Significance
Chloroplasts contain approximately 3,000 proteins, of which most are nuclear gene products synthesized in the cytosol with NH2-terminal, cleavable extensions referred to as chloroplast transit peptides for import. Chloroplasts additionally contain a set of plastid proteins lacking cleavable transit peptides. In this article we show how transit sequence-less proteins are transported from the cytosol to the inner envelope of the chloroplast. This import pathway is comparable to the mitochondrial pathway used for the import of carrier proteins and other, hydrophobic proteins without a transit sequence into the inner mitochondrial membrane.
Keywords: chloroplast biogenesis, membrane transport, protein translocation
Abstract
A family of 17 putative preprotein and amino acid transporters designated PRAT has been identified in Arabidopsis thaliana, comprising PRAT proteins in mitochondria and chloroplasts. Although some PRAT proteins, such as the translocon of the mitochondrial inner membrane (TIM) proteins TIM22 and TIM23, play decisive roles for the translocation and import of mitochondrial inner membrane proteins, little is known about the role of the different PRAT members in chloroplasts. Here we report the identification of three distinct PRAT proteins as part of a unique protein import site. One of the identified PRAT proteins is identical with a previously characterized hypothetical protein (HP) of 20 kDa designated HP20 of the outer plastid envelope membrane. The second PRAT component is represented by HP30, and the third is identical to HP30-2, a close relative of HP30. Both HP30 and HP30-2 are inner plastid envelope membrane proteins of chloroplasts. Using biochemical, cell biological, and genetic approaches we demonstrate that all three PRAT proteins cooperate during import of transit sequence-less proteins, such as the quinone oxidoreductase homolog ceQORH used as model, into the inner chloroplast envelope membrane. Our data are reminiscent of findings reported for the TIM22 translocase, which is involved in the import of carrier proteins and other, hydrophobic membrane proteins lacking cleavable transit sequences into the inner mitochondrial membrane. Together our results establish the PRAT family as a widely used system of protein translocases in different membranes of endosymbiotic origin.
Plastids represent a highly divergent family of cell organelles (1). They are ubiquitously found in plant and algal cells and provide essential metabolic and signaling functions (2, 3). The hallmark organelles of green plants are chloroplasts that contain the green pigment chlorophyll and perform photosynthesis for autotrophic growth (1–3).
Chloroplasts contain ∼3,000 proteins, of which most are nuclear gene products (4). The majority of these proteins is synthesized with NH2-terminal, cleavable extensions referred to as chloroplast transit peptides, which guide the cytosolic precursors to the outer plastid envelope membrane and initiate membrane translocation through two closely interacting multiprotein complexes, called translocon of the outer chloroplast envelope (TOC) and translocon of the inner chloroplast envelope (TIC) (5). During or shortly after membrane passage, the NH2-terminal transit peptides are cleaved off proteolytically (5).
Proteomics studies by Kleffmann et al. (6) revealed a set of plastid proteins lacking cleavable transit peptides. Therefore, the existence of alternative targeting signals and import pathways was proposed (6). Examples for inner envelope membrane proteins that lack a cleavable transit peptide are TIC32 (7) and a protein homologous to quinone oxidoreductases of bacteria, yeast, and animals, designated chloroplast-envelope quinone-oxidoreductase homolog, ceQORH (8, 9).
TIC32, a member of the TIC/TOC translocon complex (5), was shown to be imported as a mature-sized polypeptide and in a TOC159- and TOC75-independent way that required low energy amounts (<20 µM ATP) (7). TOC159 and TOC75 represent the main receptor and translocation channel components, respectively, of the outer plastid envelope membrane (5). Mutant studies revealed that the 10 most NH2-terminal amino acids are essential for chloroplast import of TIC32 (7). Moreover, a role of the outer envelope proteins OEP16, OEP21, and OEP24 could be excluded in the import of TIC32 (7). Cross-linking indicated that TIC32 closely interacts with TIC22 and TIC110 (7).
ceQORH is the second example of an inner envelope protein of chloroplasts that lacks a cleavable NH2-terminal transit peptide (8). Competition and antibody blocking experiments revealed that ceQORH import occurs independently of TOC159 and TOC75 (9). Plastid import of ceQORH was nevertheless dependent on proteinaceous receptor components exposed at the outer plastid surface and on high energy concentrations (≥2 mM ARP + 0.1 mM GTP) (9). ceQORH has an interesting tripartite structure, consisting of two membrane anchor domains that are localized at the NH2 and COOH termini, respectively, and a central domain residing between residues 60 and 100 that confers solubility to the protein (8, 9). All three domains interact to govern ceQORH’s unique import pathway. When fused to the GFP, the central domain drove import of the resulting (60-100)-ceQORH-GFP fusion protein into plastids through a TOC159- and TOC75-dependent pathway (9). Its import was inhibited by TOC75 antibodies and by the precursor of ferredoxin (pFD), a photosynthetic protein known to enter the plastids through TOC159 and TOC75. By contrast, the full-length ceQORH did not engage TOC159 and TOC75 and interacted with a protein of ∼30 kDa of unknown function (9).
Here we identified this 30-kDa protein and show that it is identical to a previously described hypothetical protein designated HP20 of the outer chloroplast envelope membrane (10, 11). HP20 belongs to the family of preprotein and amino acid transporters, PRAT, members of which are present in bacteria, mitochondria, and chloroplasts (12, 13). In addition to HP20, two additional PRAT proteins were identified, designated HP30 and HP30-2, which are present in the inner chloroplast envelope and cooperate in import of ceQORH and also are involved in import of other, transit sequence-less inner plastid envelope proteins into chloroplasts.
Results
Identification of Proteins Interacting with ceQORH.
Mitochondria and chloroplasts import a large number of proteins from the cytosol. It has been demonstrated that this import step occurs through so-called protein-conducting channels spanning the limiting borders of both organelle types (5, 14). During translocation, the translocating proteins are in such close physical proximity to the components of the import machinery that it allows formation of mixed disulfide bonds. Here we used disulfide bridge cross-linking triggered by Elman’s reagent [5,5′-dithiobis(2-nitrobenzoic acid), DTNB] (15) to identify proteins interacting with ceQORH during its import into chloroplasts.
Incubations were carried out with DTNB-activated ceQORH-(His)6 or (60-100)-ceQORH-GFP-(His)6 and Arabidopsis chloroplasts that had been isolated and energy-depleted (16). In the first set of experiments we tested the role of Mg-ATP and Mg-GTP on ceQORH import. As shown previously for ceQORH (9) and other proteins (17, 18), import is initially reversible and occurs in an equilibrium reaction unless nucleoside triphosphates are present. Low, 0.1-mM concentrations of Mg-ATP favor insertion of the precursors into the outer envelope membrane and allow first contacts with components of the import machinery of the inner envelope membrane. These interactions are stabilized in the presence of 0.1 mM Mg-GTP. The precursor then partially inserts into the inner envelope membrane but does not cross it. Membrane translocation requires ≥2-mM concentrations of Mg-ATP, powering molecular chaperons operating as import motors (5).
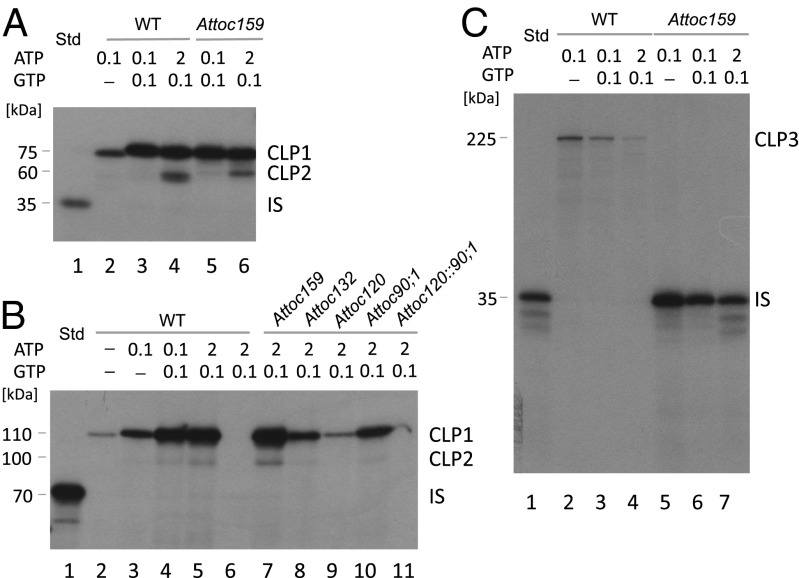
DTNB-activated ceQORH-(His)6 was added to incubation mixtures containing 0.1 mM Mg-ATP (used to study ceQORH binding to the plastids), 0.1 mM Mg-ATP and 0.1 mM Mg-GTP (used to study membrane insertion of ceQORH) or 2 mM Mg-ATP plus 0.1 mM Mg-GTP (used to study import of ceQORH). After a 15-min incubation, cross-linking was followed by nonreducing SDS/PAGE. Fig. 1 shows that one main cross-link product (CLP) of ∼75 kDa was obtained (Fig. 1A, lanes 2–4, upper band, designated CLP1). As deduced from the size difference it consists of ceQORH (∼35 kDa) and a ∼40-kDa band. The apparent molecular mass of the cross-linked ∼40-kDa band seemed larger than that reported previously for ceQORH-GFP-(His)6, giving rise to a ∼30-kDa band (9). The ∼40-kDa band appeared at 0.1 mM MgATP; its amount increased in the presence of 0.1 mM Mg-GTP, favoring insertion of ceQORH across the outer plastid envelope membrane and permitting partial insertion of ceQORH across the inner plastid envelope membrane. In addition to the ∼75-kDa band, a second, weaker cross-link product of ∼60 kDa was obtained (Fig. 1A, lane 4, lower band, designated CLP2), consisting of ceQORH (35 kDa) and a ∼25-kDa band. This ∼25-kDa band appeared only at 2 mM Mg-ATP triggering import of ceQORH into the inner plastid envelope membrane. The size of this ∼25-kDa band matched more closely that of the previously reported cross-link product of ∼30 kDa (9). Both the 75-kDa and 60-kDa bands were seen when the incubations were carried out with plastids that had been isolated from the Attoc159 mutant described by Bauer et al. (19) lacking TOC159 (Fig. 1A, lanes 5 and 6). Arabidopsis contains four TOC159 homologs, named AtTOC159, AtTOC132, AtTOC120, and AtTOC90, which display specialized functions during plant development (19, 20). AtTOC159 operates as the main import receptor for photosynthetic proteins but is not involved in import of nonphotosynthetic proteins (19, 20). Experiments using previously described Attoc120 and Attoc90 knockout mutants showed that the import of a ceQORH-GFP-(His)6 fusion protein (Fig. 1B) is dependent on AtTOC120 and AtTOC90, operating as import receptors for nonphotosynthetic proteins (20). Disulfide bridge cross-linking with DTNB-activated (60-100)-ceQORH-GFP-(His)6 yielded a completely different, unique cross-link product of ∼225 kDa that consisted of the precursor and AtTOC159, as deduced from import studies with plastids isolated from the Attoc159 mutant (Fig. 1C).
Fig. 1.
(A–C) Cross-linking of DTNB-derivatized 35S-ceQORH (A), 35S-ceQORH-GFP (B), and 35S-(60-100)-ceQORH-GFP (C) to plastid envelope proteins in Arabidopsis WT and mutant chloroplasts. Incubations were carried out at the indicated Mg-ATP and Mg-GTP concentrations; cross-link products were detected by nonreducing SDS/PAGE. IS defines the added import substrate (10% in A, 100% in B and C).
The PRAT Family of Arabidopsis thaliana.
The ∼75-kDa and ∼60-kDa disulfide-bridged cross-link products of ceQORH-(His)6 were detergent-solubilized from chloroplasts and purified by Ni-NTA chromatography (21). Protein sequencing (Fig. S1) revealed that the ∼75-kDa band consisted of ceQORH-(His)6 (35 kDa) and a previously characterized hypothetical outer plastid envelope protein (HP) of ∼20 kDa, designated HP20 (10, 11), presumably forming homodimers (40 kDa). By contrast, the weaker, ∼60-kDa cross-link product turned out to consist of ceQORH-(His)6 and two subbands that were identical to two previously described plastid proteins, designated HP30 and HP30-2 (10, 11) (Fig. S1), and each displayed a molecular mass of ∼30 kDa. Remarkably, all three HP proteins were found to be members of the PRAT family described by Rassow et al. (12).
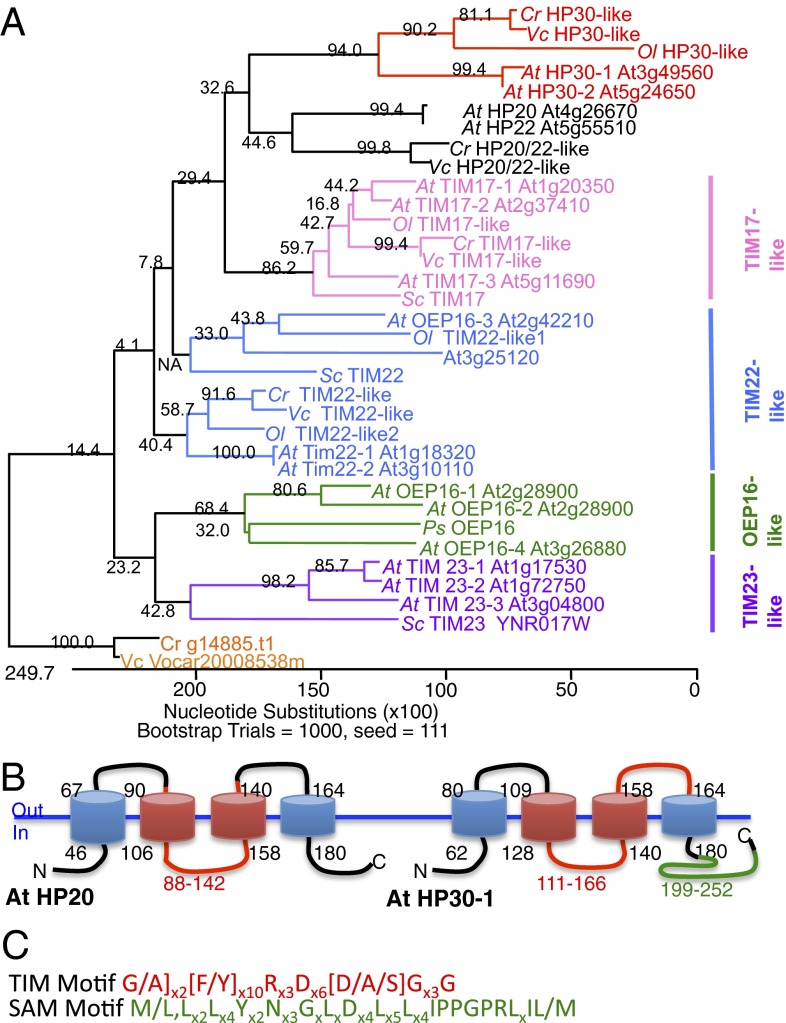
Seventeen PRAT genes have been identified for the model plant A. thaliana that form different phylogenetic clades (Fig. 2A), have different functions, and exhibit unique expression patterns (13). All PRAT proteins are predicted to be membrane proteins forming up to four trans-membrane spans (Fig. 2B), and at least some contain the family-defining PRAT motif (12, 13) (Fig. 2C). The known functions of the founding members of the PRAT family, TIM17, TIM22, TIM23, and OEP16, are in amino acid and preprotein transport. For example, TIM17, TIM22, and TIM23 are key components of the preprotein translocases in the inner mitochondrial membrane (14). For OEP16-1, a role as amino acid and preprotein transporter has been proposed (21, 22).
Fig. 2.
The PRAT family of A. thaliana, Chlamydomonas reinhardtii, Volvox carteri, and Ostreococcus lucimarinus. (A) Schematic phylogenetic tree for HP20 subfamily (black), HP30 subfamily (red), OEP16-like subfamily (green), TIM23 subfamily (purple), TIM17 subfamily (pink), and TIM22 subfamily (blue). The protein alignment was performed using the TIM motif (pfam02466) (C) as the operational taxonomic unit (OTU) (sequence coordinates are listed in Table S1). Sequences were aligned using the MUSCLE algorithm (34) using a VTML200 substitution matrix, default gap opening and extension penalties, a UPGMB cluster method [a clustering method based on a combination of both UPGMA (Unweighted Pair Group Method with Arithmetic Mean) and Neighbor Joining], Kimura % identity as distance measure, and a CLUSTALW sequence weight. The reliability of each bifurcation was estimated using bootstrap analysis (1,000 trials). (B) Predicted PRAT protein structure of AtHP20 (At4g26670) and AtHP30-1 (At3g49560) consisting of four trans-membrane domains and three hydrophilic loops connecting these segments. Transmembrane helices were predicted using the TMPRED algorithm (35). The region comprising the conserved Tim motif and used as the OTU in A is highlighted in red. A predicted SAM (cd09487) in HP30 proteins is shown in green (23). (C) Sequences of the TIM motif (pfam02466) defining the founding PRAT members and the SAM domain found in the HP30-like PRAT subfamily; x indicates variant amino acid residues (Tables S1 and S2).
HP20 was originally discovered in proteomics studies as an outer plastid envelope membrane protein in A. thaliana, annotated as Q9SZ09 (encoded by At4g26670) (10, 11). The closest relative of HP20 is HP22 (encoded by At5g55510) that shares 79% amino acid sequence identity (Fig. 2A; see also ref. 13). The second PRAT protein interacting with ceQORH is HP30 (encoded by At3g49560), and the third is HP30-2 (encoded by At5g24650) (13). HP30 and HP30-2 are closely related members of the PRAT family and exhibit 83% amino acid sequences identity (Fig. 2A and Fig. S1) (13). Phylogenetic analysis indicates that the HP20 and HP30-like proteins were already present early in the evolution of photosynthetic eukaryotes, as evidenced by the presence of single homologs in algae such as Chlamydomonas, Ostreococcus, and Volvox (Fig. 2A and Table S1), which diverged from land plants ∼1 billion years ago. The HP30 clade seems to have diverged from an HP20/22-like progenitor (Fig. 2A) through the acquisition of a predicted sterile α motif near the COOH terminus (SAM) (Fig. 2 B and C, Fig. S2, and Table S2). SAM domains have diverse functions, including a possible role in protein homo- and heterodimerization/oligomerization (23). HP20/22 and HP30 homologs were not found in the genomes of nonphotosynthetic eukaryotes.
Antibodies as Tools to Define the Localization and Functions of HP20 and HP30.
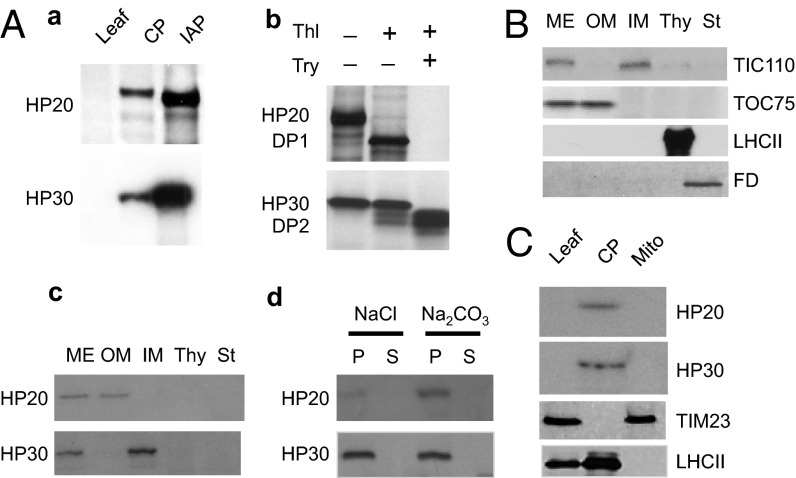
Antisera were raised against HP20 and HP30 and used to carry out biochemical tests on the localization and functions of these proteins in chloroplasts. The anti-HP20 serum did not detect a protein in total leaf extracts but revealed the presence of a ∼20-kDa band in isolated chloroplasts and complexes enriched for import intermediate-associated proteins (IAPs) formed at 0.1 mM Mg-GTP and 0.1 mM Mg-ATP with 35S-ceQORH in vitro (Fig. 3 A, a). Similarly, HP30 was at the limit of detection in total leaf extracts and enriched in chloroplasts and IAPs containing in vitro-imported 35S-ceQORH (Fig. 3 A, a). No HP20 and HP30 signals were obtained for isolated mitochondria (Fig. 3C).
Fig. 3.
Detection of HP20 and HP30 in chloroplasts and respective membrane fractions. (A, a) Identification of HP20 and HP30 in leaf extracts (leaf), purified chloroplasts (CP), and in terms of IAPs formed with in vitro-imported ceQORH. (A, b) thermolysin (Thl) and trypsin (Try) sensitivity of HP20 and HP30 in isolated chloroplasts. (A, c and d) Detection of HP20 and HP30 in mixed envelopes (ME), outer envelopes (OM), inner envelopes (IM), thylakoids (Th), and stroma (St) (c) and OM and IM membrane pellets (P) and respective supernatants (S) obtained after treatment with 1 N NaCl or 0.1 N Na2CO3 (pH 11) (d). (B) As in A, c but showing protein gel blot analysis with antibodies specific for the indicated outer and inner envelope marker proteins. (C) Western blot of HP20 and HP30 in leaf extracts (Leaf) as well as isolated chloroplasts (CP) and mitochondria (Mito). Respective marker proteins were TIM23 and LHCII. Ten micrograms of protein were loaded per lane.
Upon protease treatment, HP20 was partially sensitive to added thermolysin, but it was completely degraded by trypsin (Fig. 3 A, b). By contrast, HP30 was insensitive to thermolysin and converted into a smaller fragment upon trypsin treatment (Fig. 3 A, b). Thermolysin is known to degrade only surface-exposed plastid envelope proteins, whereas trypsin penetrates the outer envelope and breaks down inner plastid envelope proteins up to their membrane parts (24, 25). Fractionation experiments revealed that HP20 cofractionated with the outer plastid envelope membrane protein TOC75, whereas HP30 cofractionated with the inner plastid envelope membrane protein TIC110. This observation and the previous proteomics data (10, 11) are consistent with a localization of HP20 in the outer envelope membrane and HP30 in the inner envelope membrane of chloroplasts. When isolated, highly pure outer and inner envelopes (Fig. 3 A, c) were extracted with 1 N NaCl or 0.1 M Na2CO3 (pH 11), both HP20 and HP30 were recovered in the membrane pellet obtained after centrifugation (Fig. 3 A, d). These studies provide direct and compelling evidence for the localization of HP20 in the outer plastid envelope membrane and HP30 in the inner plastid envelope membrane and that both proteins are likely to represent integral membrane proteins of chloroplasts.
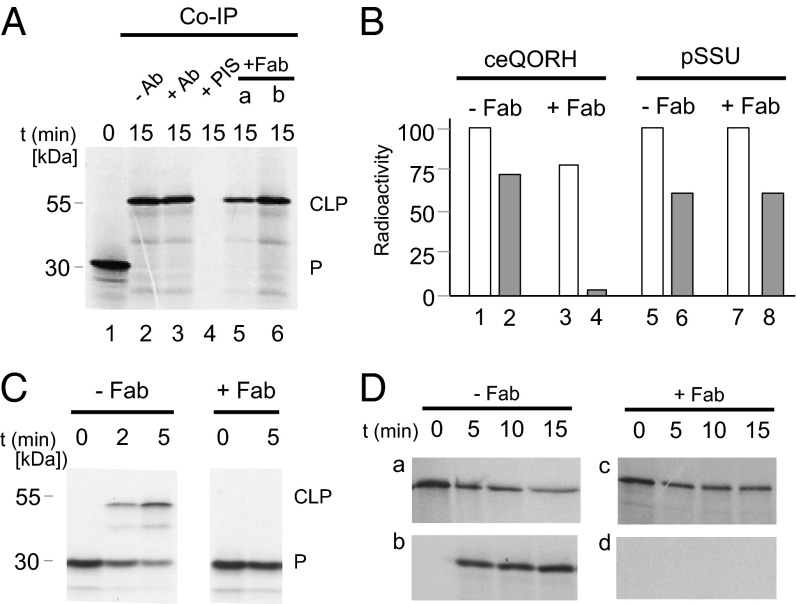
In subsequent experiments, we defined the role of HP20 during ceQORH import. DTNB-activated 35S-ceQORH was added to incubation mixtures containing 0.1 mM Mg-ATP (used to study plastid binding of ceQORH) or 2 mM Mg-ATP plus 0.1 mM Mg-GTP (used to study plastid import of ceQORH into the inner plastid envelope membrane). Fig. 4 highlights cross-linking of ceQORH to HP20 (Fig. 4A, lanes 2 and 3, as well as lanes 5 and 6). When Fab-decorated chloroplasts that had been generated during a preincubation were used, ∼20–25% reductions of 35S-ceQORH binding to the plastids were observed, compared with mock-incubated plastids (Fig. 4B, lane 3 vs. lane 1). More importantly, a block in ceQORH translocation across the outer plastid envelope membrane occurred in the presence of bound anti-HP20 Fab fragments (Fig. 4B, lane 4 vs. lane 2). In this case, no DTNB-mediated disulfide bridge cross-link product was obtained (Fig. 4C), and no ceQORH protein accumulated in chloroplasts (Fig. 4D).
Fig. 4.
Plastid binding and import of the 35S-ceQORH import substrate (IS). (A) Cross-link product (CLP) formation and coimmunoprecipitation of DTNB-activated 35S-ceQORH tested with anti-HP20 antibody (Ab)- or Fab-decorated (+Fab) as well as mock-incubated (+PIS) Arabidopsis chloroplasts. Ten microliters of HP20 antibody (lane 3), 2 µL and 5 µL of respective Fab fragments (lanes 5 and 6, respectively), or 10 µL of preimmune serum (PIS, lane 4) were used. (B) Quantitation of plastid binding and translocation of 35S-ceQORH determined for Fab-decorated and mock-incubated chloroplasts. As control, the precursor to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (SSU) was used. (C) Inhibition of cross-link product (CLP) formation of DTNB-activated 35S-ceQORH by pretreatment of chloroplast with Fab fragments against HP20. (D) Time course of 35S-ceQORH import into Fab-decorated vs. mock-treated Arabidopsis chloroplasts. (a and c) Levels of 35S-ceQORH in the supernatant obtained after centrifugation of the import assays; (b and d) amounts of imported 35S-ceQORH in thermolysin-treated chloroplasts.
Knockout Mutants to Study the Roles of HP20 and HP30 in Vitro.
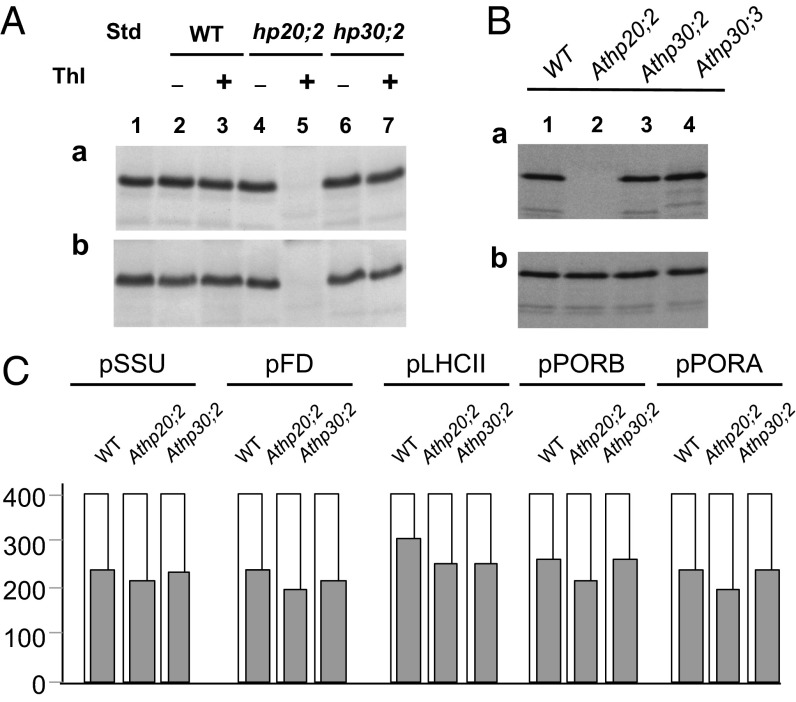
Knockout mutants referred to as Athp20;1 and Athp20;2 (SALK_020671 and SALK_125640) were obtained from the Salk Institute Genomic Analysis Laboratory collection (26) and used for performing in vivo and in vitro tests (27). Similarly, knockout mutants were obtained for HP30, referred to as Athp30;2 and Athp30;3 (SALK_112126 and SALK_046194). The detailed characterization of these various mutants is described in Fig. S3, A–D. Athp20;2 and Athp30;2 knockout plants were devoid of HP20 and HP30, respectively, but contained WT levels of TIC32 and OEP16 (Fig. S1E), two examples of inner and outer plastid envelope membrane proteins, respectively, that lack cleavable transit sequences for import (7, 21). These results underscored the observed specificity of the HP20/HP30 pathway of protein traffic and highlighted its operation in planta. In fact, in vitro import rates of 35S-ceQORH were drastically reduced for plastids isolated from light-grown and dark-grown Athp20;2 plants (Fig. 5 A, a vs. b). In Athp30;2 knockout plastids, 35S-ceQORH accumulated in a trypsin-resistant state, as found for WT chloroplasts (Fig. 5 B, a). These data revealed that HP30 and HP30-2 act redundantly and that HP30-2 can fully replace HP30 in the Athp30;2 and Athp30;3 plants. Control Western blot analyses with TIC110 confirmed the specificity of protease treatment (Fig. 5 B, b). As shown previously, TIC110 attains a trypsin-resistant state if the conditions are tightly controlled and performed properly (25). For the precursors to the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (35S-pSSU), 35S-pFD, the light harvesting chlorophyll a/b binding protein (35S-CAB), and the two NADPH:protochlorophyllide oxidoreductases PORA (35S-pPORA) and PORB (35S-pPORB), no differences in uptake were noted for Athp20;2 and Athp30;2 vs. WT chloroplasts (Fig. 5C). These various proteins are known to enter the plastids via pathways involving TOC159 and TOC75 (pSSU, pCAB, pFD, pPORB) and OEP16-1 (pPORA) (e.g., ref. 21). Along with the Western blot data, these experiments ultimately confirmed the specificity of the HP20/HP30 pathway of protein traffic for a subset of transit sequence-less inner chloroplast envelope membrane proteins and additionally demonstrate that chloroplasts lacking HP20 and/or HP30 are still viable and intact.
Fig. 5.
In vitro-import of 35S-ceQORH into isolated plastids of WT and Athp20;2 vs. Athp30;2 and Athp30;3 plants. (A) Import of 35S-ceQORH into isolated etioplasts (a) and chloroplasts (b), as assessed by thermolysin (Thl) treatment. (B) 35S-ceQORH import into isolated chloroplasts of WT and Athp20;2, Athp30;2, and Athp30;3 mutant plants, as assessed by trypsin treatment (a). For comparison, TIC110 was quantified by Western blotting (b). (C) Quantitative import data obtained for 35S-pSSU, 35S-pFD, 35S-pLHCII, 35S-pPORA, and 35S-pPORB with chloroplasts isolated from WT, Athp20;2, and Athp30;2 mutant plants. For studying import of 35S-pPORA, the assays were supplemented with 5-aminolevulinic acid, driving intraplastidic protochlorophyllide synthesis and substrate-dependent import. Gray columns define the numbers of imported, mature-sized proteins, relative to equal input radioactivities (400 precursor molecules, white columns).
RNA Interference to Drop the Expression of HP30 and HP30-2.
We next created RNA interference lines lacking both HP30 and HP30-2 (28, 29). Two independent lines were obtained and characterized further (Fig. S4 A and B). In the offspring of these lines, a large variation in the phenotype of the seedlings was observed, ranging from yellowish over pale-green to green plants (Fig. S4C). This effect seemed to correlate with changes in the HP30/HP30-2 transcript levels (Fig. S4D). In the finally obtained lines, designated RNAI-2;7#2 and -2;7#3, no HP30 and HP30-2 transcripts and proteins were detectable (Fig. S4 E and F).
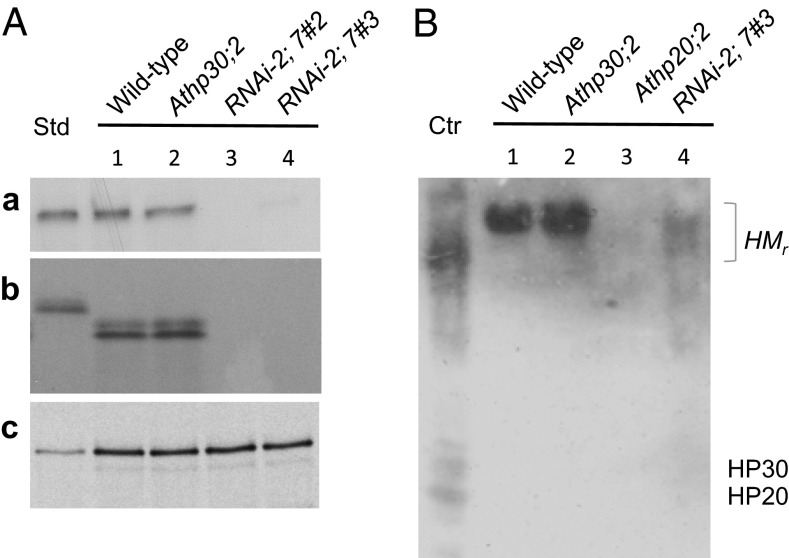
In vitro-import experiments and protease protection assays showed that the targeting of 35S-ceQORH and 35S-HP30 to the inner envelope was disturbed in HP30::HP30-2;7#2 chloroplasts, leading to an accumulation of either protein in the intermembrane space separating the outer and inner envelope membranes and their degradation by added trypsin (Fig. 6 A, a and b, lanes 3 and 4). By contrast, 35S-ceQORH and 35S-HP30 were fully and partially resistant, respectively, to trypsin in WT and Athp30;2 chloroplasts (Fig. 6 A, a and b, lanes 1 and 2), indicating that HP30-2 can functionally replace HP30. Fig. 6B demonstrates that drastically reduced amounts of HMr complexes containing 35S-ceQORH were present in RNAi-2;7 chloroplasts. By contrast, WT levels of HMr complexes were found in the Athp30;2 single knockout mutant used for comparison. Control experiments with chloroplasts from the Athp20;2 mutant failed to detect any HMr complexes containing 35S-ceQORH, underscoring that both HP20 and HP30::HP30-2 are essential for import of ceQORH into the inner envelope of chloroplasts.
Fig. 6.
(A) In vitro import of 35S-ceQORH (a) and 35S-HP30 (b) into chloroplasts isolated from WT, Athp30;2, RNAi-2;7#2, and RNAi-2;7#3 plants, as assessed by trypsin treatment. For comparison, import data are shown for 35S-TIC32 (c). Std defines input standards (100% for ceQORH and HP30; 20% for TIC32). (B) Detection of higher molecular mass (HMr) IAP complexes containing in vitro-imported, nonradiolabeled ceQORH in WT chloroplasts, Athp30;2 and Athp20;2 chloroplasts, and RNAi chloroplasts lacking HP30 and HP30:2 (RNAi-2;7#3). Ctr defines a control in which an aliquot of the IAP complexes that had been isolated from WT plants was pretreated with SDS before nondenaturing PAGE. Shown is a Western blot probed simultaneously with antisera against HP20 and HP30.
Discussion
In the present work, DTNB-mediated disulfide bridge cross-linking was used to identify three proteins as being involved in the import of transit sequence-less precursors into the inner plastid envelope of Arabidopsis chloroplasts. Using the quinone oxidoreductase homolog ceQORH as model, all three proteins were found to belong to the PRAT family (12). The first members of this family were discovered in a genetic screen in yeast approximately 20 y ago (12). From the beginning, it was clear that members of this family play a role in intracellular protein traffic, but the precise function of the proteins was a matter of debate (12). For example, the mitochondrial PRAT family members TIM23 and TIM22 were confirmed to form the core of the machinery for uptake of newly synthesized proteins into the mitochondrial inner membrane (14). By contrast, the precise function of the third mitochondrial PRAT protein, TIM17, is still unclear (14). It is well established that there are two main translocation machineries in the inner mitochondrial membrane. The machinery containing TIM23 and TIM17 is involved in the uptake of presequence-containing cytoplasmic precursors, whereas the machinery containing TIM22 is involved in the uptake of carrier proteins and other hydrophobic membrane proteins lacking cleavable presequences (14).
Our data and results obtained by others show that also chloroplasts contain at least two structurally and functionally different protein translocon complexes in their inner envelope membrane. One complex mediates uptake of presequence-containing cytoplasmic precursor proteins and has been described as the TIC machinery (30). Despite considerable progress the exact nature and composition of this complex and the topology of the postulated components have not been resolved. Some evidence suggests that the TIC machinery may have a modular arrangement consisting of core components, such as TIC110, TIC40 (30), and associated molecular chaperones of the heat shock protein HSP93-V and heat shock cognate protein HSC70 families (31), as well as auxiliary components, including TIC55 and other members of an unique Rieske, nonheme iron–sulfur protein family (32). The other inner envelope protein import machinery contains HP30 and its close relative HP30-2 and is involved in the import of transit sequence-less precursors, such as ceQORH and HP30, into the inner envelope membrane. HP30 and HP30-2 are likely to form heteromers possibly via their SAM domains and cooperate with a third member of the PRAT family, that is, HP20.
Studies in transgenic plants expressing ceQORH-GFP localized the reporter to chloroplasts in mesophyll cells of both WT as well as Athp20;1 and Athp20;2 plants (Fig. S5A). Because pretreatment of chloroplasts with Fab-fragments against HP20 had only partially inhibited binding of ceQORH to the plastid envelope, the detection of a ceQORH-GFP signal at the outer edges of chloroplasts in Athp20;1 and Athp20;2 mutant plants is not surprising and suggests the operation of other proteins that mediate the initial binding of ceQORH to the plastid surface. For guard cells, most analyses revealed a cytosolic localization but very little or no signal of ceQORH-GFP in chloroplasts of WT as well as Athp20;1 and Athp20;2 plants (Fig. S5B). These results support the observation by Miras et al. (9) that the ceQORH translocon is not present or is inactive in guard cells. Controls with transgenic plants expressing FD-GFP and GFP alone proved the expected plastidic and cytoplasmic localization, respectively, of the fluorescence markers both in mesophyll and guard cells (Fig. S6).
The path taken by ceQORH into chloroplasts is specific and also used by other transit sequence-less plastid precursors, including HP30 (Fig. S7). Mutants lacking HP20 were unable to import ceQORH and HP30 in vitro but sequestered normal amounts of other plastid proteins, such as the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase, ferredoxin, the main light harvesting chlorophyll a/b-binding protein of photosystem II, LHCII, and the two NADPH:protochlorophyllide oxidoreductases PORA and PORB. Further evidence for the specificity of the HP20- and HP30/HP30-2-mediated import pathway is provided by our Western blot studies, which demonstrated unaffected levels of TIC32 and OEP16-1, two other transit sequence-less proteins located in the outer and inner envelope membranes, respectively, in the Athp20 mutants analyzed. Expression studies with TIC32 fused to the red fluorescent protein (TIC32-RFP) proved the plastid localization of the fusion protein in mesophyll cells and cytosolic localization in guard cells in mutant Athp20;1 and the WT and thereby underscored the specificity of the ceQORH import pathway into chloroplasts (Fig. S8). Last but not least, RNA interference to simultaneously drop expression of HP30 and HP30-2 abolished the formation of import intermediates of 35S-ceQORH and drastically reduced ceQORH import into the inner envelope of chloroplasts in vitro. Marked changes occurred in the proteome of the inner plastid envelope membrane, leading to a specific depletion of basic envelope proteins, which concerned ceQORH but not TIC32 (Fig. S7). This finding ultimately proved the specificity of the HP20- and HP30/HP30-2-mediated import pathway for a subset of inner plastid envelope proteins. It is tempting to speculate that HP20 and HP30/HP30-2 may play direct roles in ceQORH import by establishing protein-conducting channels. Their predicted topology indeed seems compatible with such an idea. However, HP20 and HP30/HP30-2 also could play indirect roles. Thus, additional experiments are in progress to explore these alternative scenarios and prove the roles of HP20 and HP30/HP30-2 in chloroplast biogenesis.
Materials and Methods
Protein Import.
Protein import into isolated Arabidopsis chloroplasts and etioplasts was carried out as previously described, using cDNA-encoded, wheat germ-translated, urea-denatured, 35S-precursors (27). Plastids were treated with thermolysin (24) after import to degrade unimported precursors. Localization and topology studies using trypsin were carried out as described in ref. 25.
Cross-Linking.
Cross-linking with DTNB (15) was performed by activating the cysteine residues of 35S-ceQORH and 35S-ceQORH-GFP and incubating the derivatized, urea-denatured precursors with isolated plastids at the Mg-ATP and Mg-GTP concentrations given in the text. Sample workup is described in ref. 21.
Athp20 and Athp30 Mutant Characterization and Analysis of the Athp30/30-2 RNAi Lines.
The identification and characterization of Athp20;1 and Athp20;2 (SALK_020671 and SALK_125640), Athp30;2, and Athp30;3 (SALK_112126 and SALK_046194) mutants (26), as well as RNA interference lines (28, 29), is described in SI Materials and Methods (Fig. S4 and Table S3).
Western Blot Analyses.
Western blotting was done as described by Towbin et al. (33), using either an ECL or anti-rabbit, anti-goat, alkaline phosphatase system, using the antisera indicated in the text.
Bioinformatics Tools.
Bioinformatics tools used for sequence alignments and construction of phylogenetic trees are described in SI Materials and Methods (Tables S2 and S3).
Supplementary Material
Acknowledgments
We thank N. Roland, Commissariat à l'Energie Atomique, Grenoble, France, D. J. Schnell, University of Massachusetts, Amherst, MA, and F. Kessler, Université de Neuchatel, Neuchatel, Switzerland for gifts of cDNA clones, antisera, and mutants.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319648110/-/DCSupplemental.
References
- 1.Kirk JTO, Tilney-Basset RAE. The Plastids: Their Chemistry, Structure, Growth and Inheritance. Amsterdam: Elsevier North-Holland Biomedical; 1978. [Google Scholar]
- 2.Lopez-Juez E, Pyke KA. Plastids unleashed: Their development and their integration in plant development. Int J Dev Biol. 2005;49(5-6):557–577. doi: 10.1387/ijdb.051997el. [DOI] [PubMed] [Google Scholar]
- 3.Reinbothe C, Pollmann S, Reinbothe S. Singlet oxygen signaling links photosynthesis to translation and plant growth. Trends Plant Sci. 2010;15(9):499–506. doi: 10.1016/j.tplants.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Rolland N, et al. The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu Rev Genet. 2012;46:233–264. doi: 10.1146/annurev-genet-110410-132544. [DOI] [PubMed] [Google Scholar]
- 5.Shi LX, Theg SM. The chloroplast protein import system: From algae to trees. Biochim Biophys Acta. 2013;1833(2):314–331. doi: 10.1016/j.bbamcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Kleffmann T, et al. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr Biol. 2004;14(5):354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 7.Nada A, Soll J. Inner envelope protein 32 is imported into chloroplasts by a novel pathway. J Cell Sci. 2004;117(Pt 17):3975–3982. doi: 10.1242/jcs.01265. [DOI] [PubMed] [Google Scholar]
- 8.Miras S, et al. Non-canonical transit peptide for import into the chloroplast. J Biol Chem. 2002;277(49):47770–47778. doi: 10.1074/jbc.M207477200. [DOI] [PubMed] [Google Scholar]
- 9.Miras S, et al. Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. J Biol Chem. 2007;282(40):29482–29492. doi: 10.1074/jbc.M611112200. [DOI] [PubMed] [Google Scholar]
- 10.Ferro M, et al. Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics. 2003;2(5):325–345. doi: 10.1074/mcp.M300030-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Froehlich JE, et al. Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res. 2003;2(4):413–425. doi: 10.1021/pr034025j. [DOI] [PubMed] [Google Scholar]
- 12.Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286(1):105–120. doi: 10.1006/jmbi.1998.2455. [DOI] [PubMed] [Google Scholar]
- 13.Murcha MW, et al. Characterization of the preprotein and amino acid transporter gene family in Arabidopsis. Plant Physiol. 2007;143(1):199–212. doi: 10.1104/pp.106.090688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 15.Habeeb AFSA. Reaction of protein sulfhydryl groups with Ellman’s reagent. Methods Enzymol. 1972;25:457–464. doi: 10.1016/S0076-6879(72)25041-8. [DOI] [PubMed] [Google Scholar]
- 16.Theg SM, Bauerle C, Olsen LJ, Selman BR, Keegstra K. Internal ATP is the only energy requirement for the translocation of precursor proteins across chloroplastic membranes. J Biol Chem. 1989;264(12):6730–6736. [PubMed] [Google Scholar]
- 17.Perry SE, Keegstra K. Envelope membrane proteins that interact with chloroplastic precursor proteins. Plant Cell. 1994;6(1):93–105. doi: 10.1105/tpc.6.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kouranov A, Schnell DJ. Analysis of the interactions of preproteins with the import machinery over the course of protein import into chloroplasts. J Cell Biol. 1997;139(7):1677–1685. doi: 10.1083/jcb.139.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bauer J, et al. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;403(6766):203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- 20.Kubis S, et al. Functional specialization amongst the Arabidopsis Toc159 family of chloroplast protein import receptors. Plant Cell. 2004;16(8):2059–2077. doi: 10.1105/tpc.104.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinbothe S, Quigley F, Gray J, Schemenewitz A, Reinbothe C. Identification of plastid envelope proteins required for import of protochlorophyllide oxidoreductase A into the chloroplast of barley. Proc Natl Acad Sci USA. 2004;101(7):2197–2202. doi: 10.1073/pnas.0307284101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R. Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc Natl Acad Sci USA. 1997;94(17):9504–9509. doi: 10.1073/pnas.94.17.9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meruelo AD, Bowie JU. Identifying polymer-forming SAM domains. Proteins. 2009;74(1):1–5. doi: 10.1002/prot.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 1984;75(3):675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kessler F, Blobel G. Interaction of the protein import and folding machineries of the chloroplast. Proc Natl Acad Sci USA. 1996;93(15):7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 27.Reinbothe S, Mache R, Reinbothe C. A second, substrate-dependent site of protein import into chloroplasts. Proc Natl Acad Sci USA. 2000;97(17):9795–9800. doi: 10.1073/pnas.160242597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gleave AP. A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol. 1992;20(6):1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- 29.Ruiz-Ferrer V, Voinnet O. Roles of plant small RNAs in biotic stress responses. Annu Rev Plant Biol. 2009;60:485–510. doi: 10.1146/annurev.arplant.043008.092111. [DOI] [PubMed] [Google Scholar]
- 30.Kovács-Bogdán E, Soll J, Bölter B. Protein import into chloroplasts: The Tic complex and its regulation. Biochim Biophys Acta. 2010;1803(6):740–747. doi: 10.1016/j.bbamcr.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Flores-Pérez Ú, Jarvis P. Molecular chaperone involvement in chloroplast protein import. Biochim Biophys Acta. 2013;1833(2):332–340. doi: 10.1016/j.bbamcr.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Caliebe A, et al. The chloroplastic protein import machinery contains a Rieske-type iron-sulfur cluster and a mononuclear iron-binding protein. EMBO J. 1997;16(24):7342–7350. doi: 10.1093/emboj/16.24.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hofmann K, Stoffel W. TMbase – a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;374:166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.