Abstract
The tribe Triticeae includes the major crops wheat and barley. Within the last few years, the whole genomes of four Triticeae species—barley, wheat, Tausch’s goatgrass (Aegilops tauschii) and wild einkorn wheat (Triticum urartu)—have been sequenced. The availability of these genomic resources for Triticeae plants and innovative analytical applications using next-generation sequencing technologies are helping to revitalize our approaches in genetic work and to accelerate improvement of the Triticeae crops. Comparative genomics and integration of genomic resources from Triticeae plants and the model grass Brachypodium distachyon are aiding the discovery of new genes and functional analyses of genes in Triticeae crops. Innovative approaches and tools such as analysis of next-generation populations, evolutionary genomics and systems approaches with mathematical modeling are new strategies that will help us discover alleles for adaptive traits to future agronomic environments. In this review, we provide an update on genomic tools for use with Triticeae plants and Brachypodium and describe emerging approaches toward crop improvements in Triticeae.
Keywords: Barley, Brachypodium, Crop improvement, Next-generation sequencing, Triticeae, Wheat
Introduction
Ensuring global food security is an urgent challenge for our society. The world’s population is estimated to rise to 9 billion by 2050 (http://esa.un.org/unpd/wpp/index.htm), and global crop production will need to be increased in order to feed this population (Alexandratos and Bruinsma 2012). At the same time, the world’s agricultural systems are facing the risk of production shortfalls due to various abiotic and biotic stresses from global climate change (Wheeler and von Braun 2013). In order to develop crops that can better withstand climate change, we need to connect diverse and interdisciplinary scientific and technological advancements in crop breeding.
During the past decade, remarkable innovations in analytical platforms for omics-based research and development have provided crucial resources to promote research in model and applied plant species. A combinatorial approach using multiple omics platforms and integrating their outcomes has been an effective strategy to understand molecular systems that are critical to improving plant productivity (Mochida and Shinozaki 2010). Recent breathtaking advances in sequencing technologies have revitalized approaches in genomics and created various analytical applications (Mochida and Shinozaki 2011, Plocik and Graveley 2013).
Next-generation sequencing (NGS) technologies have accelerated whole-genome sequencing projects, sometimes in combination with the conventional Sanger sequencing methods. For instance, Phytozome, a joint project of the Department of Energy’s Joint Genome Institute and the Center for Integrative Genomics, has provided access to 41 sequenced and annotated plant genomes as of the latest release, v. 9.1 (http://www.phytozome.net/) (Goodstein et al. 2012). The latest release from Ensembl Plants, Release 20, includes information on genome sequence and associated annotations for 26 plant species, including the recently sequenced barley and wheat (http://plants.ensembl.org/index.html).
Whole-genome re-sequencing is a feasible NGS application to catalog genomic variations in natural, mapping and mutant populations, to find the association between genetics and biological consequences (Arai-Kichise et al. 2011, Ashelford et al. 2011, James et al. 2013, Miyao et al. 2012, Uchida et al. 2011). Genome-guided RNA-seq is an effective approach to identify transcription units and splice isoforms together with their expression profiles in genome-sequenced organisms (Filichkin et al. 2010, Loraine et al. 2013, Szczesniak et al. 2013). De novo RNA-seq has become a popular method to acquire initial information about transcribed sequences and gene expression profiles without a reference genome sequence, and there are many examples in various non-model organisms, including plants of particular biological and industrial interest (Fu et al. 2011, Su et al. 2011, Bai et al. 2013, Gil-Amado and Gomez-Jimenez 2013, Postnikova et al. 2013, Ramilowski et al. 2013, Tsai et al. 2013). The cDNA information reconstructed from de novo RNA-seq data sets has been useful as a proxy for the genic regions of the genome in that species (Hirsch and Buell 2013). Analyses based on high-throughput sequencing have rapidly become a significant approach in genome-wide biology for crop improvements. Herein, we focused on staple crops, wheat and barley, of the tribe Triticeae in the subfamily Pooideae. Bread wheat (Triticum aestivum L. subsp. aestivum) is one of the most important crops in the world, accounting for 20% of calories consumed by humans. Barley (Hordeum vulgare L.) is also a globally important crop that is utilized for animal feed, in the production of malt and as a human food source. In 2011, a special issue on barley was published in Plant and Cell Physiology to highlight the resources related to and achievements in Triticeae genomics at that time (Saisho and Takeda 2011). These available genomic resources in Triticeae have dramatically changed over the last 2 years, which provides us with an opportunity to design new combinations of tools to promote Triticeae genomics and to accelerate the process of gene discovery in genome-oriented breeding.
In this review, we provide an overview of recent advancements focused in Triticeae genomics. We first describe recent progress in genomic resources and their applications in Triticeae species, including wheat and barley. We then highlight the current status of genomic resources in the emerging model grass of the Pooideae subfamily, Brachypodium distachyon (L.) P. Beauv, and discuss its roles as a model plant for Triticeae species. Finally, we review several pioneering studies dealing with the association between genetic diversity and traits that are adaptive to the environment, with possible applications for future Triticeae genomics and molecular breeding. Throughout this review, we showcase recent genome-based efforts and available resources that are useful in Triticeae genomics.
Genomic resources of Triticeae crops
About 10,000 years ago, in the Fertile Crescent in western Asia, humans made the transition from the hunter–gatherer way of life to a sedentary agrarian lifestyle. Domestication of einkorn wheat, emmer wheat and barley marked the beginning of agriculture in that area (Dubcovsky and Dvorak 2007). Barley (H. vulgare; 2n = 2x = 14; HH) was domesticated there from its wild relative, H. vulgare subsp. spontaneum (K.Koch) Thell. Domesticated einkorn wheat (Triticum monococcum L.; 2n = 2x = 14; AmAm) and domesticated emmer wheat (Triticum turgidum L.; 2n = 4x = 28; AABB) were disseminated across Asia, Europe and Africa as agriculture expanded. Then, about 8,000 years ago, a spontaneous hybridization of the wild diploid grass Aegilops tauschii Coss. (2n = 14; DD) with T. turgidum produced hexaploid common wheat (T. aestivum; 2n = 6x = 42; AABBDD). Such evolution of polyploid wheat varieties has been thoroughly described and discussed in a review by Matsuoka (2011).
Repetitive DNA comprises >80% of the genome of Triticeae plants (Smith and Flavell 1975). Bread wheat and barley possess genomes estimated to be 17 Gb and 5.1 Gb in size, respectively. The large size of their genomes and their rich repetitive sequences, as well as polyploidy in wheat, have made it challenging to understand fully the genomes of the Triticeae crops. However, there is a strong need for genomic resources to facilitate the discovery of new genes and develop genome-based breeding in wheat and barley because of the agricultural importance of these crops.
Early sequence resource in Triticeae crops
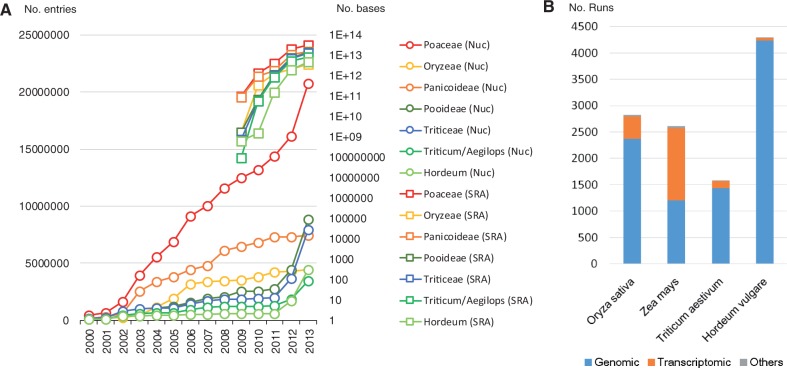
In the past decade of the 21st century, the amount of sequence data from Poaceae species available in the public domain has dramatically increased (Fig. 1A). Since 2001, large-scale collection of cDNA and expressed sequence tag (EST) analysis using Sanger sequencing have been performed in several grass species, including wheat and barley (Mochida and Shinozaki 2010). Large-scale EST analysis has facilitated comprehensive acquisition of expressed genes as primary genomic information, and has also enabled monitoring of special and temporal gene expression and interspecific and intraspecific comparative analysis of expressed genes (Mochida et al. 2003, Ogihara et al. 2003, Zhang et al. 2004, Mochida et al. 2006). It has also facilitated the design of cDNA-oriented molecular markers and probes for oligo-microarrays (Kawaura et al. 2006, Torada et al. 2006, Mochida et al. 2008, Close et al. 2009, Sato and Takeda 2009, Sato et al. 2009a). Currently, there are >1.28 million EST entries in wheat and >0.5 million in barley in the dbEST of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/dbEST/).
Fig. 1.
An overview of the publicly available sequence data in grasses. (A) The growth of sequence data in GenBank (number of entries) and the sequence read archive (number of bases). (B) Number of NGS runs of the four staple crops in Poaceae submitted to the sequence read archive (September 8, 2013).
Full-length cDNA libraries and large-scale sequence data sets have played significant roles in various life science projects (Mochida and Shinozaki 2010). The sequence resources of full-length cDNA libraries can accurately identify transcribed regions and structural features (Ohyanagi et al. 2006, Sakai et al. 2013), such as transcription units, transcription start sites and transcriptional variants in completed or draft genome sequences. Full-length cDNA libraries have also contributed to functional analyses by enabling the creation of overexpression strains. The advent of function-based gene discovery via full-length cDNA overexpressor (FOX) gene hunting has enabled high-throughput discovery of functional genes associated with phenotypic changes (Kondou et al. 2009, Sakurai et al. 2011, Hong et al. 2013). Based on the biotinylated CAP trapper method (Carninci et al. 1996), large-scale analyses of full-length cDNAs have been conducted in wheat and barley (Kawaura et al. 2009, Sato et al. 2009b, Matsumoto et al. 2011). The TriFLDB database integrates full-length coding sequence information in wheat and barley (http://trifldb.psc.riken.jp). It provides functional annotation and associated information based on comparative analysis with other plant species (Mochida et al. 2009a).
Most of the contiguous genomic sequences identified in Triticeae before 2009 were obtained through map-based gene cloning or comparative studies. The chloroplast and mitochondrial genomes of hexaploid wheat were completely sequenced in 2002 and 2005, respectively (Ogihara et al. 2002, Ogihara et al. 2005). Knowledge of these whole-genome sequences of wheat organelles has enabled the investigation of evolution and RNA editing events of genes in the organelle genomes (Matsuoka et al. 2002, Yura et al. 2009). In addition to the Sanger-based sequence data, since 2009 NGS-based data of Pooideae plants have appeared in the Sequence Read Archive (SRA; http://www.ncbi.nlm.nih.gov/sra). The amount of deposited data from NGS has rapidly increased (Fig. 1A). So far, NGS technology has mainly been used to analyze genomic sequences of wheat and barley (Fig. 1B).
Whole-genome analysis in Triticeae plants
Barley With the aim of constructing a high-quality reference sequence of the barley genome, an international consortium (International Barley Sequencing Consortium, http://barleygenome.org) was initiated in 2006 (Schulte et al. 2009). Barley genomic resources at that time were well reviewed by Sreenivasulu et al. (2008). The barley ESTs were applied to generate high-density genetic maps using innovative genotyping platforms (Stein et al. 2007, Close et al. 2009, Sato and Takeda 2009). High-density genetic maps, in turn, made it possible to compare the barley genome at the chromosome level with those of other genome-sequenced grasses (Mochida et al. 2008).
More recently, NGS technologies have been applied to accumulate information about the barley genome sequence (Wicker et al. 2006, Wicker et al. 2008, Wicker et al. 2009). An integrated approach called the genome zipper incorporates chromosome sorting, NGS technology, array hybridization and systematic exploitation of conserved synteny with model grasses and was used to build a virtual gene order of barley chromosome 1H (Mayer et al. 2009). It was possible to sort chromosome 1H by flow cytometric analysis because it was smaller than barley chromosomes 2H–7H. To sort those chromosomes, wheat–barley telosome addition lines (2HS–7HL arms) were used, and the genome zipper-based gene order in the whole-genome scale was represented in barley (Mayer et al. 2011) (http://mips.helmholtz-muenchen.de/plant/barley/gz/index.jsp).
The barley genome context was eventually published in the form of an integrated and ordered physical, genetic and functional sequence resource in 2012 (International Barley Genome Sequencing Consortium 2012). A 4.98 Gb physical map was constructed based on assembly of bacterial artificial chromosome (BAC) fingerprint contigs in the barley cultivar Morex, >3.9 Gb of which was anchored to an integrated genetic framework. Barley full-length cDNAs (Matsumoto et al. 2011) and RNA-seq data sets were used to annotate genes in the whole-genome shotgun assembly of barley. The result showed 79,379 transcript clusters, including 26,159 ‘high-confidence’ genes with homology support from other plant genes. The associated data set of the barley gene space is accessible from the MIPS barley genome database (http://mips.helmholtz-muenchen.de/plant/barley/). In the Ensembl plants, the whole-genome shotgun contigs anchored with FPC contigs in Morex barley have been displayed at genetic coordinates, which are approximations inferred from positional information in the barley physical maps (http://plants.ensembl.org/Hordeum_vulgare/Info/Index).
Wheat The International Wheat Genome Sequencing Consortium (www.wheatgenome.org) has used a chromosome by chromosome approach to mapping and sequencing 21 chromosomes of bread wheat, cultivar Chinese spring genome, using chromosome flow sorting. A physical map of wheat chromosome 3B was constructed as the first chromosome-scale physical map (Paux et al. 2008), which originated from sorted chromosome 3B (Safar et al. 2004). Double ditelosomic lines were used to construct chromosome arm-specific BAC libraries and physical contigs, for example, in chromosomes 1BL, 4A and 1A (Hernandez et al. 2012, Lucas et al. 2013, Philippe et al. 2013). Six arms of the group 1 chromosomes of bread wheat cv. Chinese spring, which were flow sorted, were sequenced by Roche/454 and compared with similar data sets from the homoeologous chromosome 1H of barley (Wicker et al. 2011). Using chromosome 3B, an improved version of the physical map was constructed and used to map about 3,000 transcripts (Rustenholz et al. 2011). Using chromosome sorting and NGS technologies, NGS-based survey sequencing analyses of each chromosome arm were performed. Data sets of NGS reads corresponding to chromosome arms are downloadable from SRA, such as ERP003210, ERP002108 and SRP007288. The online database WheatGenome.info (http://www.wheatgenome.info/) is providing an NGS-based data set derived from chromosome group 7 of wheat (Lai et al. 2012).
Brenchley et al. (2012) successfully deciphered the bread wheat genetic code using the whole-genome shotgun sequencing approach using 454 pyrosequencing and comparative analyses with diploid ancestral and progenitor genomes (Brenchley et al. 2012). In that study, Brenchley and colleagues identified around 96,000 genes and assigned two-thirds of them to the three component genomes (A, B and D) of hexaploid wheat. The associated data set of the wheat shotgun sequencing is available in the MIPS wheat genome database (http://mips.helmholtz-muenchen.de/plant/wheat/uk454survey/index.jsp).
Soon after the simultaneous publication of the compilation and analysis of the genome sequences of barley and wheat, two papers published in the same issue presented a draft genome sequences and analysis of two wheat progenitors, Triticum urartu and A. tauschii (Jia et al. 2013, Ling et al. 2013). To sequence the T. urartu genome, Ling and colleagues obtained 448.49 Gb of filtered sequence data and assembled them to scaffolds that represented 4.66 Gb, of which 66.9% comprised repetitive elements. They generated RNA-seq data from various organs and treatments and applied them to identify 34,879 protein-coding gene models (Ling et al. 2013). To sequence the A. tauschii genome, Jia et al. obtained an approximately 90-fold depth of short reads from libraries with various insert sizes and assembled them to scaffolds that represented 83.4% of the genome, of which 65.9% comprised transposable elements. They also generated RNA-seq data derived from various tissues and applied them to identify 43,150 protein-coding genes, of which 30,697 (71.1%) were anchored to chromosomes with an integrated high-density genetic map (Jia et al. 2013).
In summary, we have been able to access the deciphered genetic code of the four main Triticeae plants at the whole-genome scale (Table 1). The information from their genomes should be a primary resource for studies of their complex genomes and to make improvements in wheat and barley. Using these compilations of genomic information, genomic tools will be revitalized and new ones will be developed to accelerate the discovery of useful genes in wheat and barley. Genome-scale approaches could be applied further in Triticeae crops, coupled with NGS technologies, such as RNA-seq analysis to monitor transcriptomes with single-base resolution, chip-seq analysis to identify chromatin modifications or protein–DNA interactions, and exome sequencing to discover polymorphisms in genome scale (Mascher et al. 2013a).
Table 1.
Sequence resources of four genome-sequenced plants in Triticeae
| A. tauschii | T. urartu | T. aestivum | H. vulgare | |
|---|---|---|---|---|
| Whole-genome sequencing project | Jia et al. (2013) | Ling et al. (2013) | Brenchley et al. (2012) | International Barley Genome Sequencing Consortium (2012) |
| BioProject | PRJNA182898 | PRJNA182347 | PRJEB568 | Listed in International Barley Genome Sequencing Consortium (2012) Supplemental Note 1 |
| GenBank/EMBL/DDBJ | AOCO000000000 | AOTI00000000 | CALO00000000 | |
| AOCO010000000 | ||||
| SRA | SRA030526 | SRA030525 | ERP000319 | |
| SRA062662 | SRA066084 | |||
| SRA063175 | SRA064213 | |||
| No. of genomic scaffolds or contigs | 429,891 | 499,221 | 945,079 | 6,437 physical map contigs |
| No. of protein sequences | 33,849 | 24,169 | 94,000–96,000 (genes) | 79,379 (transcript clusters) |
| Online resources | Ensembl Plantsa | Ensembl Plantsa | MIPS Wheat Genome Databasec | MIPS barley genome databased |
| gigadbb | gigadbb | Ensembl Plantsa |
Other omics spectrums in Triticeae plants
Transcriptomics Comprehensive and high-throughput analysis of gene expression has become a significant approach for screening candidate genes, predicting gene function, discovering cis-regulatory motifs and characterizing transcriptional regulatory networks (Huang et al. 2011). A comprehensive collection of ESTs and full-length cDNA sequences was used to design probes of oligo-microarrays or GeneChips in model plants, as well as crops including wheat and barley. Rapid accumulation of large-scale gene expression data sets has provided us with an efficient and valuable resource for secondary uses, such as co-expression analyses (Hamada et al. 2011, Obayashi et al. 2011, Sasaki et al. 2011, Hirano et al. 2013a), and allow us to discover genes that are involved in certain metabolic pathways (Fukushima et al. 2013, Hirano et al. 2013b). In barley, a co-expression gene network was constructed using >1,000 sets of Affymetrix GeneChip data derived from 45 publicly available experimental series (Mochida et al. 2011a). Information resources providing co-expression gene networks of multiple plant species have been developed and are available on the web. PlaNet (http://aranet.mpimp-golm.mpg.de) provides information regarding co-expression gene networks constructed from microarray data sets of seven plant species, including wheat and barley (Mutwil et al. 2011). Microarray hybridization coupled with laser capture microdissection has been applied to monitor the transcriptome of specific cells or tissues of interest in plants (Takehisa et al. 2012, Kubo et al. 2013). In barley, laser capture microdissection microarray analysis was applied to analyze transcriptome in endosperm cells differentiate into transfer cells (Thiel et al. 2012).
Whole-transcriptome analysis using tiling array was an effective approach to explore novel transcription units, splicing variants and antisense transcripts, together with expression patterns (Matsui et al. 2008, Iida et al. 2011, Yoshimura et al. 2011, Endo et al. 2012). Because this method requires whole-genome information, it has been available only in a few genome-sequenced plants with small genomes. After NGS became available, RNA-seq rapidly became a popular approach to analyze the whole transcriptome in any species (Chang et al. 2011, Su et al. 2011, Swarbreck et al. 2011, Bai et al. 2013, Gil-Amado and Gomez-Jimenez 2013, Postnikova et al. 2013). The barley whole-genome sequencing project used 1.67 billion RNA-seq reads obtained from eight stages of barley development, and applied them to predict structures of transcription units (International Barley Genome Sequencing Consortium 2012). Dissecting the complex transcriptome in polyploidy species such as wheat in order to understand homoeolog-specific gene expression at the genome scale is challenging. Differential expression among homoeologs in the A, B and D genomes is often observed in the wheat transcriptome (Leaungthitikanchana et al. 2013, Mochida et al. 2003). RNA-seq analysis could be a promising approach to identify allele-specific and/or homoeolog-specific transcripts based on nucleotide polymorphisms in the sequenced read assembly. Recently, RNA-seq was applied to separate homoeologs in the tetraploid durum wheat transcriptome based on k-mer assembly strategies (Krasileva et al. 2013).
Metabolomics Over the last decade, metabolomics has become a powerful technique to identify metabolic features underlying complex phenotypes in biological systems (Okazaki and Saito 2012, Higashi and Saito 2013, Putri et al. 2013). Metabolome profiling provides a snapshot of the accumulation patterns of metabolites in response to various kinds of biological conditions, such as environments, developmental stages and genotypes. For example, metabolome profiling approaches have been used to evaluate stress responses in various plants, including wheat and barley (Widodo et al. 2009, Ishikawa et al. 2010, Bowne et al. 2012, Wu et al. 2013a). Recently, correlations between ionomic and metabolomic profiles in response to salt stress were investigated in wild and cultivated barley accessions (Wu et al. 2013b). Metabolome profiling has also been used to evaluate metabolic phenotypes of transgenic plants of wheat and barley (Baker et al. 2006, Kogel et al. 2010). Hormonomics is a specific, targeted use of metabolomics to analyze phytohormones. Simultaneous profiling of phytohormones and their derivatives (Kojima et al. 2009, Kanno et al. 2010) is a key approach to a holistic understanding of the plant hormone network associated with biological systems (Nomoto et al. 2012, Kamada-Nobusada et al. 2013).
The combined use of multiple omics technologies and integrated analysis of omics-based data sets have allowed us to study biological systems multilaterally. Integrated analysis of metabolome results and the microarray-based transcriptome have been widely applied to provide a multifaceted view of cellular systems in response to stress conditions, for example (Ishikawa et al. 2010, Watanabe et al. 2010, Yamakawa and Hakata 2010). More recently, the combined use of metabolome analysis and NGS-based transcriptome analysis has been an effective systems-based approach to explore gene regulatory networks superimposed with metabolic features of particular metabolic pathways (Yamazaki et al. 2013), and construct metabolic pathways (Van Moerkercke et al. 2013).
Recently, a number of information resources have been released or updated to provide tools for metabolome analysis. Recently published reviews have showcased information resources in plant metabolomics (Fukushima and Kusano 2013, Higashi and Saito 2013). In the following discussion, we will introduce two online tools for metabolomics and hormonomics that were recently released from RIKEN. PRIMe (http://prime.psc.riken.jp/), the Platform for RIKEN Metabolomics, is a website that was developed to assist systems approaches, ranging from metabolomics to transcriptomics. PRIMeLink, a newly designed tool in PRIMe, provides seamless and simultaneous access to the metabolome and transcriptome data sets (Sakurai et al. 2013). UniVIO (http://univio.psc.riken.jp/), the Uniformed Viewer for Integrative Omics, displays hormone–metabolome (hormonome) and transcriptome data in a single formatted heat map (Kudo et al. 2013).
Advanced applications of metabolomics technologies are providing new insight into molecular elucidation of plant functions. The use of metabolomics technologies combined with high-throughput and genome-scale genotyping technologies is allowing us to carry out genome-wide association studies of the accumulation patterns of metabolites in plants. These studies may provide promising links for narrowing the genotype–phenotype gap for complex agronomic traits (Riedelsheimer et al. 2012). Plant lipidomics is a new area in plant metabolomics focused on identifying novel and important roles played in plants by hydrophobic compounds (Okazaki et al. 2013). Metabolite imaging, a promising technology to image the distribution of metabolites among cells and tissues of plants, is expected to increase its power of resolution to provide unprecedented data on the distribution of metabolites at the subcellular level (Lee et al. 2012).
Changing the resolution and scale in genome-wide approaches, from single-cell resolution to life-scale and population-scale analyses, is expected to become an important solution to reveal biological systems in higher resolution. For single-cell or cell type-specific transcriptome analysis, linear amplification methods of RNAs or cDNAs that are compatible with NGS platforms have been developed and applied to various species (Osaka et al. 2013). Recently, cell type-specific metabolome analysis was demonstrated in Arabidopsis root (Moussaieff et al. 2013). Furthermore, thanks to an increase in throughput in analytical platforms, it has become possible to acquire omics profiles at a population scale and/or over time, throughout the life cycle of an organism. These advancements will significantly upgrade our molecular understanding of biological systems in plants and could be rapidly applied not only in model plants but also in economically important plants, including wheat and barley.
Study of Brachypodium and its roles in Pooideae genomics
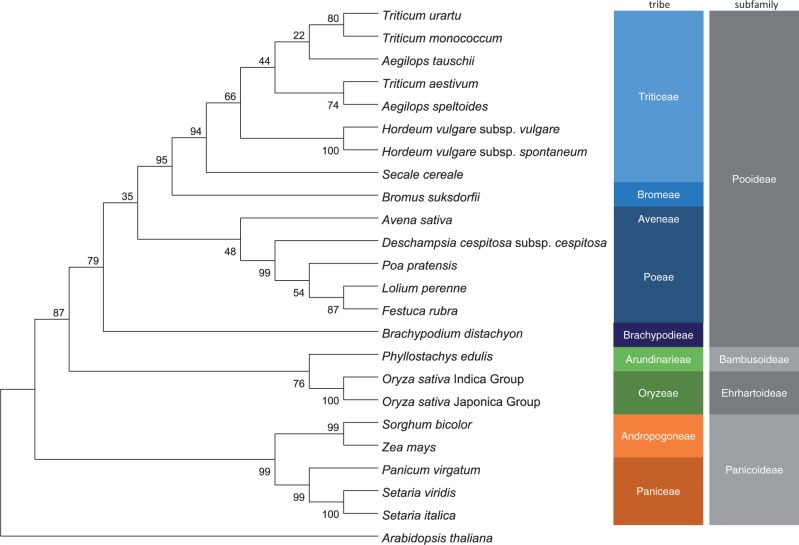
Arabidopsis thaliana (L.) Heynh has been used as the model organism in plant biology for almost 30 years. The success of Arabidopsis research provides proof of the importance of a model organism. Brachypodium distachyon was proposed as a model system by Draper et al. in 2001, for analyzing genetic functions and biological systems in temperate grasses, cool-season cereals and dedicated biofuel crops (Draper et al. 2001, Mur et al. 2011). The species has tractable features, a short life cycle, small plant size, facile transformation, simple growth requirements and a small genome. It belongs to the Pooideae subfamily (Fig. 2) and serves as a model system for major crops, such as wheat, barley, rye and oats (Bevan et al. 2010). In 2010, the whole-genome sequence of the inbred line Bd21 was published, making it the first completely genome-sequenced species of the Pooideae subfamily (International Brachypodium Initiative 2010).
Fig. 2.
A phylogenetic tree of grass species based on the ndhF gene: YP 008239279 (T. urartu), YP 008239142 (T. monococcum), YP 008474350 (A. tauschii), AGP51333 (T. aestivum), YP 008474440 (A. speltoides), AGP50802 (H. vulgare subsp. vulgare), AGP50881 (H. vulgare subsp. spontaneum), YP 008239219 (Secale cereale), ABH02660 (Bromus suksdorfii), AAA64204 (Avena sativa), ABH02669 (Deschampsia cespitosa subsp. cespitosa), AAA64698 (Poa pratensis), YP 001531329 (Lolium perenne), ABH02677 (Festuca rubra), YP 002000532 (B. distachyon), YP 004733625 (Phyllostachys edulis), YP 654249 (Oryza sativa Indica group), NP 039441 (O. sativa Japonica group), YP 899454 (Sorghum bicolor), NP 043084 (Zea mays), YP 004841996 (Panicum virgatum), AAA64841 (Setaria viridis), AAM22087 (S. italica) and NP 051106 (A. thaliana). The phylogenetic tree was constructed with the aligned NdhF protein sequences by MEGA [version 5.2; http://www.megasoftware.net/ (Tamura et al. 2011)] using the Neighbor–Joining method with the following parameters: Poisson model, complete deletion and bootstrap method. The bootstrap values from 1,000 replicates are given at each node. The NdhF protein sequences were aligned by ClustalW implemented in MEGA.
Brachypodium resources
Brachypodium has garnered attention as a model system, and a number of projects to develop genomic resources have been initiated at various institutions. Already, several reviews have described available resources related to Brachypodium (Brkljacic et al. 2011, Mur et al. 2011). Here, we focused on recently published Brachypodium resources (Table 2).
Table 2.
Online resources for Brachypodium research
| Description | URLs | |
|---|---|---|
| Brachypodium.org | Genome sequence | http://brachypodium.org/ |
| Gene annotation | ||
| ESTs | ||
| RNA-seq data sets | ||
| Resequencing data sets | ||
| Phytozome | Genome sequence | http://www.phytozome.net/ |
| Gene annotation | ||
| Brachypodium genome database–MIPS | Genome sequence | http://mips.helmholtz-muenchen.de/plant/brachypodium/ |
| Gene annotation | ||
| Protein doman | ||
| Comparative genome view | ||
| NCBI RefSeq | Gene annotation as RefSeq entry | http://www.ncbi.nlm.nih.gov/refseq/ |
| Ensembl Plants | Genome sequence | http://plants.ensembl.org/Brachypodium_distachyon/Info/Index |
| Gene annotation | ||
| Wheat transcriptome alignments | ||
| Gramene | Gene annotation | http://www.gramene.org/ |
| Comparative maps | ||
| Pathways (BrachyCyc) | ||
| RIKEN Brachypodium Full-length cDNA Database | Gene annotation | http://brachy.bmep.riken.jp/ver.1/index.pl |
| Full-length cDNAs | ||
| Triticeae transcriptome alignment | ||
| PlaNet | Gene expression profiles based on the GeneChip | http://aranet.mpimp-golm.mpg.de/ |
| Co-expression analysis | ||
| BrachyTAG | T-DNA tag lines | http://www.brachytag.org/ |
| FTS on the Bd21 genome | ||
| Western Regional Research Center Brachypodium | T-DNA tag lines | http://brachypodium.pw.usda.gov/ |
| FTS on the Bd21 genome | ||
| BRACHYTIL | TILLING lines | http://urgv.evry.inra.fr/UTILLdb |
| GramineaeTFDB | Transcription factors | http://gramineaetfdb.psc.riken.jp/ |
| PlantTFDB | Transcription factors | |
| BrachyCyc | Pathways | http://pathway.gramene.org/BRACHY/class-tree?object=Pathways |
| Gene Ontology | ||
| KEGG | Pathways | http://www.genome.jp/kegg/ |
| Gene annotation | ||
| Mapman | Mapping to MapMan Ontology | http://mapman.gabipd.org/ |
| Plant Genome Duplication Database | Syntenic relationships | http://chibba.agtec.uga.edu/duplication/ |
| PLAZA | Syntenic relationships | http://bioinformatics.psb.ugent.be/plaza/ |
| Gene annotation | ||
| Gene Ontology | ||
| E-TALEN | Web service to design TALENs | http://www.e-talen.org/E-TALEN/ |
Genome sequence and annotations The current version of the reference genome sequence of Bd21 is the Joint Genome Institute v1.0 8x assembly with approximately 272 Mb and a 99.6% deciphered genome, arranged in five chromosomes with 78 unmapped scaffolds (International Brachypodium Initiative 2010). The sequence data set of the Bd21 genome is available from a genomic portal, Brachypodium.org, as well as from Phytozome (http://www.phytozome.net/brachy.php), MIPS (http://mips.helmholtz-muenchen.de/plant/brachypodium/) and Ensembl Plants (http://plants.ensembl.org/Brachypodium_distachyon/Info/Index). The high-quality reference genome sequence of Bd21 could be an essential resource for every genome-oriented application using Brachypodium. The representative gene structural annotations of Bd21 are available from Phytozome and from MIPS, and each contains the same number of annotated gene models on identical loci, with slightly different annotations, mainly in the untranslated regions. More recently, we updated gene structure annotations of the Bd21 genome by comparing full-length cDNA sequences with the publicly available annotations from the Phytozome and MIPS. About 10,000 non-redundant gene models were supported by the full-length cDNAs and about 6,000 showed some modifications in the gene model. We also found about 580 novel gene models, including 362 newly identified transcription units (Mochida et al. 2013a).
Whole-genome re-sequencing data have recently appeared in the public domain, representing the progress of the re-sequencing project in the Joint Genome Institute-Department of Energy. In the Brachypodium.org data set, Bd21-3, Bd3-1, Bd30-1, Bd1-1, Koz-3 and BdTR12C are currently available with analyzed data sets. In addition to those, the row data sets of 20 other accessions, BdTR8i, Mur1, Foz1, Mig3, ABR5, Mon3, ABR9, ABR2, BdTR13a, BdTR7a, BdTR1i, Tek-2, Koz-1, Gaz-8, BdTR2G, BdTR10C, Adi-10, Arn1, ABR6 and ABR9, released from JGI, have been deposited in the public sequence read archives (September 24, 2013).
Transcriptome The National Center for Biotechnology Information database dbEST contains 128,092 Sanger ESTs from B. distachyon. Brachypodium.org provides EST data sets from Bd21 and some other accessions (ftp://brachypodium.org/brachypodium.org/ESTs/). The data set of the ‘454 sequencing of Brachypodium distachyon EST project’ are available from the National Center for Biotechnology Information’s SRA (SRX014502–SRX014507). We recently constructed a mixed full-length cDNA library from 21 different tissues of Bd21 and obtained 78,163 ESTs from both the 5′ and 3′ ends of about 40,000 clones, which were applied to update structural annotations, as described above. We integrated the Brachypodium full-length cDNAs and updated gene structures with available sequence resources in wheat and barley in a web-accessible database, the RIKEN Brachypodium FL cDNA database (RBFLDB, http://brachy.bmep.riken.jp/ver.1/index.pl).
The Affimetrix GeneChip is a combination of genome tiling and exon scanning designed for multiple uses, such as expression profiling and ChIP-chip studies. Information related to the probe design of the GeneChip is available from Brachypodium.org (ftp://brachypodium.org/brachypodium.org/Affymetrix/Current/). Expression profiles of Brachypodium genes in 36 tissues or conditions, and co-expression networks, which are based on hybridization experiments using the GeneChip, are available from the PlaNet website http://aranet.mpimp-golm.mpg.de/. RNA-seq transcriptome data based on Illumina sequencing were generated to annotate gene structure for the sequencing project of Bd21, available from the National Center for Biotechnology Information’s SRA (SRA010177) and from Brachypodium.org (ftp://brachypodium.org/brachypodium.org/Bd21_Original_RNaseq/) as the Bd21 original RNA-seq data set. More recently, RNA-seq analysis was performed to compare the transcriptome in species representing three Poaceae subgroups, i.e. the Pooideae (B. distachyon), the Panicoideae (sorghum) and the Ehrhartoideae (rice) (Davidson et al. 2012).
Genetic resources Comprehensive collections of mutant lines are essential bioresources to promote forward and reverse genetics. Using the efficient Agrobacterium tumefaciens-mediated transformation method, a project to create a Brachypodium T-DNA collection was initiated. The BrachyTAG collection (http://www.brachytag.org) and the Western Regional Research Center Brachypodium insertional mutant population (http://brachypodium.pw.usda.gov) are available from searchable websites, providing Brachypodium T-DNA lines with flanking sequence tags (Thole et al. 2010, Bragg et al. 2012). TILLING (Targeting Induced Local Lesions In Genomes), a useful platform to identify an allelic series of mutants, has been widely developed in model organisms and crops (Saito et al. 2011, Wang et al. 2012). A TILLING platform called BRACHYTIL, created for the inbred line Bd21-3 mutagenized by NaN3, was released, and phenotypes were organized in a phenotypic tree available at http://urgv.evry.inra.fr/UTILLdb (Dalmais et al. 2013).
Brachypodium genetic linkage maps serve as important tools for a range of molecular genetic analyses. A genetic linkage map was developed with an F2 population from a cross between the lines Bd3-1 and Bd21 using simple sequence repeat (SSR)-based markers (Garvin et al. 2010). A genetic linkage map of an F2 mapping population of 476 individuals from the same cross combination was generated using single nucleotide polymorphism (SNP) markers with the GoldenGate assay (Huo et al. 2011). Then an F(6:7) recombinant inbred line population from the same cross combination was generated and used to conduct fine mapping of the Bsr1, Barley stripe mosaic virus (BSMV) resistance gene (Cui et al. 2012).
Site-specific nuclease technologies, zinc finger nucleases, transcription activator-like effector nucleases (TALENs) and clustered regulatory interspaced short palindromic repeat/Cas-based RNA-guided DNA endonucleases are innovative tools for genome engineering (Gaj et al. 2013). Recently, efficient genome modification using the TALEN method was demonstrated at eight loci in Brachypodium (Shan et al. 2013). The latest update of E-TALEN (http://www.e-talen.org/), which is a tool to design and evaluate TALEN, supported Brachypodium as one of the additional organisms (Heigwer et al. 2013).
Integration of genomic resources in Pooideae
The recent progress in genomic resources and analytical tools for use in Triticeae crops and Brachypodium facilitates gene discovery studies and functional analysis of genes involved in specific properties of Pooideae grass species. Comparative genomics and integration of available genomic instances in the Pooideae species should synergistically enable us to use genomic features to accelerate the gene discovery process, as well as complement and enrich our knowledge of structures and functions of orthologous genes in this subfamily.
Comparative genomics in Pooideae Conserved synteny is an important genomic property in genome evolution and is helpful in molecular marker development and positional gene cloning. Synteny among genome-sequenced Poaceae species, including rice, sorghum and Brachypodium, was used to develop a virtual gene order map (‘genome zipper’) for barley (H. vulgare), as mentioned earlier. The genome zipper approach has now become a key genomic technology to unlock complex genomes in the Pooideae subfamily (Spannagl et al. 2013), and its use has been applied in other Pooideae species, such as in wheat chromosome 4A (Hernandez et al. 2012), rye chromosome B (Martis et al. 2012) and Lolium (Pfeifer et al. 2013). The synteny between Brachypodium and Triticeae crops has assisted scientists in narrowing down candidate causal genes of mutants and quantitative trait loci (QTLs) in wheat and barley (Griffiths et al. 2006, Mueller et al. 2012, Liu et al. 2013). In the whole-genome shotgun sequencing of bread wheat, the wheat 454 reads, SNPs and genetic maps of wheat were anchored to the Brachypodium genome (Brenchley et al. 2012). Comparative use of the high-quality genome sequence of Brachypodium Bd21 has helped us to reconstruct a syntenically ordered draft genome in Pooideae species. These examples of comparative analysis in Pooideae show the significance of integrating genomic instances of Triticeae crops with those in Brachypodium.
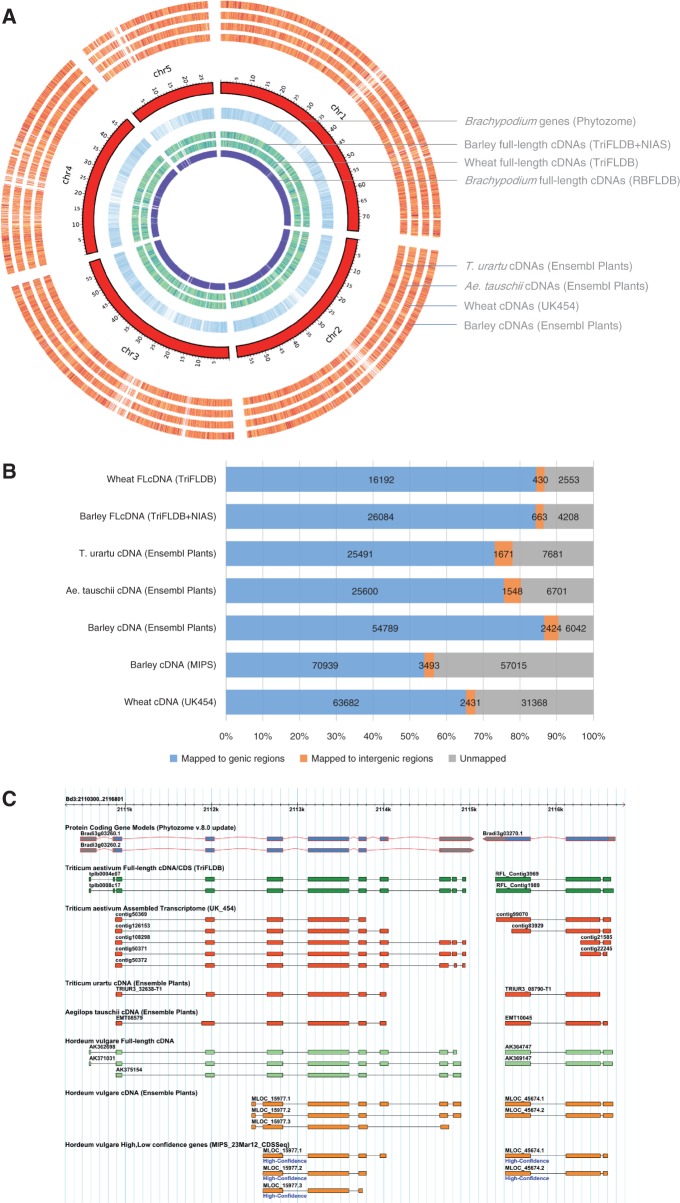
Comparative mapping analysis to identify closest counterparts is one of the practical approaches to achieve significant data integration of sequence resources in Pooideae. We performed a comparative mapping analysis of Brachypodium, wheat and barley, based on cDNAs and genomic assemblies. To establish the inter-relationships between Pooideae transcripts and genomic sequences, comparative mapping analysis was performed using available comprehensive cDNA sequences from each species of wheat, barley and Brachypodium as queries against the genomic sequences of Brachypodium and barley. We demonstrated that the data sets allowed us immediately to identify full-length cDNAs of putative orthologous genes in counter-species of Pooideae, and to compare gene structures conserved among putative orthologs (Mochida et al. 2013a). As a further extension, we also mapped transcript sequences of annotated genes in A. tauschii and T. urartu genes to the Brachypodium genome. Fig. 3A represents an overall image of the mapping results of cDNAs of wheat, barley and Brachypodium, as well as annotated gene models of barley, A. tauschii and T. urartu with annotated Brachypodium genes located on its five chromosomes. Most of the queried transcripts were allocated to the annotated gene regions of the Brachypodium genome; some were allocated to the non-annotated regions. These results suggest that there are potential transcription units of Brachypodium with conserved homology to cDNAs in Triticeae plants that have not yet been annotated (Mochida et al. 2013a) (Fig. 3B). The Brachypodium genome browser in RBFLDB (http://brachy.bmep.riken.jp/ver.1/tools.pl?t=gbrowse&sp=Bdi) consists of annotation tracks for the results of the comparative mapping, which should help to confirm conserved gene structures among orthologs in the Pooideae species (Fig. 3C). The integration of Triticeae sequence resources with the Brachypodium genome should be useful for annotating the genomes of the Pooideae, performing comparative studies and yielding putative orthologs among Pooideae species.
Fig. 3.
Comparative mapping of Triticeae cDNAs to the Brachypodium genome. A data set of full-length cDNAs of barley (GenBank accession Nos. AK353559–AK377172 deposited by NIAS, and available in TriFLDB) and wheat retrieved from TriFLDB, shotgun genome analysis of wheat by Roche 454 sequencing (downloaded from MIPS), cDNAs of T. urartu and A. taucschii (downloaded from Ensembl Plants) and barley cDNAs annotated on the cv. Morex genome (downloaded from MIPS and from Ensembl Plants) were used to the comparative mapping analysis. Cross-species mapping using cDNA data sets for querying the Brachypodium Bd21 genome was performed by sim4 with default parameter settings, followed by a BLASTN search, with e-value cut-offs of <1e-20. (A) An overview of the mapping results represented by a circular image. (B) Numbers of queries of wheat full-length cDNAs [Wheat FLcDNA (TriFLDB) and those of barley (Barley FLcDNA (TriFLDB + NIAS)], cDNAs annotated in the draft sequences of T. urartu [T. urartu cDNA (Ensembl Plants)] and of A. tauschii [Ae. tauschii cDNA (Ensembl Plants)], gene models annotated in the barley Morex genome of MIPS [Barley cDNA (MIPS)] and those of Ensembl Plants [Barley cDNA (Ensembl Plants)] and wheat cDNAs from the shotgun genome analysis [Wheat cDNA (UK454)], respectively, mapped to the genic or intergenic regions of the Brachypodium genome, or not mapped, are represented. (C) An example screen shot of a genome browser for mapped cDNAs to the Brachypodium genome aligned with annotated genes of Brachypodium. The mapping results were implemented in the Gbrowse interface of the RBFLDB web site and are available from http://brachy.bmep.riken.jp/ver.1/tools.pl?t=gbrowse&sp=Bdi.
The identification of orthologous groups is another practical approach to integrate genomic knowledge among related species. This work encompasses useful genome annotation, studies of gene/protein evolution and the identification of taxonomically restricted sequences (Li et al. 2003). Orthologous identification using OrthoMCL has been widely used to determine the distribution of orthologous gene families in genome-sequenced species compared with other species. The Brachypodium proteome data set was analyzed using OrthoMCL to detect Brachypodium- and Pooideae-specific genes (International Brachypodium Initiative 2010). Proteome data sets of A. tauschii and T. urartu were used, respectively, to analyze the distribution of orthologous gene families in the Poaceae family, based on the OrthoMCL analysis of Brachypodium, sorghum, rice and barley, and of Brachypodium, sorghum, rice and maize (Jia et al. 2013, Ling et al. 2013). Genome-wide assignments of protein-coding genes to homologous gene families provide us with a foundation to perform subsequent evolutionary studies for functional predication of genes.
After completing genome sequencing and full-length cDNA projects in human, the mouse and rice, ‘jamboree-style’ annotation meetings were orchestrated, such as H-inv (the Human Invitational Annotation project) (Imanishi et al. 2004), FANTOM (Functional Annotation Of Mouse) (Maeda et al. 2006) and RAP (Rice Annotation Projects) (Ohyanagi et al. 2006, Sakai et al. 2013). Accurate gene annotations of these species have played essential roles in their various post-genome projects, and associated databases have become hubs of information resources for each species. In the Pooideae subfamily, a high-quality genome sequence of Brachypodium Bd21 and enriched gene annotations will provide a firm foothold in comparative genomics with Triticeae crops. Integrating and cross-referencing information resources between Brachypodium and Triticeae plants could improve gene annotation work and allow researchers to infer shared orthologs and specific or divergent genes in this subfamily.
Some function-related annotations of genes, such as ontology, pathways and conserved sequence features, also play important roles in the prediction of biological functions of genes, as well as allowing us to grasp an overview of genome-scale instances. The Gene Ontology (GO) project has been widely used for functional classification of genes in various situations. Mapping results of Brachypodium genes to the GO terms are available in some online resources, Plaza (ftp://ftp.psb.ugent.be/pub/plaza/plaza_public_02_5/GO/go.bdi.csv.gz) and RBFLDB (http://brachy.bmep.riken.jp/ver.1/static/download/iprscan.results.tab.txt.gz). Regarding metabolic pathways, BrachyCyc version 2.0 is available from the Gramene website (http://pathway.gramene.org/gramene/brachycyc.shtml). With the availability of complete genome sequences, we have been able to compile catalogs describing the function and organization of regulatory systems of transcription factors (TFs) and further integrate information on TFs with knowledge related to their functions (Mochida et al. 2009b, Mochida et al. 2010, Mochida et al. 2013b, Naika et al. 2013). PlantTFDB (http://planttfdb_v2.cbi.edu.cn/index.php?sp=Bdi) and GramineaeTFDB (http://gramineaetfdb.psc.riken.jp/) provide information about genes encoding TFs in the Brachypodium genome (Mochida et al. 2011b, Zhang et al. 2011). Information about putative cis-elements located in the promoter regions (–3,000 bp from the TU start) in the Brachypodium genome is available from RBFLDB (http://brachy.bmep.riken.jp/ver.1/dl.pl). These resources could function as contextual filters for meta-analysis in omics-based approaches to narrow down candidate genes and build hypotheses for further analysis in Brachypodium and in Triticeae plants.
Brachypodium to Triticeae crops
Comparative genomics studies among Poaceae species have suggested that Brachypodium is a better, more closely related model than rice for Triticeae crops. Brachypodium possesses all the features of a modern model organism. It is therefore advantageous to accelerate gene discovery and translational research to improve the Triticeae crops. Recently, Brachypodium has become an accepted model plant for research related to various physiological phenomena, particularly those observed in monocots; many citations demonstrate the utility of Brachypodium as a model system. Here, we introduce several typical examples recently studied using Brachypodium, which could provide a useful model system for better understanding of the biological systems and transferring knowledge to Triticeae crops.
Triticeae crops suffer severe yield losses from biotic stresses such as pathogens and pests. For example, viruses such as BSMV and the Wheat streak mosaic virus, which infect the Triticeae crops, have been studied using dicot host plants to dissect viral processes and determine how the pathogens cause disease. However, the underlying mechanisms of immunity remain largely unknown, especially the molecular basis of disease in monocot plants. Pathogens infecting B. distachyon and Brachypodium sylvaticum were listed by Mandadi and Scholthof (2013), and the reported bacterial, fungal and viral pathogens that infect Brachypodium species will assist further studies in better understanding the molecular basis of disease in monocots (Mandadi and Scholthof 2013). For example, to analyze the interaction of the Panicum mosaic virus (PMV) and its satellite virus (SPMV) with host plants, Mandadi and Scholthof investigated changes in the transcriptome of control Brachypodium plants and those infected by both viruses, using the custom-designed Affymetrix GeneChip (Mandadi and Scholthof 2012). More recently, the Puccinia graminis–Brachypodium pathosystem was examined for its potential to demonstrate incompatibility and non-host resistance in P. graminis using eight inbred Brachypodium lines (Figueroa et al. 2013). The Fusarium pathogen is the causal agent of Fusarium head blight (FHB) of small grain cereals including wheat and barley, which is a global problem due to the reduced yield and quality as well as to the contamination of mycotoxins. Peraldi et al (2011) showed disease symptoms on Brachypodium spikes and the accumulation of a mycotoxin within floral tissues, which were highly similar to those in wheat (Peraldi et al. 2011). Hence, as the number of Triticeae crop pathogens that also can infect Brachypodium species increases, Brachypodium will provide a significant model system to elucidate the molecular basis of interactions with those pathogens and will accelerate translational studies to wheat and barley.
For temperate grass species such as wheat and barley, Brachypodium provides a good model system to investigate the responses of biological systems to temperature stresses. Li et al. (2012) examined potential uses of Brachypodium as a model for cold stress responses in the Pooideae subfamily. They described the significance of using Brachypodium in studies of cold stress responses in Pooideae based on comparative genomics. They also noted that the evolutionary history of key genes for low temperature responses differed among Brachypodium and core Pooideae species (Li et al. 2012). Recently, the thermal response of Brachypodium was analyzed based on transcriptomics using the Affymetrix GeneChip, chromatin modification with Chromatin-IP analysis and molecular genetics with RNA interference (RNAi). In this study, the authors indicated that H2A.Z-nucleosomes are important to coordinate sensitivity to increased temperature during grain development in Brachypodium (Boden et al. 2013). Plant response to drought has been extensively studied in Arabidopsis and in a few grass species (Hirayama and Shinozaki 2010). Brachypodium may possess specific attributes to adapt to or tolerate drought because of its geographic origin, and these attributes may be translated to related crop species, such as wheat and barley. Transcriptome profiles using the Affymetrix GeneChip of different developmental leaf zones illustrated different responses to drought in Brachypodium (Verelst et al. 2013).These examples represent the usefulness of Brachypodium as a model system in abiotic stress studies of temperate grass species with critical genomic tools and tractable biological features. In previous studies, the function of genes in wheat and barley has often been studied in transgenic plants such as Arabidopsis, tobacco and rice (Brini et al. 2011, Hu et al. 2012). However, the physiologically similar properties of Brachypodium and Triticeae crops provide opportunities to promote the functional characterization of genes in wheat and barley, including those involved in abiotic stress responses and tolerance of harsh conditions.
Brachypodium is a promising model system in some additional areas. It has already been widely used in studies of cellulosic biomass production and cell wall biosynthesis (Rancour et al. 2012). Brachypodium has provided a model system to understand biogenesis of type II cell wall in grasses, a type of cell wall that is structurally different from type I in dicots (Yokoyama and Nishitani 2004). Based on a phylogenetic and expression analysis of genes encoding Yellow Stripe-Like (YSL) transporters, Brachypodium was also proposed as a promising species for studies of iron homeostasis in grasses (Yordem et al. 2011). Recently, a similarity search in a barley EST database, using HvYS1 as the query in barley, identified HvYSL5 and HvYSL2 (Araki et al. 2011, Zheng et al. 2011). The Brachypodium orthologs of the YSL family offer a model system in comparative genomics of Fe distribution and translocation in grasses. Furthermore, in a recent review article by Chochois et al. (2012), the authors proposed Brachypodium as a model system for root research and to accelerate the identification of genes for wheat root improvement. Brachypodium and wheat have very similar root anatomies, distinct from the roots of rice, which are specialized to overcome anaerobic conditions associated with submerged roots (Chochois et al. 2012).
Future prospects for crop improvements
As climate change progresses, creating varieties that can adapt to environmental stresses is one of the most important targets of crop breeding. Although our understanding of plant systems’ responses to environmental stresses have greatly increased in the last decade, applying the knowledge from basic research to improve the environmental stress tolerance of crops will be a challenge (Qin et al. 2011). Recently, a number of genes involved in responses to and tolerance of environmental stresses were identified and characterized in wheat and barley (Conde et al. 2011, Horie et al. 2011). Rapid advances in genomic tools with NGS applications in Triticeae and in Brachypodium will definitely accelerate gene discovery research in Triticeae crops. In addition to the sequence resources at the whole-genome scale, some important areas for crop improvement are being advanced by new experimental and computational approaches that use omics and mathematics.
One of the important areas is plant epigenetics, which has recently gained attention as a genomic regulatory system, not only for developmental controls but also for responses to environmental stresses and as a possible new source of beneficial traits for plant breeding (Mirouze and Paszkowski 2011, Kim et al. 2012b, Kinoshita and Jacobsen 2012). For example, histone modifications were shown to be correlated with the inactivation of drought-inducible genes, such as RD20, RD29A and AtGOLS2, during recovery by rehydration in Arabidopsis (Kim et al. 2012a). The epigenomics approach, involving whole-genome-scale epigenetics analysis, should elucidate genome-wide chromatin regulation in abiotic stress responses (Ikeda 2012). The data set of whole-genome sequences of Triticeae plants, at least for the gene space of their genomes, will provide opportunities to study epigenomic phenomena in wheat and barley and will potentially allow epigenetic knowledge to be applied to their improvements in the near future.
Another important area is mining alleles for beneficial traits in plants. Whole-genome reference sequences and platforms for high-throughput genotyping are providing opportunities to improve our understanding of the history of plant domestication and to apply evolutionary knowledge of the crop genome to crop improvements (Morrell et al. 2011). Chip-based and NGS-based platforms for whole-genome-scale genotyping are available for a number of genome-sequenced organisms (Chen et al. 2013) and also for wheat and barley (Sato et al. 2011, Cavanagh et al. 2013). High-throughput genotyping with dense genome-wide markers has allowed us to identify genomic regions associated with complex agronomic traits. Genome-wide association studies have been performed not only in Arabidopsis but also in various crops, including wheat and barley, using NGS-based whole-genome re-sequencing or microarray-based SNP genotyping (Huang et al. 2010, Pasam et al. 2012, Cavanagh et al. 2013, Morris et al. 2013). For species with large complex genomes, such as barley and wheat, reduced genomic representation using restriction enzymes has been an efficient approach, coupled with NGS-based DNA polymorphism discovery. Genotyping-by-sequencing (GBS) and restriction-associated DNA sequencing (RAD-seq) were developed and rapidly popularized for whole-genome profiling of DNA polymorphisms in many species, including barley and wheat (Poland et al. 2012, Mascher et al. 2013b).
The application of NGS technology to next-generation populations could provide an efficient strategy to identify loci that contribute to a complex trait. The nested association mapping (NAM) population, which is based on crossing diverse strains to a reference parent, has successfully been applied to a number of traits in maize (Kump et al. 2011, Tian et al. 2011). The multiparent advanced generation intercross (MAGIC) population or the recombinant inbred advanced intercross line (RIAIL) population also will help improve genetic mapping and advance breeding programs of crops, including barley and wheat (Cavanagh et al. 2008, Kover et al. 2009, Huang et al. 2012).
Based on the evolutionary history of a target crop, the discovery of loci responsible for adaptive evolution has been a promising approach to identify loci contributing to adaptation in the agronomic environment. Geographic distribution of the target locus represents patterns of historical selection of alleles that are adaptive to a local agronomic environment (Saisho et al. 2011). The geographic distribution of genotypes superimposed on information about local environmental conditions may inspire us to discover allelic combinations that would be of value in forecasted environments. Furthermore, the integration of genotype and epigenomic profiles of each strain (epigenotype) will provide important genomic information for dissecting the contribution of epigenetic and genetic variations to phenotypic changes in responses to the environment (Schmitz et al. 2013).
Crops in the field are continuously exposed to various environmental signals and must respond to fluctuating conditions. In order to translate knowledge more effectively from laboratories to field conditions in agriculture, we must understand the omics dynamics of crops in response to endogenous and exogenous perturbations under fluctuating field conditions. Recent advances in high-throughput technology allow us to measure changes in the omics of plants, not only under controlled conditions but also under natural conditions (Izawa et al. 2011, Richards et al. 2012, Dal Santo et al. 2013). In addition to the omics analysis in natural environments, to gain a mechanistic understanding of biological systems, mathematical modeling and simulation approaches are increasingly used in studies of plants’ cellular metabolism, growth, developmental processes and responses to the environment (Aikawa et al. 2010, De Vos et al. 2012, Katsuragi et al. 2013). In rice, a successful model and prediction of genome-wide transcriptional changes under fluctuating field conditions was recently represented based on transcriptome data and the corresponding meteorological data (Nagano et al. 2012). As we work to improve crops, we must take ongoing climate change into consideration. The omics-based data set of plants in natural environments may be a substantial resource to generate models for predicting how a plant system will react under field conditions it has not experienced, such as warmer, more rainy or less rainy. The combination of such a systems-based approach and genome-wide evolutionary analyses to identify beneficial genes will provide an opportunity to apply our wisdom of the past to crop breeding for the future. It may be a pre-emptive strategy to ensure sustainable food production under the changed environments of the future.
Conclusions
Available tools for Triticeae genomics have dramatically advanced in recent years. Rapidly increasing sequence data sets and associated resources for Pooideae will potentially accelerate the gene discovery process and improve our understanding of their biological functions. We urgently need to integrate fragmented knowledge and translate genome-based wisdom to achieve practical improvements in the Triticeae crops. Integration of genomic information and resources from the model grass Brachypodium and those from Triticeae crops should make a huge contribution to gene discovery and improved understanding of gene functions, as well as transferring knowledge to the Triticeae crops. Recent advanced analytical platforms and applications based on NGS technologies have provided opportunities to advance radically the discovery of genes and alleles for beneficial traits despite the genome complexity of Triticeae species. The promising combination of these approaches with systems approaches and mathematical modeling will help us achieve crop improvements to ensure a sustainable agriculture in the future.
Funding
This work was supported by a grant from a strategic research project in RIKEN Biomass engineering program.
Acknowledgments
The authors thank Tetsuya Sakurai, Takuhiro Yoshida of the RIKEN Center for Sustainable Resource Science for their data generation of cDNA mapping among Pooideae plants. The authors also thank Daisuke Saisho of Okayama University for his valuable comments and discussions on this manuscript.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- BAC
bacterial artificial chromosome
- BSMV
Barley stripe mosaic virus
- EST
expressed sequence tag
- FOX
full-length cDNA overexpressor
- GO
Gene Ontology
- NGS
next-generation sequencing
- QTL
quantitative trait locus
- SNP
single nucleotide polymorphism
- SRA
sequence read archive
- TF
transcription factor
- TILLING
Targeting Induced Local Lesions IN Genomes
- TALEN
transcription activator-like effector nuclease
References
- Aikawa S, Kobayashi MJ, Satake A, Shimizu KK, Kudoh H. Robust control of the seasonal expression of the Arabidopsis FLC gene in a fluctuating environment. Proc. Natl Acad. Sci. USA. 2010;107:11632–11637. doi: 10.1073/pnas.0914293107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandratos N, Bruinsma J. World Agriculture Towards 2030/2050: The 2012 Revision. 2012 ESA Working paper No. 12.03. [Google Scholar]
- Arai-Kichise Y, Shiwa Y, Nagasaki H, Ebana K, Yoshikawa H, Yano M, et al. Discovery of genome-wide DNA polymorphisms in a landrace cultivar of Japonica rice by whole-genome sequencing. Plant Cell Physiol. 2011;52:274–282. doi: 10.1093/pcp/pcr003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki R, Murata J, Murata Y. A novel barley yellow stripe 1-like transporter (HvYSL2) localized to the root endodermis transports metal–phytosiderophore complexes. Plant Cell Physiol. 2011;52:1931–1940. doi: 10.1093/pcp/pcr126. [DOI] [PubMed] [Google Scholar]
- Ashelford K, Eriksson ME, Allen CM, D’Amore R, Johansson M, Gould P, et al. Full genome re-sequencing reveals a novel circadian clock mutation in Arabidopsis. Genome Biol. 2011;12:R28. doi: 10.1186/gb-2011-12-3-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Saito T, Sakamoto D, Ito A, Fujii H, Moriguchi T. Transcriptome analysis of Japanese pear (Pyrus pyrifolia Nakai) flower buds transitioning through endodormancy. Plant Cell Physiol. 2013;54:1132–1151. doi: 10.1093/pcp/pct067. [DOI] [PubMed] [Google Scholar]
- Baker JM, Hawkins ND, Ward JL, Lovegrove A, Napier JA, Shewry PR, et al. A metabolomic study of substantial equivalence of field-grown genetically modified wheat. Plant Biotechnol. J. 2006;4:381–392. doi: 10.1111/j.1467-7652.2006.00197.x. [DOI] [PubMed] [Google Scholar]
- Bevan MW, Garvin DF, Vogel JP. Brachypodium distachyon genomics for sustainable food and fuel production. Curr. Opin. Biotechnol. 2010;21:211–217. doi: 10.1016/j.copbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Boden SA, Kavanova M, Finnegan EJ, Wigge PA. Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 2013;14:R65. doi: 10.1186/gb-2013-14-6-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowne JB, Erwin TA, Juttner J, Schnurbusch T, Langridge P, Bacic A, et al. Drought responses of leaf tissues from wheat cultivars of differing drought tolerance at the metabolite level. Mol. Plant. 2012;5:418–429. doi: 10.1093/mp/ssr114. [DOI] [PubMed] [Google Scholar]
- Bragg JN, Wu J, Gordon SP, Guttman ME, Thilmony R, Lazo GR, et al. Generation and characterization of the Western Regional Research Center Brachypodium T-DNA insertional mutant collection. PLoS One. 2012;7:e41916. doi: 10.1371/journal.pone.0041916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley R, Spannagl M, Pfeifer M, Barker GL, D’Amore R, Allen AM, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491:705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini F, Yamamoto A, Jlaiel L, Takeda S, Hobo T, Dinh HQ, et al. Pleiotropic effects of the wheat dehydrin DHN-5 on stress responses in Arabidopsis. Plant Cell Physiol. 2011;52:676–688. doi: 10.1093/pcp/pcr030. [DOI] [PubMed] [Google Scholar]
- Brkljacic J, Grotewold E, Scholl R, Mockler T, Garvin DF, Vain P, et al. Brachypodium as a model for the grasses: today and the future. Plant Physiol. 2011;157:3–13. doi: 10.1104/pp.111.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- Cavanagh C, Morell M, Mackay I, Powell W. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 2008;11:215–221. doi: 10.1016/j.pbi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Cavanagh CR, Chao S, Wang S, Huang BE, Stephen S, Kiani S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl Acad. Sci. USA. 2013;110:8057–8062. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Chu YW, Chen CW, Leu WM, Hsu HF, Yang CH. Characterization of Oncidium ‘Gower Ramsey’ transcriptomes using 454 GS-FLX pyrosequencing and their application to the identification of genes associated with flowering time. Plant Cell Physiol. 2011;52:1532–1545. doi: 10.1093/pcp/pcr101. [DOI] [PubMed] [Google Scholar]
- Chen H, He H, Zhou F, Yu H, Deng XW. Development of genomics-based genotyping platforms and their applications in rice breeding. Curr. Opin. Plant Biol. 2013;16:247–254. doi: 10.1016/j.pbi.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Chochois V, Vogel JP, Watt M. Application of Brachypodium to the genetic improvement of wheat roots. J. Exp. Bot. 2012;63:3467–3474. doi: 10.1093/jxb/ers044. [DOI] [PubMed] [Google Scholar]
- Close TJ, Bhat PR, Lonardi S, Wu Y, Rostoks N, Ramsay L, et al. Development and implementation of high-throughput SNP genotyping in barley. BMC Genomics. 2009;10:582. doi: 10.1186/1471-2164-10-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde A, Chaves MM, Geros H. Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant Cell Physiol. 2011;52:1583–1602. doi: 10.1093/pcp/pcr107. [DOI] [PubMed] [Google Scholar]
- Cui Y, Lee MY, Huo N, Bragg J, Yan L, Yuan C, et al. Fine mapping of the Bsr1 barley stripe mosaic virus resistance gene in the model grass Brachypodium distachyon. PLoS One. 2012;7:e38333. doi: 10.1371/journal.pone.0038333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Santo S, Tornielli GB, Zenoni S, Fasoli M, Farina L, Anesi A, et al. The plasticity of the grapevine berry transcriptome. Genome Biol. 2013;14:r54. doi: 10.1186/gb-2013-14-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, Wang Y, Darracq O, d’Yvoire MB, et al. A TILLING platform for functional genomics in Brachypodium distachyon. PLoS One. 2013;8:e65503. doi: 10.1371/journal.pone.0065503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, et al. Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J. 2012;71:492–502. doi: 10.1111/j.1365-313X.2012.05005.x. [DOI] [PubMed] [Google Scholar]
- De Vos D, Dzhurakhalov A, Draelants D, Bogaerts I, Kalve S, Prinsen E, et al. Towards mechanistic models of plant organ growth. J. Exp. Bot. 2012;63:3325–3337. doi: 10.1093/jxb/ers037. [DOI] [PubMed] [Google Scholar]
- Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, et al. Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 2001;127:1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J, Dvorak J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Tatematsu K, Hanada K, Duermeyer L, Okamoto M, Yonekura-Sakakibara K, et al. Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol. 2012;53:16–27. doi: 10.1093/pcp/pcr171. [DOI] [PubMed] [Google Scholar]
- Figueroa M, Alderman S, Garvin DF, Pfender WF. Infection of Brachypodium distachyon by formae speciales of Puccinia graminis: early infection events and host–pathogen incompatibility. PLoS One. 2013;8:e56857. doi: 10.1371/journal.pone.0056857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CH, Chen YW, Hsiao YY, Pan ZJ, Liu ZJ, Huang YM, et al. OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 2011;52:238–243. doi: 10.1093/pcp/pcq201. [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M. Recent progress in the development of metabolome databases for plant systems biology. Front. Plant Sci. 2013;4:73. doi: 10.3389/fpls.2013.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima EO, Seki H, Sawai S, Suzuki M, Ohyama K, Saito K, et al. Combinatorial biosynthesis of legume natural and rare triterpenoids in engineered yeast. Plant Cell Physiol. 2013;54:740–749. doi: 10.1093/pcp/pct015. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvin DF, McKenzie N, Vogel JP, Mockler TC, Blankenheim ZJ, Wright J, et al. An SSR-based genetic linkage map of the model grass Brachypodium distachyon. Genome. 2010;53:1–13. doi: 10.1139/g09-079. [DOI] [PubMed] [Google Scholar]
- Gil-Amado JA, Gomez-Jimenez MC. Transcriptome analysis of mature fruit abscission control in olive. Plant Cell Physiol. 2013;54:244–269. doi: 10.1093/pcp/pcs179. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, et al. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- Hamada K, Hongo K, Suwabe K, Shimizu A, Nagayama T, Abe R, et al. OryzaExpress: an integrated database of gene expression networks and omics annotations in rice. Plant Cell Physiol. 2011;52:220–229. doi: 10.1093/pcp/pcq195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, Walther N, Glaeser K, Pelz O, Breinig M, et al. E-TALEN: a web tool to design TALENs for genome engineering. Nucleic Acids Res. 2013;41:e190. doi: 10.1093/nar/gkt789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez P, Martis M, Dorado G, Pfeifer M, Galvez S, Schaaf S, et al. Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 2012;69:377–386. doi: 10.1111/j.1365-313X.2011.04808.x. [DOI] [PubMed] [Google Scholar]
- Higashi Y, Saito K. Network analysis for gene discovery in plant-specialized metabolism. Plant Cell Environ. 2013;36:1597–1606. doi: 10.1111/pce.12069. [DOI] [PubMed] [Google Scholar]
- Hirano K, Aya K, Morinaka Y, Nagamatsu S, Sato Y, Antonio BA, et al. Survey of genes involved in rice secondary cell wall formation through a co-expression network. Plant Cell Physiol. 2013a;54:1803–1821. doi: 10.1093/pcp/pct121. [DOI] [PubMed] [Google Scholar]
- Hirano K, Kondo M, Aya K, Miyao A, Sato Y, Antonio BA, et al. Identification of transcription factors involved in rice secondary cell wall formation. Plant Cell Physiol. 2013b;54:1791–1802. doi: 10.1093/pcp/pct122. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hirsch CN, Buell CR. Tapping the promise of genomics in species with complex, nonmodel genomes. Annu. Rev. Plant Biol. 2013;64:89–110. doi: 10.1146/annurev-arplant-050312-120237. [DOI] [PubMed] [Google Scholar]
- Hong JP, Takeshi Y, Kondou Y, Schachtman DP, Matsui M, Shin R. Identification and characterization of transcription factors regulating Arabidopsis HAK5. Plant Cell Physiol. 2013;54:1478–1490. doi: 10.1093/pcp/pct094. [DOI] [PubMed] [Google Scholar]
- Horie T, Kaneko T, Sugimoto G, Sasano S, Panda SK, Shibasaka M, et al. Mechanisms of water transport mediated by PIP aquaporins and their regulation via phosphorylation events under salinity stress in barley roots. Plant Cell Physiol. 2011;52:663–675. doi: 10.1093/pcp/pcr027. [DOI] [PubMed] [Google Scholar]
- Hu W, Yuan Q, Wang Y, Cai R, Deng X, Wang J, et al. Overexpression of a wheat aquaporin gene, TaAQP8, enhances salt stress tolerance in transgenic tobacco. Plant Cell Physiol. 2012;53:2127–2141. doi: 10.1093/pcp/pcs154. [DOI] [PubMed] [Google Scholar]
- Huang BE, George AW, Forrest KL, Kilian A, Hayden MJ, Morell MK, et al. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 2012;10:826–839. doi: 10.1111/j.1467-7652.2012.00702.x. [DOI] [PubMed] [Google Scholar]
- Huang MD, Hsing YI, Huang AH. Transcriptomes of the anther sporophyte: availability and uses. Plant Cell Physiol. 2011;52:1459–1466. doi: 10.1093/pcp/pcr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Huo N, Garvin DF, You FM, McMahon S, Luo MC, Gu YQ, et al. Comparison of a high-density genetic linkage map to genome features in the model grass Brachypodium distachyon. Theor. Appl. Genet. 2011;123:455–464. doi: 10.1007/s00122-011-1598-4. [DOI] [PubMed] [Google Scholar]
- Iida K, Kawaguchi S, Kobayashi N, Yoshida Y, Ishii M, Harada E, et al. ARTADE2DB: improved statistical inferences for Arabidopsis gene functions and structure predictions by dynamic structure-based dynamic expression (DSDE) analyses. Plant Cell Physiol. 2011;52:254–264. doi: 10.1093/pcp/pcq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y. Plant imprinted genes identified by genome-wide approaches and their regulatory mechanisms. Plant Cell Physiol. 2012;53:809–816. doi: 10.1093/pcp/pcs049. [DOI] [PubMed] [Google Scholar]
- Imanishi T, Itoh T, Suzuki Y, O’Donovan C, Fukuchi S, Koyanagi KO, et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Barley Genome Sequencing Consortium. A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491:711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Ishikawa T, Takahara K, Hirabayashi T, Matsumura H, Fujisawa S, Terauchi R, et al. Metabolome analysis of response to oxidative stress in rice suspension cells overexpressing cell death suppressor Bax inhibitor-1. Plant Cell Physiol. 2010;51:9–20. doi: 10.1093/pcp/pcp162. [DOI] [PubMed] [Google Scholar]
- Izawa T, Mihara M, Suzuki Y, Gupta M, Itoh H, Nagano AJ, et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell. 2011;23:1741–1755. doi: 10.1105/tpc.111.083238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James GV, Patel V, Nordstrom KJ, Klasen JR, Salome PA, Weigel D, et al. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013;14:R61. doi: 10.1186/gb-2013-14-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496:91–95. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- Kamada-Nobusada T, Makita N, Kojima M, Sakakibara H. Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: the role of glutamine metabolism as an additional signal. Plant Cell Physiol. 2013;54:1881–1893. doi: 10.1093/pcp/pct127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y, Jikumaru Y, Hanada A, Nambara E, Abrams SR, Kamiya Y, et al. Comprehensive hormone profiling in developing Arabidopsis seeds: examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010;51:1988–2001. doi: 10.1093/pcp/pcq158. [DOI] [PubMed] [Google Scholar]
- Katsuragi T, Ono N, Yasumoto K, Altaf-Ul-Amin M, Hirai MY, Sriyudthsak K, et al. SS-mPMG and SS-GA: tools for finding pathways and dynamic simulation of metabolic networks. Plant Cell Physiol. 2013;54:728–739. doi: 10.1093/pcp/pct052. [DOI] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Enju A, Totoki Y, Toyoda A, Sakaki Y, et al. Assessment of adaptive evolution between wheat and rice as deduced from full-length common wheat cDNA sequence data and expression patterns. BMC Genomics. 2009;10:271. doi: 10.1186/1471-2164-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaura K, Mochida K, Yamazaki Y, Ogihara Y. Transcriptome analysis of salinity stress responses in common wheat using a 22k oligo-DNA microarray. Funct. Integr. Genomics. 2006;6:132–142. doi: 10.1007/s10142-005-0010-3. [DOI] [PubMed] [Google Scholar]
- Kim JM, To TK, Ishida J, Matsui A, Kimura H, Seki M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012a;53:847–856. doi: 10.1093/pcp/pcs053. [DOI] [PubMed] [Google Scholar]
- Kim JM, To TK, Seki M. An epigenetic integrator: new insights into genome regulation, environmental stress responses and developmental controls by histone deacetylase 6. Plant Cell Physiol. 2012b;53:794–800. doi: 10.1093/pcp/pcs004. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Jacobsen SE. Opening the door to epigenetics in PCP. Plant Cell Physiol. 2012;53:763–765. doi: 10.1093/pcp/pcs061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogel KH, Voll LM, Schafer P, Jansen C, Wu Y, Langen G, et al. Transcriptome and metabolome profiling of field-grown transgenic barley lack induced differences but show cultivar-specific variances. Proc. Natl Acad. Sci. USA. 2010;107:6198–6203. doi: 10.1073/pnas.1001945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Kamada-Nobusada T, Komatsu H, Takei K, Kuroha T, Mizutani M, et al. Highly sensitive and high-throughput analysis of plant hormones using MS-probe modification and liquid chromatography-tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 2009;50:1201–1214. doi: 10.1093/pcp/pcp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–894. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasileva KV, Buffalo V, Bailey P, Pearce S, Ayling S, Tabbita F, et al. Separating homeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 2013;14:R66. doi: 10.1186/gb-2013-14-6-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Fujita M, Takahashi H, Nakazono M, Tsutsumi N, Kurata N. Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol. 2013;54:750–765. doi: 10.1093/pcp/pct029. [DOI] [PubMed] [Google Scholar]
- Kudo T, Akiyama K, Kojima M, Makita N, Sakurai T, Sakakibara H. UniVIO: a multiple omics database with hormonome and transcriptome data from rice. Plant Cell Physiol. 2013;54:e9. doi: 10.1093/pcp/pct003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat. Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- Lai K, Berkman PJ, Lorenc MT, Duran C, Smits L, Manoli S, et al. WheatGenome.info: an integrated database and portal for wheat genome information. Plant Cell Physiol. 2012;53:e2. doi: 10.1093/pcp/pcr141. [DOI] [PubMed] [Google Scholar]
- Leaungthitikanchana S, Fujibe T, Tanaka M, Wang S, Sotta N, Takano J, et al. Differential expression of three BOR1 genes corresponding to different genomes in response to boron conditions in hexaploid wheat (Triticum aestivum L.) Plant Cell Physiol. 2013;54:1056–1063. doi: 10.1093/pcp/pct059. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Perdian DC, Song Z, Yeung ES, Nikolau BJ. Use of mass spectrometry for imaging metabolites in plants. Plant J. 2012;70:81–95. doi: 10.1111/j.1365-313X.2012.04899.x. [DOI] [PubMed] [Google Scholar]
- Li C, Rudi H, Stockinger EJ, Cheng H, Cao M, Fox SE, et al. Comparative analyses reveal potential uses of Brachypodium distachyon as a model for cold stress responses in temperate grasses. BMC Plant Biol. 2012;12:65. doi: 10.1186/1471-2229-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- Liu S, Sehgal SK, Li J, Lin M, Trick HN, Yu J, et al. Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics. 2013;195:263–273. doi: 10.1534/genetics.113.152330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loraine AE, McCormick S, Estrada A, Patel K, Qin P. RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol. 2013;162:1092–1109. doi: 10.1104/pp.112.211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas SJ, Akpinar BA, Kantar M, Weinstein Z, Aydinoglu F, Safar J, et al. Physical mapping integrated with syntenic analysis to characterize the gene space of the long arm of wheat chromosome 1A. PLoS One. 2013;8:e59542. doi: 10.1371/journal.pone.0059542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda N, Kasukawa T, Oyama R, Gough J, Frith M, Engstrom PG, et al. Transcript annotation in FANTOM3: mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof KB. Characterization of a viral synergism in the monocot Brachypodium distachyon reveals distinctly altered host molecular processes associated with disease. Plant Physiol. 2012;160:1432–1452. doi: 10.1104/pp.112.204362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandadi KK, Scholthof KB. Plant immune responses against viruses: how does a virus cause disease? Plant Cell. 2013;25:1489–1505. doi: 10.1105/tpc.113.111658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martis MM, Klemme S, Banaei-Moghaddam AM, Blattner FR, Macas J, Schmutzer T, et al. Selfish supernumerary chromosome reveals its origin as a mosaic of host genome and organellar sequences. Proc. Natl Acad. Sci. USA. 2012;109:13343–13346. doi: 10.1073/pnas.1204237109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Richmond TA, Gerhardt DJ, Himmelbach A, Clissold L, Sampath D, et al. Barley whole exome capture: a tool for genomic research in the genus Hordeum and beyond. Plant J. 2013a;76:494–505. doi: 10.1111/tpj.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M, Wu S, Amand PS, Stein N, Poland J. Application of genotyping-by-sequencing on semiconductor sequencing platforms: a comparison of genetic and reference-based marker ordering in barley. PLoS One. 2013b;8:e76925. doi: 10.1371/journal.pone.0076925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–1149. doi: 10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, et al. Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 2011;156:20–28. doi: 10.1104/pp.110.171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y. Evolution of polyploid triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011;52:750–764. doi: 10.1093/pcp/pcr018. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K. Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals. Mol. Biol. Evol. 2002;19:2084–2091. doi: 10.1093/oxfordjournals.molbev.a004033. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Martis M, Hedley PE, Simkova H, Liu H, Morris JA, et al. Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell. 2011;23:1249–1263. doi: 10.1105/tpc.110.082537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KF, Taudien S, Martis M, Simkova H, Suchankova P, Gundlach H, et al. Gene content and virtual gene order of barley chromosome 1H. Plant Physiol. 2009;151:496–505. doi: 10.1104/pp.109.142612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 2011;14:267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- Miyao A, Nakagome M, Ohnuma T, Yamagata H, Kanamori H, Katayose Y, et al. Molecular spectrum of somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol. 2012;53:256–264. doi: 10.1093/pcp/pcr172. [DOI] [PubMed] [Google Scholar]
- Mochida K, Kawaura K, Shimosaka E, Kawakami N, Shin IT, Kohara Y, et al. Tissue expression map of a large number of expressed sequence tags and its application to in silico screening of stress response genes in common wheat. Mol. Genet. Genomics. 2006;276:304–312. doi: 10.1007/s00438-006-0120-1. [DOI] [PubMed] [Google Scholar]
- Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K. TriMEDB: a database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae. BMC Plant Biol. 2008;8:72. doi: 10.1186/1471-2229-8-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol. 2010;51:497–523. doi: 10.1093/pcp/pcq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Shinozaki K. Advances in omics and bioinformatics tools for systems analyses of plant functions. Plant Cell Physiol. 2011;52:2017–2038. doi: 10.1093/pcp/pcr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Uehara-Yamaguchi Y, Takahashi F, Yoshida T, Sakurai T, Shinozaki K. Large-scale collection and analysis of full-length cDNAs from Brachypodium distachyon and integration with Pooideae sequence resources. PLoS One. 2013a;8:e75265. doi: 10.1371/journal.pone.0075265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Uehara-Yamaguchi Y, Yoshida T, Sakurai T, Shinozaki K. Global landscape of a co-expressed gene network in barley and its application to gene discovery in Triticeae crops. Plant Cell Physiol. 2011a;52:785–803. doi: 10.1093/pcp/pcr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yamazaki Y, Ogihara Y. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Genet. Genomics. 2003;270:371–377. doi: 10.1007/s00438-003-0939-7. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Ogihara Y, Shinozaki K. TriFLDB: a database of clustered full-length coding sequences from Triticeae with applications to comparative grass genomics. Plant Physiol. 2009a;150:1135–1146. doi: 10.1104/pp.109.138214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009b;16:353–369. doi: 10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. LegumeTFDB: an integrative database of Glycine max, Lotus japonicus and Medicago truncatula transcription factors. Bioinformatics. 2010;26:290–291. doi: 10.1093/bioinformatics/btp645. [DOI] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. In silico analysis of transcription factor repertoires and prediction of stress-responsive transcription factors from six major gramineae plants. DNA Res. 2011b;18:321–332. doi: 10.1093/dnares/dsr019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K, Yoshida T, Sakurai T, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. TreeTFDB: an integrative database of the transcription factors from six economically important tree crops for functional predictions and comparative and functional genomics. DNA Res. 2013b;20:151–162. doi: 10.1093/dnares/dss040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell PL, Buckler ES, Ross-Ibarra J. Crop genomics: advances and applications. Nat. Rev. Genet. 2011;13:85–96. doi: 10.1038/nrg3097. [DOI] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, et al. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl Acad. Sci. USA. 2013;110:453–458. doi: 10.1073/pnas.1215985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, et al. High-resolution metabolic mapping of cell types in plant roots. Proc. Natl Acad. Sci. USA. 2013;110:E1232–E1241. doi: 10.1073/pnas.1302019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AH, Dockter C, Gough SP, Lundqvist U, von Wettstein D, Hansson M. Characterization of mutations in barley fch2 encoding chlorophyllide a oxygenase. Plant Cell Physiol. 2012;53:1232–1246. doi: 10.1093/pcp/pcs062. [DOI] [PubMed] [Google Scholar]
- Mur LA, Allainguillaume J, Catalan P, Hasterok R, Jenkins G, Lesniewska K, et al. Exploiting the Brachypodium Tool Box in cereal and grass research. New Phytol. 2011;191:334–347. doi: 10.1111/j.1469-8137.2011.03748.x. [DOI] [PubMed] [Google Scholar]
- Mutwil M, Klie S, Tohge T, Giorgi FM, Wilkins O, Campbell MM, et al. PlaNet: combined sequence and expression comparisons across plant networks derived from seven species. Plant Cell. 2011;23:895–910. doi: 10.1105/tpc.111.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano AJ, Sato Y, Mihara M, Antonio BA, Motoyama R, Itoh H, et al. Deciphering and prediction of transcriptome dynamics under fluctuating field conditions. Cell. 2012;151:1358–1369. doi: 10.1016/j.cell.2012.10.048. [DOI] [PubMed] [Google Scholar]
- Naika M, Shameer K, Mathew OK, Gowda R, Sowdhamini R. STIFDB2: an updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in Arabidopsis and rice. Plant Cell Physiol. 2013;54:e8. doi: 10.1093/pcp/pcs185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T. Circadian clock- and PIF4-controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 2012;53:1950–1964. doi: 10.1093/pcp/pcs137. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol. 2011;52:213–219. doi: 10.1093/pcp/pcq203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, Shiina T, et al. Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol. Genet. Genomics. 2002;266:740–746. doi: 10.1007/s00438-001-0606-9. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Mochida K, Nemoto Y, Murai K, Yamazaki Y, Shin IT, et al. Correlated clustering and virtual display of gene expression patterns in the wheat life cycle by large-scale statistical analyses of expressed sequence tags. Plant J. 2003;33:1001–1011. doi: 10.1046/j.1365-313x.2003.01687.x. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyanagi H, Tanaka T, Sakai H, Shigemoto Y, Yamaguchi K, Habara T, et al. The Rice Annotation Project Database (RAP-DB): hub for Oryza sativa ssp. japonica genome information. Nucleic Acids Res. 2006;34:D741–D744. doi: 10.1093/nar/gkj094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Kamide Y, Hirai MY, Saito K. Plant lipidomics based on hydrophilic interaction chromatography coupled to ion trap time-of-flight mass spectrometry. Metabolomics. 2013;9:121–131. doi: 10.1007/s11306-011-0318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y, Saito K. Recent advances of metabolomics in plant biotechnology. Plant Biotechnol. Rep. 2012;6:1–15. doi: 10.1007/s11816-011-0191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka M, Matsuda T, Sakazono S, Masuko-Suzuki H, Maeda S, Sewaki M, et al. Cell type-specific transcriptome of Brassicaceae stigmatic papilla cells from a combination of laser microdissection and RNA sequencing. Plant Cell Physiol. 2013;54:1894–1906. doi: 10.1093/pcp/pct133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, Kilian B, et al. Genome-wide association studies for agronomical traits in a world wide spring barley collection. BMC Plant Biol. 2012;12:16. doi: 10.1186/1471-2229-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paux E, Sourdille P, Salse J, Saintenac C, Choulet F, Leroy P, et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science. 2008;322:101–104. doi: 10.1126/science.1161847. [DOI] [PubMed] [Google Scholar]
- Peraldi A, Beccari G, Steed A, Nicholson P. Brachypodium distachyon: a new pathosystem to study Fusarium head blight and other Fusarium diseases of wheat. BMC Plant Biol. 2011;11:100. doi: 10.1186/1471-2229-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Martis M, Asp T, Mayer KF, Lubberstedt T, Byrne S, et al. The perennial ryegrass GenomeZipper: targeted use of genome resources for comparative grass genomics. Plant Physiol. 2013;161:571–582. doi: 10.1104/pp.112.207282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe R, Paux E, Bertin I, Sourdille P, Choulet F, Laugier C, et al. A high density physical map of chromosome 1BL supports evolutionary studies, map-based cloning and sequencing in wheat. Genome Biol. 2013;14:R64. doi: 10.1186/gb-2013-14-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plocik AM, Graveley BR. New insights from existing sequence data: generating breakthroughs without a pipette. Mol. Cell. 2013;49:605–617. doi: 10.1016/j.molcel.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. 2012;7:e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikova OA, Shao J, Nemchinov LG. Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol. 2013;54:1041–1055. doi: 10.1093/pcp/pct056. [DOI] [PubMed] [Google Scholar]
- Putri SP, Nakayama Y, Matsuda F, Uchikata T, Kobayashi S, Matsubara A, et al. Current metabolomics: practical applications. J. Biosci. Bioeng. 2013;115:579–589. doi: 10.1016/j.jbiosc.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- Ramilowski JA, Sawai S, Seki H, Mochida K, Yoshida T, Sakurai T, et al. Glycyrrhiza uralensis transcriptome landscape and study of phytochemicals. Plant Cell Physiol. 2013;54:697–710. doi: 10.1093/pcp/pct057. [DOI] [PubMed] [Google Scholar]
- Rancour DM, Marita JM, Hatfield RD. Cell wall composition throughout development for the model grass Brachypodium distachyon. Front. Plant Sci. 2012;3:266. doi: 10.3389/fpls.2012.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CL, Rosas U, Banta J, Bhambhra N, Purugganan MD. Genome-wide patterns of Arabidopsis gene expression in nature. PLoS Genet. 2012;8:e1002662. doi: 10.1371/journal.pgen.1002662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl Acad. Sci. USA. 2012;109:8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustenholz C, Choulet F, Laugier C, Safar J, Simkova H, Dolezel J, et al. A 3,000-loci transcription map of chromosome 3B unravels the structural and functional features of gene islands in hexaploid wheat. Plant Physiol. 2011;157:1596–1608. doi: 10.1104/pp.111.183921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safar J, Bartos J, Janda J, Bellec A, Kubalakova M, Valarik M, et al. Dissecting large and complex genomes: flow sorting and BAC cloning of individual chromosomes from bread wheat. Plant J. 2004;39:960–968. doi: 10.1111/j.1365-313X.2004.02179.x. [DOI] [PubMed] [Google Scholar]
- Saisho D, Ishii M, Hori K, Sato K. Natural variation of barley vernalization requirements: implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell Physiol. 2011;52:775–784. doi: 10.1093/pcp/pcr046. [DOI] [PubMed] [Google Scholar]
- Saisho D, Takeda K. Barley: emergence as a new research material of crop science. Plant Cell Physiol. 2011;52:724–727. doi: 10.1093/pcp/pcr049. [DOI] [PubMed] [Google Scholar]
- Saito T, Ariizumi T, Okabe Y, Asamizu E, Hiwasa-Tanase K, Fukuda N, et al. TOMATOMA: a novel tomato mutant database distributing Micro-Tom mutant collections. Plant Cell Physiol. 2011;52:283–296. doi: 10.1093/pcp/pcr004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, et al. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:e6. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, et al. RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011;52:265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Yamada Y, Sawada Y, Matsuda F, Akiyama K, Shinozaki K, et al. PRIMe Update: innovative content for plant metabolomics and integration of gene expression and metabolite accumulation. Plant Cell Physiol. 2013;54:e5. doi: 10.1093/pcp/pcs184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki E, Takahashi C, Asami T, Shimada Y. AtCAST, a tool for exploring gene expression similarities among DNA microarray experiments using networks. Plant Cell Physiol. 2011;52:169–180. doi: 10.1093/pcp/pcq185. [DOI] [PubMed] [Google Scholar]
- Sato K, Close TJ, Bhat P, Munoz-Amatriain M, Muehlbauer GJ. Single nucleotide polymorphism mapping and alignment of recombinant chromosome substitution lines in barley. Plant Cell Physiol. 2011;52:728–737. doi: 10.1093/pcp/pcr024. [DOI] [PubMed] [Google Scholar]
- Sato K, Nankaku N, Takeda K. A high-density transcript linkage map of barley derived from a single population. Heredity. 2009a;103:110–117. doi: 10.1038/hdy.2009.57. [DOI] [PubMed] [Google Scholar]
- Sato K, Shin IT, Seki M, Shinozaki K, Yoshida H, Takeda K, et al. Development of 5006 full-length cDNAs in barley: a tool for accessing cereal genomics resources. DNA Res. 2009b;16:81–89. doi: 10.1093/dnares/dsn034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Takeda K. An application of high-throughput SNP genotyping for barley genome mapping and characterization of recombinant chromosome substitution lines. Theor. Appl. Genet. 2009;119:613–619. doi: 10.1007/s00122-009-1071-9. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Urich MA, Nery JR, Pelizzola M, Libiger O, et al. Patterns of population epigenomic diversity. Nature. 2013;495:193–198. doi: 10.1038/nature11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, et al. The International Barley Sequencing Consortium—at the threshold of efficient access to the barley genome. Plant Physiol. 2009;149:142–147. doi: 10.1104/pp.108.128967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, et al. Rapid and efficient gene modification in rice and brachypodium using TALENs. Mol. Plant. 2013;6:1365–1368. doi: 10.1093/mp/sss162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Flavell RB. Characterization of wheat genome by renaturation kinetics. Chromosoma. 1975;50:223–242. [Google Scholar]
- Spannagl M, Martis MM, Pfeifer M, Nussbaumer T, Mayer KF. Analysing complex Triticeae genomes—concepts and strategies. Plant Methods. 2013;9:35. doi: 10.1186/1746-4811-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasulu N, Graner A, Wobus U. Barley genomics: an overview. Int. J. Plant Genomics. 2008;2008:486258. doi: 10.1155/2008/486258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, et al. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theor. Appl. Genet. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. [DOI] [PubMed] [Google Scholar]
- Su CL, Chao YT, Alex Chang YC, Chen WC, Chen CY, Lee AY, et al. De novo assembly of expressed transcripts and global analysis of the Phalaenopsis aphrodite transcriptome. Plant Cell Physiol. 2011;52:1501–1514. doi: 10.1093/pcp/pcr097. [DOI] [PubMed] [Google Scholar]
- Swarbreck SM, Lindquist EA, Ackerly DD, Andersen GL. Analysis of leaf and root transcriptomes of soil-grown Avena barbata plants. Plant Cell Physiol. 2011;52:317–332. doi: 10.1093/pcp/pcq188. [DOI] [PubMed] [Google Scholar]
- Szczesniak MW, Kabza M, Pokrzywa R, Gudys A, Makalowska I. ERISdb: a database of plant splice sites and splicing signals. Plant Cell Physiol. 2013;54:e10. doi: 10.1093/pcp/pct001. [DOI] [PubMed] [Google Scholar]
- Takehisa H, Sato Y, Igarashi M, Abiko T, Antonio BA, Kamatsuki K, et al. Genome-wide transcriptome dissection of the rice root system: implications for developmental and physiological functions. Plant J. 2012;69:126–140. doi: 10.1111/j.1365-313X.2011.04777.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel J, Riewe D, Rutten T, Melzer M, Friedel S, Bollenbeck F, et al. Differentiation of endosperm transfer cells of barley: a comprehensive analysis at the micro-scale. Plant J. 2012;71:639–655. doi: 10.1111/j.1365-313X.2012.05018.x. [DOI] [PubMed] [Google Scholar]
- Thole V, Worland B, Wright J, Bevan MW, Vain P. Distribution and characterization of more than 1000 T-DNA tags in the genome of Brachypodium distachyon community standard line Bd21. Plant Biotechnol. J. 2010;8:734–747. doi: 10.1111/j.1467-7652.2010.00518.x. [DOI] [PubMed] [Google Scholar]
- Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat. Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- Torada A, Koike M, Mochida K, Ogihara Y. SSR-based linkage map with new markers using an intraspecific population of common wheat. Theor. Appl. Genet. 2006;112:1042–1051. doi: 10.1007/s00122-006-0206-5. [DOI] [PubMed] [Google Scholar]
- Tsai WC, Fu CH, Hsiao YY, Huang YM, Chen LJ, Wang M, et al. OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 2013;54:e7. doi: 10.1093/pcp/pcs187. [DOI] [PubMed] [Google Scholar]
- Uchida N, Sakamoto T, Kurata T, Tasaka M. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 2011;52:716–722. doi: 10.1093/pcp/pcr029. [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A, Fabris M, Pollier J, Baart GJ, Rombauts S, Hasnain G, et al. CathaCyc, a metabolic pathway database built from Catharanthus roseus RNA-Seq data. Plant Cell Physiol. 2013;54:673–685. doi: 10.1093/pcp/pct039. [DOI] [PubMed] [Google Scholar]
- Verelst W, Bertolini E, De Bodt S, Vandepoele K, Demeulenaere M, Pe ME, et al. Molecular and physiological analysis of growth-limiting drought stress in Brachypodium distachyon leaves. Mol. Plant. 2013;6:311–322. doi: 10.1093/mp/sss098. [DOI] [PubMed] [Google Scholar]
- Wang TL, Uauy C, Robson F, Till B. TILLING in extremis. Plant Biotechnol. J. 2012;10:761–772. doi: 10.1111/j.1467-7652.2012.00708.x. [DOI] [PubMed] [Google Scholar]
- Watanabe CK, Hachiya T, Takahara K, Kawai-Yamada M, Uchimiya H, Uesono Y, et al. Effects of AOX1a deficiency on plant growth, gene expression of respiratory components and metabolic profile under low-nitrogen stress in Arabidopsis thaliana. Plant Cell Physiol. 2010;51:810–822. doi: 10.1093/pcp/pcq033. [DOI] [PubMed] [Google Scholar]
- Wheeler T, von Braun J. Climate change impacts on global food security. Science. 2013;341:508–513. doi: 10.1126/science.1239402. [DOI] [PubMed] [Google Scholar]
- Wicker T, Mayer KF, Gundlach H, Martis M, Steuernagel B, Scholz U, et al. Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell. 2011;23:1706–1718. doi: 10.1105/tpc.111.086629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Narechania A, Sabot F, Stein J, Vu GT, Graner A, et al. Low-pass shotgun sequencing of the barley genome facilitates rapid identification of genes, conserved non-coding sequences and novel repeats. BMC Genomics. 2008;9:518. doi: 10.1186/1471-2164-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Schlagenhauf E, Graner A, Close TJ, Keller B, Stein N. 454 sequencing put to the test using the complex genome of barley. BMC Genomics. 2006;7:275. doi: 10.1186/1471-2164-7-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, et al. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 2009;59:712–722. doi: 10.1111/j.1365-313X.2009.03911.x. [DOI] [PubMed] [Google Scholar]
- Widodo, Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U. Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J. Exp. Bot. 2009;60:4089–4103. doi: 10.1093/jxb/erp243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Cai S, Chen M, Ye L, Chen Z, Zhang H, et al. Tissue metabolic responses to salt stress in wild and cultivated barley. PLoS One. 2013a;8:e55431. doi: 10.1371/journal.pone.0055431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Shen Q, Cai S, Chen ZH, Dai F, Zhang G. Ionomic responses and correlations between elements and metabolites under salt stress in wild and cultivated barley. Plant Cell Physiol. 2013b;54:1976–1988. doi: 10.1093/pcp/pct134. [DOI] [PubMed] [Google Scholar]
- Yamakawa H, Hakata M. Atlas of rice grain filling-related metabolism under high temperature: joint analysis of metabolome and transcriptome demonstrated inhibition of starch accumulation and induction of amino acid accumulation. Plant Cell Physiol. 2010;51:795–809. doi: 10.1093/pcp/pcq034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Mochida K, Asano T, Nakabayashi R, Chiba M, Udomson N, et al. Coupling deep transcriptome analysis with untargeted metabolic profiling in Ophiorrhiza pumila to further the understanding of the biosynthesis of the anti-cancer alkaloid camptothecin and anthraquinones. Plant Cell Physiol. 2013;54:686–696. doi: 10.1093/pcp/pct040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R, Nishitani K. Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol. 2004;45:1111–1121. doi: 10.1093/pcp/pch151. [DOI] [PubMed] [Google Scholar]
- Yordem BK, Conte SS, Ma JF, Yokosho K, Vasques KA, Gopalsamy SN, et al. Brachypodium distachyon as a new model system for understanding iron homeostasis in grasses: phylogenetic and expression analysis of Yellow Stripe-Like (YSL) transporters. Ann. Bot. 2011;108:821–833. doi: 10.1093/aob/mcr200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Mori T, Yokoyama K, Koike Y, Tanabe N, Sato N, et al. Identification of alternative splicing events regulated by an Arabidopsis serine/arginine-like protein, atSR45a, in response to high-light stress using a tiling array. Plant Cell Physiol. 2011;52:1786–1805. doi: 10.1093/pcp/pcr115. [DOI] [PubMed] [Google Scholar]
- Yura K, Sulaiman S, Hatta Y, Shionyu M, Go M. RESOPS: a database for analyzing the correspondence of RNA editing sites to protein three-dimensional structures. Plant Cell Physiol. 2009;50:1865–1873. doi: 10.1093/pcp/pcp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, et al. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39:D1114–D1117. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, et al. Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J. 2004;40:276–290. doi: 10.1111/j.1365-313X.2004.02209.x. [DOI] [PubMed] [Google Scholar]
- Zheng L, Fujii M, Yamaji N, Sasaki A, Yamane M, Sakurai I, et al. Isolation and characterization of a barley yellow stripe-like gene, HvYSL5. Plant Cell Physiol. 2011;52:765–774. doi: 10.1093/pcp/pcr009. [DOI] [PubMed] [Google Scholar]