *P < 0.05, **P < 0.01, ***P < 0.001.
To identify specific up-regulated Mup loci in promiscuous-line males, we used reverse transcription quantitative qPCR (rt-qPCR) with nine primer sets from published studies that amplify most Mup transcripts (19, 29) (Table S1). Because of extremely high sequence identity, some primer pairs were able to discriminate only subsets of paralogous Mup loci (29). Total sequence identity of the Mup locus in our congenic wild mice with that of reference strains (C57 and BALB/c) is unknown; however, sequence analysis of PCR products confirmed amplification of similar Mups in our mice. We found that fourth generation promiscuous-line males had higher liver expression of Mups in five out of nine primer sets evaluated (Fig. 1C): Mup2 [t (15) = 2.63, P = 0.019], Mup15 [t (15) = 2.78, P = 0.014], Mup11 [t (16) = 3.18, P = 0.0058], Mup13,14,17 [t (16) = 3.37, P = 0.0039], and Mup3,7,9,12,16 [t (16) = 4.30, P = 0.0006]. All other differences, including Darcin (30), were nonsignificant.
We predict that promiscuous-line males excrete a greater mass of MUPs per volume of urine, and this value could be influenced by water consumption. Urinary MUP expression is often normalized as a ratio with creatinine, a metabolite of muscular creatine that is converted by a nonenzymatic process proceeding at a constant rate in similarly sized individuals (17, 21). However, MUP/creatinine ratios should be interpreted with some caution as we are unaware of experiments determining whether this ratio correlates with total 24-h excreted protein, or whether glomerular and tubular filtration affects MUPs and creatinine similarly (31). Creatinine itself responds to environmental challenges (32), including by transgenerational inheritance (described in the following section). Nevertheless, consistent with absolute measurements, F4 promiscuous-line males had greater MUP/creatinine ratios than monogamous-line males [t (27) = 2.57, P = 0.016]. Differences between F5 ratios were consistent but did not reach significance [t (37) = 1.91, P = 0.064]. Together, these results show that promiscuous-line males (who are conceived in enclosures but born in cages) have marginally higher MUP expression at adulthood, and significantly higher expression upon exposure to nonkin females.
Social Competition Triggers Maternal and Paternal Effects on MUP Expression in Sons.
Because promiscuous-line males were conceived (but not born) in seminatural enclosures, the inheritance of increased MUP expression could be due to genetic selection on parental alleles encoding high MUP expression, or it could be a result of transgenerational effects due to social competition. To address the first hypothesis, we relaxed selection by breeding the promiscuous line in cages under enforced monogamy. When fifth-generation promiscuous-line mice were bred in cages, their sons did not have higher MUP expression than the sons of fifth-generation monogamous-line mice [t (23) = −0.36, P = 0.73]. This result suggested that parental exposure to social competition is necessary for up-regulation of MUPs in sons and raised the question of whether a single parental exposure to social competition is sufficient to increase MUP expression in sons.
To identify parental (maternal and paternal) effects on MUP expression following a single exposure to a socially competitive environment, we used a reciprocal breeding design with naïve mice from the laboratory colony. Adult “parents” experienced one of two treatments for roughly 8 wk: competition (C) or noncompetition (NC). Using the same protocol to generate promiscuous-line and monogamous-line mice (14), C males and females were introduced to enclosures where they competed for resources, mates, and social dominance (12, 14) whereas NC males and females experienced 8 wk in monogamous breeding cages. Following a brief intermission period (during which pups from pregnant females were removed), females and males were assigned, respectively, to monogamous breeding in the following four-way reciprocal breeding design: C × C; C × NC; NC × C; and NC × NC. Males were removed from breeding cages after 8 d.
Behavioral analysis of parents showed that, strikingly, C males had over twofold lower creatinine levels than NC males (Fig. S4A), suggesting that they increase water consumption to meet the demands of continuous scent marking and counter marking of territories (Fig. S4B) (33). C males had equivalent urinary MUP expression as NC males and therefore had over twofold greater normalized MUP expression (Fig. S4C). C and NC females had equivalent creatinine excretion and normalized MUP expression (Fig. S4 D and E). Notably, analysis of reproductive success in reciprocal breeding cages found a significant, positive effect of C maternity on litter size (mean litter size: C = 8.55 ± 0.13; NC = 7.06 ± 0.15) (Table S2).
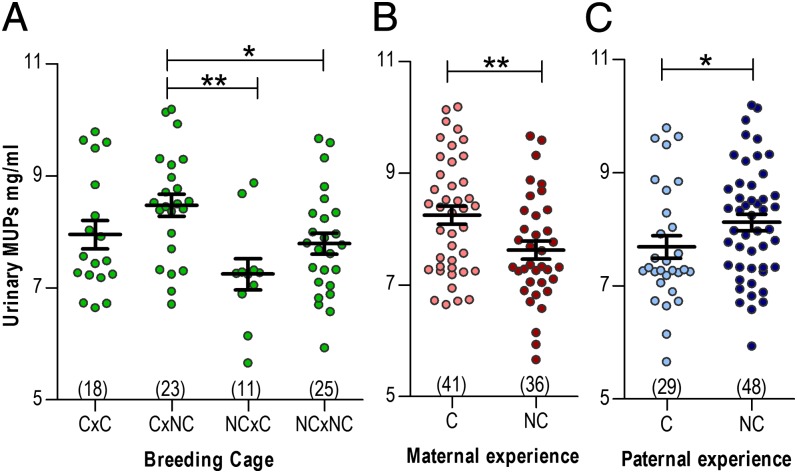
Analysis of individually housed adult male offspring indicated heterogeneous parental effects on MUP expression (Fig. 2A), including a strong C maternal-driven up-regulation (Fig. 2B), and, unexpectedly, a C paternal-driven down-regulation (Fig. 2C). Using a general linear mixed model (GLMM) with restricted maximum likelihood estimation, we analyzed the independent effects of maternal and paternal treatment, maternal × paternal treatment interaction, and creatinine concentration (to control for hydration). Litter of origin was added to the model as a random blocking effect. We also calculated the variance components of maternal, paternal, and litter effects on MUP expression (Table S3). GLMM results, summarized in Table 1, confirmed a significant maternal effect (accounting for over 14% of total variation in MUP expression), and a marginally significant paternal effect (accounting for 7% of total variation). Litter effects accounted for 16% of the variance in MUP expression and were significant (Loglinear test: df = 29, χ2 = 44.3, P = 0.03), indicating substantial genetic variation for this trait. Unexpectedly, GLMM analysis of urinary creatinine in sibling-housed adolescent males revealed a C maternal-driven increase in excretion (Table S4); it is unclear whether this effect was due to lower water consumption or higher muscle mass in sons. No parental effects on creatinine were found in individually housed adult males.
Fig. 2.
Competition-driven maternal and paternal effects on MUP expression. After 8 wk of competition (C) or noncompetition (NC), male and female mice were assigned to a reciprocal breeding design, and urinary MUP expression in their adult sons (age 170 d) was measured. Shown are maternal x paternal interactions (A), maternal effects (B), and paternal effects (C). General linear model (GLM), *P < 0.05, **P < 0.01.
Table 1.
Sources of variation in male MUP expression following parental experience of competition (C) or noncompetition
| Term | Estimate | Std error | DFDen | t Ratio | P value |
| Intercept | 6,927.12 | 402.13 | 69.95 | 17.23 | <0.0001 |
| Father experience (C) | −272.99 | 134.00 | 27.11 | −2.04 | 0.051 |
| Mother experience (C) | 307.32 | 134.13 | 27.34 | 2.29 | 0.029 |
| Father x mother experience (C x C) | −68.24 | 134.71 | 27.53 | −0.51 | 0.616 |
| Creatinine, μg/mL | 1.582 | 0.672 | 69.04 | 2.35 | 0.021 |
GLMM, n = 75 offspring. Litter of origin was added as a random effect and was significant (P = 0.03). DFDen, degrees of freedom in denominator of F statistic.
This reciprocal breeding experiment revealed that the experience of social competition triggers complex parental effects. Females experiencing social competition did not up-regulate MUP expression, but programmed their male offspring to up-regulate. Males experiencing social competition did up-regulate MUP expression, but programmed their sons to down-regulate. These results are suggestive of parental conflict and parent–offspring conflict over pheromone signaling (Discussion).
Transcriptional Mechanisms of Mup Up-Regulation.
STAT5b is a GH-regulated transcription factor critical for expression of sexually dimorphic genes in the liver, including Mups and Igf1. This pathway involves activation of growth hormone receptor (GHR) and phosphorylation of JAK2 and STAT5b; STAT5b translocates to the nucleus, binds DNA, and regulates transcription (34). The involvement of this pathway in Mup expression has been confirmed in Stat5b-deleted mice, which show decreased Mup expression (35). Using rt-qPCR, we probed F4 liver expression of Ghr and Igf1, the latter of which serves as a reliable proxy for the activity of the GHR–STAT5b transduction pathway (35). We found no differences in expression of Ghr [t (15) = 1.07, P = 0.30] or Igf1 [t (15) = 0.15, P = 0.88]. Together with testosterone results, these data suggest that MUP up-regulation in promiscuous-line males is not driven by changes in hormone-mediated transcription.
Differential DNA methylation of Mups occurs in unmanipulated sperm and oocytes, and, unlike most genes, this methylation is not completely erased during gametogenesis and embryogenesis (36). Also, mice born from manipulated zygotes (nucleocytoplasmic parthenogenetic hybrids) show increased Mup-specific DNA methylation and associated transcriptional repression in the liver relative to genetically identical controls (37), and this phenotype is heritable (24). The first mammalian genes found to exhibit transgenerational epigenetic inheritance were Mups and Obp (odorant binding protein) (24), but the adaptive significance of this regulation is unknown. To test whether sociality triggers transgenerational epigenetic regulation of Mups, we analyzed promoter methylation of Mup11 (up-regulated in promiscuous-line males and virtually silent in monogamous-line males) using bisulfite sequencing of liver genomic DNA from F4 adult males. We targeted four CpG dinucleotides within the promoter and five-prime untranslated region (Fig. S5). Promiscuous-line males had twofold reduced methylation at site 4 [P = 0.016, Fisher's exact test (FET)] relative to monogamous-line males (Fig. 3). All other differences were nonsignificant (P > 0.27, FET). Urinary MUP expression and methylation at site 4 were negatively correlated and marginally significant [t (4) = −3.20, P = 0.0853]. Sequencing of PCR products revealed several amplicons that were highly similar to Mup11. To confirm that DNA methylation of these sites was associated with liver-specific expression, we measured methylation in tail epithelial cells (where Mups are not expressed) and found that all sites were more methylated than in the liver (n = 74, P = 0.012, Mann–Whitney Test), including site 4 (P = 0.013, FET). Thus, loss of methylation in the 5′ UTR of Mups in promiscuous-line males could contribute to higher MUP expression.
Fig. 3.
DNA methylation of Mup promoter in F4 P line and M line males. Open and filled circles represent unmethylated and methylated CpG dinucleotides, respectively. Promiscuous-line males had significantly less methylation at site 4, a putative binding site for USF1 (furthest right); P < 0.05 Fisher’s exact test. n = 8 individuals, 66 sequences.
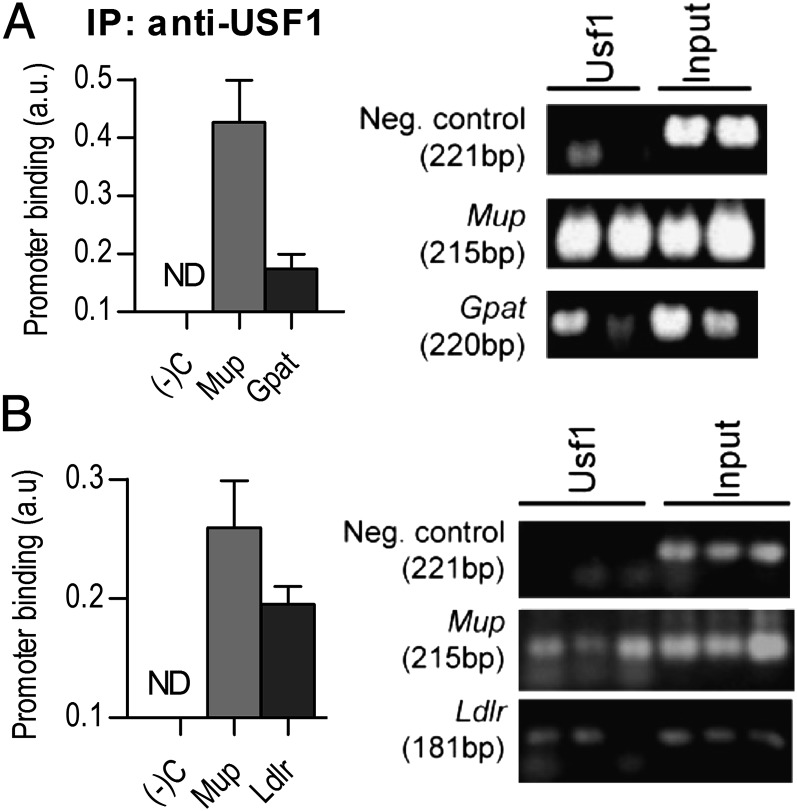
To identify whether reduction in promoter methylation at site 4 in promiscuous-line males was biologically relevant, we identified transcription-factor candidates (38). Upstream stimulatory factor 1 (USF1) was selected because it is a mammalian factor active in the liver and is methylation-sensitive (i.e., DNA methylation inhibits binding) (39). A putative binding motif of this factor (AAGACGTG) was conserved in 19 out of 21 Mup coding sequences analyzed, and located 37 bp upstream from the start codon and ∼60 bp downstream from the transcriptional start site. Using chromatin immunoprecipitation (ChIP) on liver extracts, we interrogated USF1-bound DNA with primers flanking the putative binding site and sharing sequence identity with 11 Mup loci. Glycerol-3-phosphate acyltransferase (Gpat) (40) and low-density lipoprotein receptor (Ldlr) (41) are constitutive hepatic targets of USF1 and were used as positive controls. A highly conserved gene desert (42) served as a negative control (Table S1). Analysis found support for occupancy of the Mup promoter by USF1; the Mup amplicon was enriched relative to Gpat by 2.45-fold (Fig. 4A) and Ldlr by 1.39-fold (Fig. 4B). Therefore, a reduction of methylation in site 4 of the Mup promoter could contribute to increased Mup expression via enhanced binding and activity of USF1.
Fig. 4.
USF1 binding to Mup promoter in vivo. ChIP was performed on hepatocytes from naïve mice and assessed with rt-qPCR (Left) and gel electrophoresis (Right). Two positive controls were used: Gpat (A) and Ldlr (B). ND, none detected. Means ± SEM.
Female Odor Preference for Up-Regulation of MUPs.
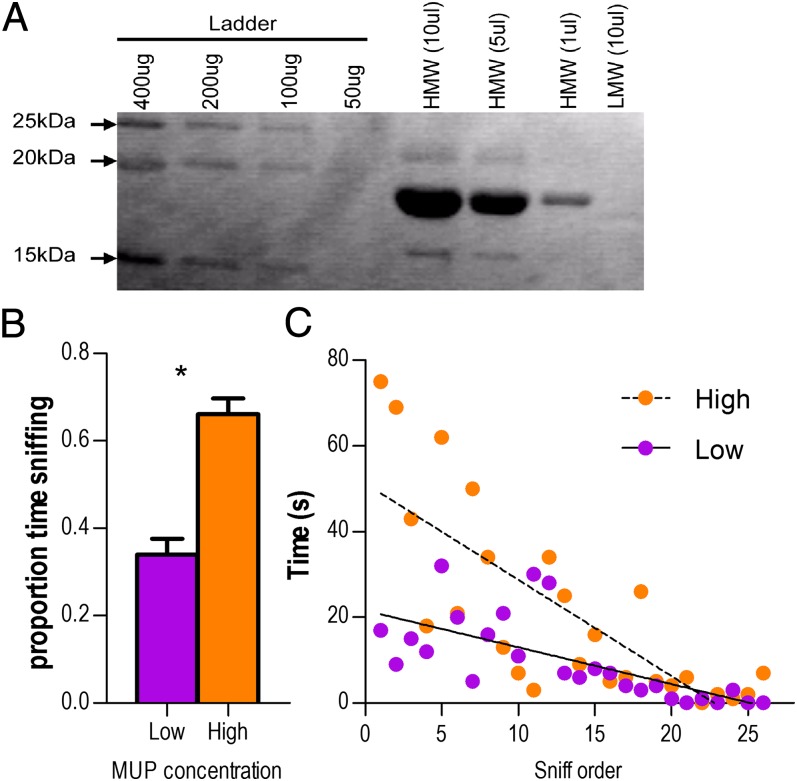
Our model assumes that up-regulated MUPs in promiscuous-line males contribute to their enhanced mating success, but the quantitative role of MUP expression in mate choice has not been evaluated. Mice respond to some pheromones at subpicomolar concentrations (43), yet some males excrete molar concentrations of MUPs (44). Are MUPs analogous to a sexually selected peacock's tail? To address this question, we measured female attraction to artificial scent marks that were identical except for MUP concentration. Using the same preference arena used to identify female attraction to promiscuous-line males (14), one female per trial was given a choice between high concentration (20 mg/mL) vs. low concentration (3 mg/mL) urine spots derived from a single male, thereby controlling for potential preferences based on the identity of the male (Fig. 5A).
Fig. 5.
Female odor preference for increased MUP concentration. (A) SDS/PAGE of manipulated urine samples; shown are protein ladder, HMW (>10 kDa), and LMW (<10 kDa) fractions. HMW bands are MUPs. (B) Females spent significantly more time sniffing high-concentration than low-concentration spots. Means ± SE. (C) Females were more likely to visit high-concentration spots first (see Female Odor Preference for Up-Regulation of MUPs), and for a longer period, than low concentration spots. n = 12 trials.
GLM analysis found that females spent a greater proportion of time in direct contact with the high-concentration marks than the low concentration marks (Fig. 5B) (S = −37.0; P = 0.0015). Additionally, females were more likely to sniff high-concentration odors first (10/12 trials, χ2 = 5.3, P = 0.02), and for a longer time, than low-concentration odors (Fig. 5C); the slopes of these lines were significantly different (high, β = −0.45; low β = −1.165; F1,14 = 10.8, P = 0.002). Thus, increasing urinary MUP concentration serves as a female attractant.
Discussion
Vertebrate social networks are established within territorial boundaries and regulate access to mating success in nature (7). In captivity, this social context is missing, and lack of social selection is hypothesized to favor phenotypic traits that erode fitness in natural conditions (6). We found that laboratory mice reintroduced to the opportunity for social selection adapted by producing highly attractive males with otherwise impaired viability. There were no major female effects, suggesting selection on male secondary sexual traits (14). Here, we identify a role of transgenerational regulation of MUP pheromones contributing to this male phenotype. Although MUPs are a primary constituent of mouse urine, they also release several volatile pheromones (45), and additional odorants could be associated with the effects reported here. Indeed, we probed MUPs using a candidate-gene approach, and it is possible that other physiological and behavioral mechanisms underlie this male phenotype.
Mammalian maternal effects can be adaptive, and modulation of offspring development in response to fluctuating resource availability has been described (46). Very few studies, however, have identified adaptive maternal effects under contrasting social conditions. Recent work in free-ranging red squirrels found that experimental increases in population density triggered elevated glucocorticoids in breeding females, which in turn led to an adaptive increase in offspring growth rate (47). We add to this knowledge by showing that female mice respond to sociality by producing larger litters and, importantly, by programming their sons to up-regulate MUPs (whereas they themselves do not up-regulate). This maternal effect appears to be uniquely independent of hormonal signals (46). Although both growth hormone and testosterone regulate MUP expression, these systems were unaffected in our experiment. Both hormones undergo pulsatile, bimodal release from endocrine glands, and we may have lacked power to detect testosterone differences. Igf1, however, is a direct target of the GH–STAT5b signaling pathway and is stably correlated with growth hormone; we found no differences in Ghr or Igf1 expression in promiscuous-line and monogamous-line males.
Sociality-driven paternal effects had unexpected consequences for MUP expression: sons of C males had lower MUP expression than sons of NC males. The divergent maternal and paternal effects on MUPs reported here are suggestive of intriguing conflicts of interest. First, when male mice breed in a defended social territory, maturing sons become competitors and will often supplant their father from the territory (48). Urinary pheromones are a medium by which males compete (45), and MUPs instigate male–male aggression in a concentration-dependent fashion (49). Thus, fathers and sons are potentially in a conflict of interest over the paternal territory, and fathers might benefit by repressing MUP expression in sons. Consistent with this hypothesis, promiscuous line males had elevated MUP expression as individually housed adults but not as juveniles, a timepoint coinciding with onset of dispersal behaviors and father–son aggression (44, 48). Second, these results are suggestive of parental conflict. Females may benefit from producing sons with up-regulated MUPs because, as shown here, those sons have better success in attracting mates. Thus, mothers and fathers might be at conflict over MUP expression in offspring. Our data indicate that mothers exert a roughly twofold stronger influence on offspring MUP expression than fathers.
Transgenerational epigenetic inheritance (8) might have contributed to the parental effects described here. Mups in house mice cover an approximately 2 megabase region of chromosome four and are interspersed with many endogenous retrotransposons (29, 50). Epigenetic gene regulation often involves interactions with the silencing of neighboring transposable elements and includes response to environmental stressors (51) and transgenerational inheritance (8). Site-specific, intronic DNA methylation has been linked to Mup expression, and altered methylation profiles have been shown to be heritable across generations (24). We found a reduction in methylation at a CpG dinucleotide in the Mup promoter in hepatocytes of promiscuous-line males. This reduction in methylation could enhance Mup expression by promoting the binding of the stimulatory transcription factor USF1, which has been shown to be methylation-sensitive (39). In summary, these results provide insight as to why MUPs are under transgenerational regulation and motivate further investigations of parental effects on USF1-mediated modulation of Mup expression in a methylation-dependent manner. Our paradigm of enforced monogamy vs. promiscuous breeding simulates the dramatic fluctuations in population density experienced by mice in nature (14), and we hypothesize that mice in high-density populations will gain more benefits from up-regulating MUPs than mice in low-density populations.
Materials and Methods
All experimental procedures are detailed in SI Material and Methods. The University of Utah Institutional Animal Care and Use Committee approved all experiments (08-10017). Mice were derived from a cross between wild-caught and MHC-congenic mice carrying five known haplotypes (14). Plasma testosterone from 15 males of each treatment was measured by RIA. Males were exposed to estrus females (identified by vaginal cytology) for 30 min. Urine, collected by scruffing mice over a clean Plexiglas sheet, was immediately flash frozen and stored at −70 °C. Immediately following a 1:20 dilution, urinary protein concentration (of which >95% is MUPs) (52) was determined with a Bradford Assay. Creatinine was measured colorimetrically using Jaffe’s picrate method (Stanbio Liquicolor Kit). Rt-qPCR was performed on 20 ng of liver-extracted RNA (RNeasy; Qiagen) with the Lightcycler SYBR reaction kit (Roche). Gapdh was used as a housekeeper comparison.
Liver and tail DNA was sodium bisulfite converted as described (53). Seminested bisulfite PCR fragments were islolated from gels (Qiagen), phenol-chloroform purified, cloned (Promega), and plasmids were isolated with minipreps (Qiagen). Individually sequenced clones were analyzed with QUMA (RIKEN Institute). ChIP was performed on livers from naïve males. Samples were cross-linked in 1% formaldehyde, neutralized with glycine, Dounce homogenized, and chromatin-immunoprecipated as described (54).
For odor preference assays, protein fractions from individual male urine samples were separated using 10-kDa molecular weight cutoff centrifugal filters (Millipore). HMW concentrate was added back to two equal volumes of LMW filtrate to achieve 20 mg/mL or 3 mg/mL MUP concentration samples. Aliquots of samples were pipetted onto a grid of 18 spots in either side of the arena. Using Timescience video analysis software, preference was determined by time spent in direct contact with urine spots for 30 min.
Supplementary Material
Acknowledgments
We thank John Sampinos, Christine Sembrano, Linda Morrison, and Jon Gale for help with experiments. We also thank Catherine Dulac for support. This work was funded by National Science Foundation Grants 0909801 (to A.C.N.), 0914244 (to A.C.N.), and 0918969 (to W.K.P.), and by National Institutes of Health Genetics Training Grant T32 (to A.C.N.) and R01-GM039578 and R01-GM109500 (to W.K.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310427110/-/DCSupplemental.
References
- 1.Frankham R. Genetic adaptation to captivity in species conservation programs. Mol Ecol. 2008;17(1):325–333. doi: 10.1111/j.1365-294X.2007.03399.x. [DOI] [PubMed] [Google Scholar]
- 2.Araki H, Cooper B, Blouin MS. Genetic effects of captive breeding cause a rapid, cumulative fitness decline in the wild. Science. 2007;318(5847):100–103. doi: 10.1126/science.1145621. [DOI] [PubMed] [Google Scholar]
- 3.Christie MR, Marine ML, French RA, Blouin MS. Genetic adaptation to captivity can occur in a single generation. Proc Natl Acad Sci USA. 2012;109(1):238–242. doi: 10.1073/pnas.1111073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnett SA, Dickson RG. Wild mice in the cold: Some findings on adaptation. Biol Rev Camb Philos Soc. 1989;64(4):317–340. doi: 10.1111/j.1469-185x.1989.tb00679.x. [DOI] [PubMed] [Google Scholar]
- 5.Heath DD, Heath JW, Bryden CA, Johnson RM, Fox CW. Rapid evolution of egg size in captive salmon. Science. 2003;299(5613):1738–1740. doi: 10.1126/science.1079707. [DOI] [PubMed] [Google Scholar]
- 6.Wedekind C. Sexual selection and life-history decisions: Implications for supportive breeding and the management of captive populations. Conserv Biol. 2002;16(5):1204–1211. [Google Scholar]
- 7.West Eberhard MJ. Sexual selection, social competition and speciation. Q Rev Biol. 1983;58:155–183. [Google Scholar]
- 8.Youngson NA, Whitelaw E. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet. 2008;9:233–257. doi: 10.1146/annurev.genom.9.081307.164445. [DOI] [PubMed] [Google Scholar]
- 9.Uller T. Developmental plasticity and the evolution of parental effects. Trends Ecol Evol. 2008;23(8):432–438. doi: 10.1016/j.tree.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Krebs JR, Davies NB. Behavioural Ecology: An Evolutionary Approach. UK: Wiley-Blackwell, Oxford; 1997. [Google Scholar]
- 11.Ruff JS, Nelson AC, Kubinak JL, Potts WK. MHC signaling during social communication. Adv Exp Med Biol. 2012;738:290–313. doi: 10.1007/978-1-4614-1680-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll LS, Potts WK. Sexual selection: Using social ecology to determine fitness differences. In: Wolff JO, Sherman PW, editors. Rodent Societies. Chicago: University of Chicago Press; 2007. pp. 57–67. [Google Scholar]
- 13.Rice WR. Dangerous liaisons. Proc Natl Acad Sci USA. 2000;97(24):12953–12955. doi: 10.1073/pnas.97.24.12953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson AC, Colson KE, Harmon S, Potts WK. Rapid adaptation to mammalian sociality via sexually selected traits. BMC Evol Biol. 2013;13(1):81. doi: 10.1186/1471-2148-13-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurst JL, et al. Individual recognition in mice mediated by major urinary proteins. Nature. 2001;414(6864):631–634. doi: 10.1038/414631a. [DOI] [PubMed] [Google Scholar]
- 16.Cheetham SA, et al. The genetic basis of individual-recognition signals in the mouse. Curr Biol. 2007;17(20):1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Cheetham SA, Smith AL, Armstrong SD, Beynon RJ, Hurst JL. Limited variation in the major urinary proteins of laboratory mice. Physiol Behav. 2009;96(2):253–261. doi: 10.1016/j.physbeh.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Gosling LM, Roberts SC, Thornton EA, Andrew MJ. Life history costs of olfactory status signalling in mice. Behav Ecol Sociobiol. 2000;48:328–332. [Google Scholar]
- 19.Stopková R, Stopka P, Janotová K, Jedelský PL. Species-specific expression of major urinary proteins in the house mice (Mus musculus musculus and Mus musculus domesticus) J Chem Ecol. 2007;33(4):861–869. doi: 10.1007/s10886-007-9262-9. [DOI] [PubMed] [Google Scholar]
- 20.Garratt M, Stockley P, Armstrong SD, Beynon RJ, Hurst JL. The scent of senescence: Sexual signalling and female preference in house mice. J Evol Biol. 2011;24(11):2398–2409. doi: 10.1111/j.1420-9101.2011.02367.x. [DOI] [PubMed] [Google Scholar]
- 21.Janotova K, Stopka P. The level of major urinary proteins is socially regulated in wild Mus musculus musculus. J Chem Ecol. 2011;37(6):647–656. doi: 10.1007/s10886-011-9966-8. [DOI] [PubMed] [Google Scholar]
- 22.Garratt M, et al. Tissue-dependent changes in oxidative damage with male reproductive effort in house mice. Funct Ecol. 2012;26(2):423–433. [Google Scholar]
- 23.Sampsell BM, Held WA. Variation in the major urinary protein multigene family in wild-derived mice. Genetics. 1985;109(3):549–568. doi: 10.1093/genetics/109.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roemer I, Reik W, Dean W, Klose J. Epigenetic inheritance in the mouse. Curr Biol. 1997;7(4):277–280. doi: 10.1016/s0960-9822(06)00124-2. [DOI] [PubMed] [Google Scholar]
- 25.Reed WL, et al. Physiological effects on demography: A long-term experimental study of testosterone’s effects on fitness. Am Nat. 2006;167(5):667–683. doi: 10.1086/503054. [DOI] [PubMed] [Google Scholar]
- 26.Litvinova EA, Kudaeva OT, Mershieva LV, Moshkin MP. High level of circulating testosterone abolishes decline in scent attractiveness in antigen-treated male mice. Anim Behav. 2005;69(3):511–517. [Google Scholar]
- 27.Knopf JL, Gallagher JF, Held WA. Differential, multihormonal regulation of the mouse major urinary protein gene family in the liver. Mol Cell Biol. 1983;3(12):2232–2240. doi: 10.1128/mcb.3.12.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James PJ, Nyby JG, Saviolakis GA. Sexually stimulated testosterone release in male mice (Mus musculus): Roles of genotype and sexual arousal. Horm Behav. 2006;50(3):424–431. doi: 10.1016/j.yhbeh.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 29.Logan DW, Marton TF, Stowers L. Species specificity in major urinary proteins by parallel evolution. PLoS ONE. 2008;3(9):e3280. doi: 10.1371/journal.pone.0003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts SA, et al. Darcin: A male pheromone that stimulates female memory and sexual attraction to an individual male’s odour. BMC Biol. 2010;8:75. doi: 10.1186/1741-7007-8-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grauer GF, Thomas CB, Eicker SW. Estimation of quantitative proteinuria in the dog, using the urine protein-to-creatinine ratio from a random, voided sample. Am J Vet Res. 1985;46(10):2116–2119. [PubMed] [Google Scholar]
- 32.Heymsfield SB, Arteaga C, McManus C, Smith J, Moffitt S. Measurement of muscle mass in humans: Validity of the 24-hour urinary creatinine method. Am J Clin Nutr. 1983;37(3):478–494. doi: 10.1093/ajcn/37.3.478. [DOI] [PubMed] [Google Scholar]
- 33.Beynon RJ, Hurst JL. Urinary proteins and the modulation of chemical scents in mice and rats. Peptides. 2004;25(9):1553–1563. doi: 10.1016/j.peptides.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 34.Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20(11):2613–2629. doi: 10.1210/me.2006-0007. [DOI] [PubMed] [Google Scholar]
- 35.Davey HW, et al. STAT5b is required for GH-induced liver IGF-I gene expression. Endocrinology. 2001;142(9):3836–3841. doi: 10.1210/endo.142.9.8400. [DOI] [PubMed] [Google Scholar]
- 36.Sanford JP, Clark HJ, Chapman VM, Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes Dev. 1987;1(10):1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- 37.Reik W, et al. Adult phenotype in the mouse can be affected by epigenetic events in the early embryo. Development. 1993;119(3):933–942. doi: 10.1242/dev.119.3.933. [DOI] [PubMed] [Google Scholar]
- 38.Messeguer X, et al. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18(2):333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 39.Fujii G, et al. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem J. 2006;395(1):203–209. doi: 10.1042/BJ20051802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshida M, et al. Identification of cis-acting promoter sequences required for expression of the glycerol-3-phosphate acyltransferase 1 gene in mice. Biochim Biophys Acta. 2009;1791(1):39–52. doi: 10.1016/j.bbalip.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Latasa M-J, Griffin MJ, Moon YS, Kang C, Sul HS. Occupancy and function of the -150 sterol regulatory element and -65 E-box in nutritional regulation of the fatty acid synthase gene in living animals. Mol Cell Biol. 2003;23(16):5896–5907. doi: 10.1128/MCB.23.16.5896-5907.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nóbrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431(7011):988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- 43.Leinders-Zufall T, Ishii T, Mombaerts P, Zufall F, Boehm T. Structural requirements for the activation of vomeronasal sensory neurons by MHC peptides. Nat Neurosci. 2009;12(12):1551–1558. doi: 10.1038/nn.2452. [DOI] [PubMed] [Google Scholar]
- 44.Rusu AS, Krackow S, Jedelsky PL, Stopka P, Konig B. A qualitative investigation of major urinary proteins in relation to the onset of aggressive behavior and dispersive motivation in male wild house mice (Mus musculus domesticus) J Ethol. 2008;26(1):127–135. [Google Scholar]
- 45.Hurst JL, Beynon RJ. Scent wars: The chemobiology of competitive signalling in mice. Bioessays. 2004;26(12):1288–1298. doi: 10.1002/bies.20147. [DOI] [PubMed] [Google Scholar]
- 46.Maestripieri D, Mateo JM. Maternal Effects in Mammals. Chicago: Univ Chicago Press; 2009. [Google Scholar]
- 47.Dantzer B, et al. Density triggers maternal hormones that increase adaptive offspring growth in a wild mammal. Science. 2013;340(6137):1215–1217. doi: 10.1126/science.1235765. [DOI] [PubMed] [Google Scholar]
- 48.Gerlach G. Emigration mechanisms in feral house mice: A laboratory investigation of the influence of social structure, population density, and aggression. Behav Ecol Sociobiol. 1996;39(3):159–170. [Google Scholar]
- 49.Chamero P, et al. Identification of protein pheromones that promote aggressive behavior. Nature. 2007;450(7171):899–902. doi: 10.1038/nature05997. [DOI] [PubMed] [Google Scholar]
- 50.Mudge JM, et al. Dynamic instability of the major urinary protein gene family revealed by genomic and phenotypic comparisons between C57 and 129 strain mice. Genome Biol. 2008;9(5):R91. doi: 10.1186/gb-2008-9-5-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8(4):272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 52.Beynon RJ, et al. Polymorphism in major urinary proteins: Molecular heterogeneity in a wild mouse population. J Chem Ecol. 2002;28(7):1429–1446. doi: 10.1023/a:1016252703836. [DOI] [PubMed] [Google Scholar]
- 53.Arnaud P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12(9):1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 54.Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100(25):14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.