Abstract
Global spread and genetic monomorphism are hallmarks of Mycobacterium tuberculosis, the agent of human tuberculosis. In contrast, Mycobacterium canettii, and related tubercle bacilli that also cause human tuberculosis and exhibit unusual smooth colony morphology, are restricted to East-Africa. Here, we sequenced and analyzed the genomes of five representative strains of smooth tubercle bacilli (STB) using Sanger (4-5x coverage), 454/Roche (13-18x coverage) and/or Illumina DNA sequencing (45-105x coverage). We show that STB are highly recombinogenic and evolutionary early-branching, with larger genome sizes, 25-fold more SNPs, fewer molecular scars and distinct CRISPR-Cas systems relative to M. tuberculosis. Despite the differences, all tuberculosis-causing mycobacteria share a highly conserved core genome. Mouse-infection experiments revealed that STB are less persistent and virulent than M. tuberculosis. We conclude that M. tuberculosis emerged from an ancestral, STB-like pool of mycobacteria by gain of persistence and virulence mechanisms and we provide genome-wide insights into the molecular events involved.
Mycobacterium tuberculosis is a pervasive human pathogen, currently estimated to infect two billion people throughout the world1. The bacterial population size resulting from this massive spread is very large, yet the genetic diversity within the classical members of the M. tuberculosis complex (MTBC), comprising Mycobacterium africanum, Mycobacterium bovis, Mycobacterium microti, Mycobacterium pinnipedii, and M. tuberculosis is very limited. Tuberculosis is therefore assumed to be a recent human disease2,3 linked to clonal expansion of its causative organism4-6.
In contrast to MTBC, smooth tubercle bacilli (STB), defined as clinical isolates displaying a distinctive smooth colony phenotype on culture media, named Mycobacterium canettii and/or Mycobacterium prototuberculosis7-10, are less genetically restricted. Initial genotyping analysis suggested that these isolates possess a higher diversity with traces of intraspecies horizontal gene transfer (HGT) and might therefore represent early-branching lineages of tuberculosis-causing mycobacteria. Since their first isolation by Georges Canetti in 1969, less than one hundred strains of STB have been identified. All STB have been obtained from human tuberculosis patients, mostly from (or with connection to) East-Africa8,11. Thus, a collection of a few tens of STB strains from a geographically restricted region appears to contain greater genetic diversity than the worldwide population of MTBC strains. This observation raises intriguing questions about the origin of tuberculosis and provided an opportunity to examine the molecular and evolutionary events involved in the emergence of M. tuberculosis. Herein, we describe and compare complete genome sequences of five diverse STB isolates and the physiopathological properties of these mycobacteria relative to M. tuberculosis as well as whole genome shotgun (WGS) sequences of four additional STB strains for secondary screening and confirmation purposes.
Ancestral features of STB genomes
We applied multilocus sequence typing (MLST) based on 12 house-keeping genes to a panel of 55 available STB isolates and identified a total of 13 sequence types among them (Fig. 1 and Supplementary Tables 1 and 2). From analyses of the concatenated sequences we inferred a highly reticulated phylogeny, suggestive of conflicting phylogenetic signals and possible HGT among the target genes. We then selected five representative isolates for the principal comprehensive genomic analysis. This selection included the original strain isolated by George Canetti, of sequence type A and an isolate from the most prevalent group of sequence type D (both belonging to the M. canettii cluster), as well as strains from the most distant sequence types L, J and K (Fig. 1 and Supplementary Fig. 1)9.
Figure 1. Selection and genome features of analyzed strains.
(a) multilocus sequence typing of 56 STB and 10 MTBC reference isolates. Phylogenetic positions based on split decomposition analysis of concatenated sequences of 12 house-keeping gene segments are represented. The scale bar represents Hamming distance. Numbers indicate the percent of bootstrap support of the splits obtained after 1,000 replicates. Arrows and stars indicate isolates selected for complete genome sequence and genome shotgun analyses, respectively. (b) pairwise, linear genomic comparisons of M. tuberculosis H37Rv, M. bovis AF2122/97, five selected STB strains, and two non-tuberculous mycobacterial species, M. marinum M and M. smegmatis mc2155. Red and blue lines indicate co-linear blocks of DNA:DNA similarity, and inverted matches, respectively. M. tub., M. tuberculosis; M. mar, M. marinum; M. smeg., M. smegmatis. (c) numbers of SNPs in pairwise comparisons between the indicated genomes. (d) Network phylogeny inferred among the five STB isolates subjected to complete genome sequence analysis and MTBC by NeighborNet analysis, based on pairwise alignments of whole genome SNP data. ‘*’ indicates 90% bootstrap support, while all other nodes had 100% support, 1000 iterations. (e) Histogram showing the respective numbers of SNPs between the aligned M. tuberculosis H37Rv and M. bovis or STB genomes (depicted in panel b).
Comparison of these five STB genomes with those of M. tuberculosis H37Rv12 and other MTBC members13 revealed a very similar overall organization between STB and MTBC, with a high percentage of syntenic genes (from 96% for strain A and 93% for strain K, compared to only 77% between M. tuberculosis H37Rv and Mycobacterium marinum, one of the phylogenetically closest non-tuberculous mycobacterial species14). No major chromosomal rearrangements or plasmids were detected (Fig. 1). Pairwise analyses between the conserved STB and MTBC genome sequences showed that all combinations had average nucleotide identities of at least 97.3%, above the 95% threshold proposed for classification into the same species15. However, the genomes of STB are 10-115 kb larger than those of the MTBC members and thus represent the largest genomes known for tubercle bacilli, although they are still much smaller than those of M. marinum (6.6 Mb)16 and the other most closely related, non-tuberculous species M. kansasii (6.4 Mb)17. Excluding repetitive sequences such as PE_PGRS- and PPE_MPTR-encoding regions, which account for ~ 8% of the coding capacity of M. tuberculosis12, STB and MTBC share > 89.3% of their genomes, representing a core genome for tubercle bacilli of > 3.938 Mb. This core comprises 96.3% of the 774 M. tuberculosis H37Rv genes predicted as being essential for in vitro growth, and all 194 genes required for mycobacterial survival during mouse infection18-20, further reflecting the close affiliations of STB and M. tuberculosis. The accessory genomes of individual STB strains harbor from 124 (strain A) to 366 genes (strains K and J) not present in MTBC members that enlarge the known pan-genome of tubercle bacilli by 890 predicted coding sequences (CDS), representing a supplement of more than 20% relative to the gene pool of M. tuberculosis (Supplementary Table 3 and Supplementary Figs 2a, b). Interestingly, only nine of these CDS were common to all five STB genomes analyzed (Supplementary Table 3 and Supplementary Fig. 2c). Conversely, 51 genes partially overlapping with genomic islands21 present in MTBC were not found in any of the STB strains (Supplementary Table 4). These genes encode derivatives of mobile elements, such as the phiRv1 and phiRv2 prophage-like regions (24 CDS), 3 transposases, 5 unique members of a glycine-rich protein family (e.g. PE_PGRS33; Supplementary Fig. 3a) and 19 other hypothetical proteins (Supplementary Fig. 3b). It is noteworthy that Rv1989c-Rv1990c from one such MTBC specific region showed around 90% identity with proteins encoded on a plasmid from Mycobacterium gilvum and Mycobacterium sp. KMS, raising intriguing questions about possible transmission routes of the corresponding genes into the MTBC. Several other MTBC-specific hypothetical proteins had no or only weak amino-acid similarity with other mycobacterial proteins (Supplementary Table 4), suggesting HGT into MTBC from distant donors after the separation from the STB lineages.
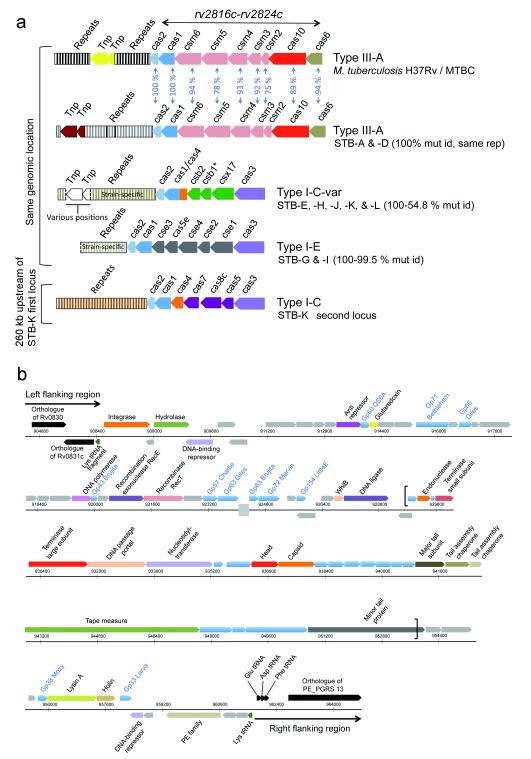
We also identified prominent, HGT-related differences in clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins (CRISPR-Cas) systems between STB and MTBC. These systems may confer adaptive immunity against phages and plasmids in bacteria and archea via repeat/spacer-derived short RNAs22. The genomes of STB strains A and D contain a single CRISPR-Cas locus encoding a system of major type III-A that is similar to that of MTBC genomes, but with a few crispr spacers in common7,10 and substantially lower sequence similarities of their Cas proteins (down to 75%) than those of the core proteins (98%-100 %) (Fig. 2.). The same genomic region in the more distant K, L and J strains is occupied by a completely different CRISPR-Cas system of a rare type-Ic variant (Fig. 2), most closely related to those of environmental actinobacteria such as Gordonia amarae or purple sulfur bacteria Thioalkalivibrio sp.. Furthermore, in strain K, the presence of a second CRISPR-Cas module of a different type Ic was identified 260 kb upstream of the other locus (Fig. 2), whose Cas proteins were most similar to those of Moorella sp. or Thiorhodovibrio sp. Finally, screening of WGS-derived sequences from STB strains E, G, H, and I, located at well-distributed intermediate positions of the STB MLST-based network (Fig. 1), revealed the existence of yet another type I-E module in strains G and I that was most closely related to those of environmental actinobacteria such as Saccharomonospora sp., while in the two remaining E and H strains, a type Ic variant similar to those of STB-J, -K and -L was found (Fig. 2a). As CRISPR-Cas systems have not been identified in non-tuberculous mycobacterial species, these different systems were most likely acquired by independent HGT events that occurred after the divergence of STB and MTBC. While it is not known whether the CRISPR systems in tubercle bacilli are functional, their disparate origins suggest that the distinct, respective crispr spacer sets might not necessarily reflect genetic records of recent encounters of tubercle bacilli with distinct phage transgressors but, instead, older traces of interaction of the respective CRISPR-Cas donor organisms with non-mycobacterial phages. The identification of a 55 kb prophage region in the WGS-derived sequence of STB-I that is large enough to encode a potentially complete virion23 (Fig. 2), which to our knowledge represents the first such finding in tubercle bacilli, provides a promising future model for testing the functionality of mycobacterial CRISPR-Cas systems on adaptive immunity against phages.
Figure 2. CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated proteins) systems and prophages in STB and MTBC genomes.
(a) Gene content of different CRISPR-Cas systems in MTBC and STB strains. Spacers are color-coded according to sequence similarities. Percentages of protein sequence identities are indicated between type III-A systems of M. tuberculosis H37Rv and STB A and D. The various combinations of identities between ubiquitous proteins (e.g. Cas2) of different CRISPR-Cas types are much lower (below 40%) and are not indicated. A star indicates a potential csb1 pseudogene in the system of STB-H. A broken line denotes ends of DNA sequence contigs variably delimiting the identified repeat zones of type I-E systems of STB-F, -G and -I. Mut id, mutual protein sequence identities; rep, repeats; Tnp, transposon; cas, csm, csb, csx, cse, various Cas gene families. (b) Schematic representation of a 55 kb spanning genomic region that encodes a putative prophage in STB strain I. STB-I genomic positions are marked on horizontal scales in bp. Brackets indicate a portion homologous to a prophage region in the M. marinum genome. Predicted coding sequences are shown above or below scales, corresponding to rightward and leftward transcription, respectively. Color-coding define features of predicted encoded products as follows. Gray, phage protein without database match or homologous to non-mycobacteriophage proteins of unknown function; blue, phage protein homologous to other mycobacteriophage proteins of unknown function (names of homologs are written in blue text, except for the portion homologous to the M. marinum prophage region); black, STB-I coding sequences and tRNA genes (conserved in other STB strains and M. tuberculosis H37Rv) flanking the phage insertion site corresponding to the Lys tRNA gene; all other colors, phage proteins with a predicted function (indicated in black text). A gray box on the second horizontal scale indicates a sequence contig break. Functional annotations of the predicted genes were made based on comparisons of the encoded products via the Genbank database, detection of protein domain signatures, and expert annotation of 374 other mycobacteriophage genomes retrieved from the PhagesDB database.
Progressive genome downsizing is a hallmark of mycobacterial pathogen evolution17,24. Therefore, the larger genome sizes of STB compared with MTBC argue for their ancestral status. Further evidence for ancestrality of STB genome structures comes from inspection of interrupted coding sequence (ICDS) orthologs, thought to reflect molecular scars inherited during pseudogenization of the MTBC genomes25,26. Among the 81 reported ICDS in MTBC, most were found to be also interrupted both in STB and more distantly related mycobacteria, suggesting evolutionary ancient mycobacterial scars (Supplementary Table 5). However, we identified four ICDS, e.g. pks8 belonging to the pks multi-gene family encoding polyketide synthases that are involved in the biosynthesis of important cell envelope lipids16,27, which were intact in the genomes of STB (in one case - rv3741/42, the region was absent from STB-J) and from the M. marinum and/or M. kansasii outgroup genomes (Supplementary Fig. 4). Thus, these scars occurred in the most recent common ancestor of the MTBC after divergence from STB-like progenitors. The opposite situation, i.e. ICDS shared by the STB genomes corresponding to intact CDS in MTBC, was never observed, further supporting the ancestral status of the STB genome structure. In addition, we detected four independent loci (narX, pks5, pknH, lppV), where a likely ancestral gene organization present both in the mycobacterial outgroups M. marinum and/or M. kansasii and in STB, was rearranged to result in a single hybrid gene and loss of intervening gene(s) in MTBC genomes (Supplementary Fig. 5), similar to what has been observed for pknH in M. africanum28.
Ancient branching of STB lineages is also consistent with the much higher numbers of single nucleotide polymorphisms (SNPs) detected among STB genomes compared to MTBC. Pairwise comparisons of the STB-D, -A, -L, -J, and -K genome sequences with the M. tuberculosis H37Rv reference uncovered 16,168 - 61,228 SNPs (Fig. 1). This amount is within the 9,525-65,744 SNP range observed among the group of STB strains alone, and up to 25-fold higher than the 741-2437 SNPs previously observed among members of the MTBC13,29,30. Consistent with MLST data (here and ref. 9), a NeighborNet analysis based on pairwise comparisons of the genome-wide SNP data showed that MTBC forms a single compact group within a much larger, reticulated network of the STB genotypes (Fig. 1). Consistently, this reticulation was even increased when WGS-derived sequence data from four additional STB strains were included (Supplementary Fig. 6), further confirming the MLST-derived phylogeny at the genome level. The Phi test for recombination was highly significant (p=10−6). Importantly, the relative compactness of the MTBC branch is additionally confirmed by the structure of the phylogenetic tree, obtained after exclusion of the genome portions affected by recombination/HGT (see further and Fig. 3a). These results thus firmly demonstrate that the worldwide MTBC population only represents a genetically homogeneous subset branching from the larger diversity of recombinogenic STB isolates. Taken together with independent lines of evidence pointing to an earlier branching, they suggest that STB lineages diverged from the common ancestor of all tubercle bacilli well before the successful clonal radiation of MTBC began.
Figure 3. Inter-strain recombination segments between STB and MTBC genomes.
(a) phylogenetic tree inferred by using Neighbor-Joining algorithm on nucleotide p-distances, after concatenation of sequence alignments of 2,047 genes of the predicted clonal portion of the STB-MTBC core genome (i.e. after exclusion of the genes affected by recombination- see text- and of gapped regions). (b) SNP distribution among STB and MTBC aligned genome segments, showing probable recombination regions involving genes rv1936-rv1937 between STB-J and M. tuberculosis. Each of the three panels shows a comparison of two STB or M. bovis strains (top, bottom) relative to M. tuberculosis H37Rv (middle). Red lines indicate individual SNPs identified between pairwise compared genomes. Thicker or uneven red lines result from multiple SNPs in close proximity or shifts due to small insertions/deletions. Note the SNP-free, identical genome segments between STB-J and H37Rv (boxed) conflict with their distant respective positions on the clonal core genome-based tree. M. tub. H37Rv, M. tuberculosis H37Rv.
Impact of selection and recombination
In order to compare the impact of selection on the evolution of the STB and MTBC genomes, we calculated global ratios of non-synonymous vs synonymous SNPs (dN/dS). The genome-wide dN/dS ratio is unusually high in MTBC, which has been suggested to reflect relaxed purifying selection against non-synonymous changes that are in general slightly deleterious31. The dN/dS ratios in the different gene categories among the STB strains were only about a third of those found in the MTBC (Table 1), and are thus compatible with a much longer time of exposure of STB to purifying selection, given the time dependence of dN/dS for closely related bacteria32-34 and assuming that purifying selection pressures were the same for STB as for MTBC.
Table 1.
Ratios of non-synonymous versus synonymous SNPs in gene categories
|
|
|||||
|---|---|---|---|---|---|
| dN/dS in gene category | |||||
|
|
|||||
| Strain | All | essential | Nonessential | T-cell antigens | T-cell epitopes |
| STB-A | 0.19/0.15 | 0.14/0.11 | 0.21/0.17 | 0.14/0.12 | 0.18/0.14 |
| STB-J | 0.18/ 0.13 | 0.14/0.11 | 0.19/0.15 | 0.14/0.11 | 0.13/0.09 |
| STB-D | 0.20/0.16 | 0.16/0.12 | 0.22/0.17 | 0.15/0.12 | 0.10/0.08 |
| STB-L | 0.19/ 0.15 | 0.16/0.12 | 0.21/0.17 | 0.14/0.12 | 0.15/0.11 |
| STB-K | 0.17/0.13 | 0.14/ 0.10 | 0.19/0.15 | 0.15/0.12 | 0.13/0.09 |
|
| |||||
| MTBCa | ND | 0.53 | 0.66 | 0.50 | 0.53-0.25b |
ND, not done. dN/dS ratios were calculated on orthologs conserved in the 5 STB strains subjected to complete genome sequence analysis and M. tuberculosis H37Rv, based on pairwise, concatenated codon alignments and using SNAP (value on the left)35 and PAML maximum likelihood methods (value on the right)36. M. tuberculosis H37Rv T-cell antigen, essential and non-essential gene categories, as well as T cell epitope codon concatenates were constructed as in Comas et al.30.
dN/dS ratios calculated by Comas et al.30 from SNPs identified across 21 MTBC strains.
Lower value obtained after exclusion of epitopes of three antigens considered as outliers.
As an important exception, protective human CD4+ and CD8+ T-cell antigens and epitopes of M. tuberculosis have been described to be under purifying selection, suggesting that MTBC members do not use T-cell antigen variation to escape human immune responses but, instead, might benefit from recognition by T-cells30. Similarly, we found that the dN/dS ratios based on pairwise, concatenated codon alignments35,36 of the 65 T-cell antigen-encoding STB genes conserved across all STB genomes were on average lower than those of the 2,300 genes classified as non essential and similar to slightly lower than the 710 essential genes18 conserved among all STB (Table 1). Overall similar results were also obtained when only the epitope regions of the T-cell antigens were considered. Thus, like the subset of essential proteins, human T-cell antigens tend to be more conserved in STB relative to the rest of the proteome. Following the argument of Comas and colleagues30, this sequence conservation suggests that STB and MTBC might have inherited a common strategy of immune subversion of the human host that predates the clonal emergence of the MTBC. However, there may be alternative explanations, as most of these low dN/dS antigens are also highly conserved in the environmental, facultative pathogens M. marinum and/or M. kansasii and/or other mycobacteria. For example, the 6-kD early secreted antigenic target (ESAT-6, Rv3875) and the 34.6-kD secreted antigen 85B (Ag85B; Rv1886c), that both show 100% amino-acid conservation in MTBC and STB, have orthologues in M. marinum that show 91% (ESAT-6) and 89% (Ag85B) amino-acid identity, which is above the average overall pairwise identity of 85.2%16. The conservation of these proteins might thus also be explained by their role in host-pathogen interaction such as phagosomal rupture37, cell envelope stability38 or other functions that are not necessarily linked to interactions with human T-cells.
Extensive recombination among STB, revealed by our comparative genome analysis, might also have played a role in the discrepancy in dN/dS between the MTBC and STB groups, as it could more efficiently oppose fixation of slightly deleterious mutations than in the more clonal MTBC population39. Consistent with this contention, strong variations in the local distribution of SNPs were observed throughout the aligned STB and MTBC genomes, suggestive of numerous recombination events. Approximately one-third of the core genome alignment consists of zones with significantly lower or higher SNP density compared to expectations for predicted recombination-free nucleotide differences between each pair of genomes. A stringent selection of informative regions among the predicted recombination-free blocks led to a minimal clonal backbone of 1,794,643 characters (~33% of the core genome), which was used to infer a phylogenetic tree (Fig. 3a). Inspection of the genomic regions with unexpected SNP densities allowed us to identify > 110 blocks, of up to 14 kb and including each from 1 to 5 complete genes (Supplementary Table 6), with homoplasic SNP distributions (relative to the tree), indicative of likely inter-strain recombination events among STB and/or between STB and MTBC strains (Fig. 3b and Supplementary Fig. 7a, b). The extensive impact of recombination was independently confirmed by the finding that ~8% to ~15% of the protein coding sequence alignments from the core genome has mosaic structures indicative of inter-strain intragenic recombination events. In contrast, the influence of exogenous importation from more distant mycobacterial species on the core genome sequence diversity is apparently minimal, as inferred by the detection of only few regions with unexpectedly high SNP densities in STB strains, yielding BLAST best hits closer to non-tuberculous mycobacteria than to STB and MTBC (Supplementary Fig. 7c).
Remarkably, the gene blocks in M. tuberculosis whose sequences perfectly match those of one or more STB strains, showed SNPs in the orthologous region in M. bovis and/or other MTBC strains (Fig. 3b), suggesting that gene fluxes between M. tuberculosis and the STB strain pool existed even well after the divergence of the MTBC, and perhaps still exist. We also found intermediate situations, where the SNP distribution clearly suggests recombination events that were more ancient and likely followed by accumulation of a few mutations in the recipient or the donor strains (Supplementary Fig. 7a). These data provide new, solid evidence to the question of inter-strain gene flux in M. tuberculosis40,41. Our findings also raise puzzling questions on the (micro-) environments and mechanisms favoring or having favored such extensive DNA exchanges. The high number of apparently recent recombination episodes, as suggested by numerous perfect large sequence matches detected among sequences from different STB lineages together with the almost exclusive isolation of STB strains from patients around the Horn of Africa strongly suggests a common local source. Aquatic environments rich in mycobacteria, potentially residing in protozoan hosts24,42, are one possible opportunity for genetic exchange to occur, as suggested by a recent report on detection of MTBC DNA in rural water sources in Ethiopia (E. Wellington, personal communication, Abstract, 16th International Symposium on the Biology of Actinomycetes). The presence of a 55 kb genomic segment corresponding to a putative complete phage-encoding region inserted into the Lys tRNA gene of strain STB-I (Fig. 2) suggests a possible mediation by phages, although alternative mechanisms such as DNA transfer by conjugation, reported for Mycobacterium smegmatis under biofilm conditions43, could also be involved.
STB persist less during infection than M. tuberculosis
To determine whether the genome differences between the STB and MTBC strains impact on host-pathogen interactions, we first measured their growth in in vitro cultures. Most STB grew 2 to 3 times faster than M. tuberculosis both in liquid (Supplementary Fig. 8) and on solid media (data not shown) at 30°c and 37°c, in line with previous observations10,11. Upon infection of BALB/c mice (Fig. 4) and C57BL6 mice (Supplementary Fig. 9) by aerosol, the STB strains effectively multiplied in lungs and disseminated to the spleens during the acute infection phase, but consistently persisted less well during the chronic infection phase compared to M. tuberculosis. While the latter was able to persist in the lungs for up to 30 weeks at levels close to those of the acute phase (peaking at 3 weeks with around 107.7 colony-forming units (CFUs)), the infection levels of all STB strains dropped by at least 1 log at all (and by 2 to 3 logs at most) later time points in these organs (p=0.05 by Mann-Whitney test, except for day 130 for strains D, L and K). The strongest difference with M. tuberculosis was observed for strain K, the strain phylogenomically most distant from MTBC and for which bacterial counts were undetectable after 30 weeks in BALB/c mice (Fig. 4c, d). Similar trends were observed in spleens, with strain K also almost completely cleared at day 210. In parallel, histopathological analyses revealed less intense lung lesions and inflammation 128 days after infection with the STB strains compared to M. tuberculosis infection, with strain K showing the least damages (Fig. 4 and Supplementary Table 7). Furthermore, C57BL/6 mice intravenously infected with high doses of STB survived in contrast to controls infected with M. tuberculosis strains of different lineages (data not shown), confirming decreased virulence of STB.
Figure 4. Virulence and persistence of smooth tubercle bacilli (STB) and M. tuberculosis.
(a) colony forming units (CFUs) recovered from lungs (a, c) and spleens (b, d) of BALB/c mice after intranasal infection with 103 CFUs. Panels a/b and c/d depict two independent experiments. The results are the median and range of CFUs from four mice. e, f, histopathological sections of lungs of BALB/c infected mice, 128 days post-intranasal infection with 103 CFUs. Blue circles show bronchi, “A” indicates alveoli, and “V” indicates blood-vessels.
Finally, we determined whether these variations could be correlated to differences in innate or adaptive immune responses elicited by infection. The STB and M. tuberculosis strains were similarly able to induce maturation of innate immunity cells in vitro, such as dentritic cells derived from C57BL/6, tlr2°/°, tlr4°/°, or double KO mice (data not shown), suggesting shared major Pathogen-Associated Molecular Patterns (PAMPs)44. Consistently, substantial recruitment of activated innate immune cells, i.e., CD11b+ BST-2+ (Bone Marrow Stromal Cell Antigen-2)+ and CD11c+ MHC-IIhi, was observed in vivo in the lung parenchyma of SCID mice after 3 weeks of infection by STB, but to a lower extent as compared to M. tuberculosis infection (data not shown). Concerning adaptive responses, massive recruitment of activated CD4+ and CD8+ T-cells, displaying CD44 modulation and CD45RB, CD27, CD62L downregulation, was detected in the lungs of C57BL/6 mice after 13 weeks of infection by smooth strains. Again, the responses were overall quantitatively lower for STB compared to M. tuberculosis strains, especially for STB-K (Supplementary Fig. 10), in line with the lower virulence and persistence of STB.
Concluding remarks
With the larger pan genome reflecting the ancestral, wider gene pool of tubercle bacilli, their, lower virulence and faster growth especially at temperatures below 37°C, plausibly reflecting broader environmental adaptability, STB strains might thus come nearer to the as yet unknown missing link between the obligate pathogen M. tuberculosis and environmental mycobacteria. We propose that M. tuberculosis has evolved its so successful widespread, pathogenic lifestyle starting from a pool of STB-like mycobacteria by gaining additional virulence and persistence mechanisms through a potential combination of i) loss of gene function, ii) acquisition of novel genes via HGT, iii) inter-strain recombination of gene clusters and (iv) fixation of SNPs. From the data presented here, a rational experimental design to elucidate which of these genetic events were involved can now be undertaken. Primary candidates are MTBC-specific genes (Supplementary Table 4), including prophage-like phiRv1 / phiRv2 encoding regions reported to be important for late infection45, genes encoding PE_PGRS33 or other MTBC-acquired PE/PPE proteins known to enhance cellular toxicity46, polyketide synthase Pks8/17, the large prophage region in STB-I and/or CRISPR-cas systems. The insights gained through our analysis thus open novel perspectives to identify new targets to combat tuberculosis infection and disease.
Online-Methods
Bacterial strains and multi-locus sequence typing
The 55 STB and 10 reference MTBC isolates are described in Supplementary Table 1. Twelve house-keeping genes were selected for MLST47 (Supplementary Table 2). Phylogenetic groupings were identified by split decomposition analysis48 on the concatenated target sequences .
Genome sequencing
Genomic DNA was extracted from cultured single bacterial colonies12. For genome sequencing of STB-D, -J, -K and -L, Sanger reads from 10-kb fragment shotgun libraries at 4- 4.9 fold coverages were assembled with contigs obtained from Newbler assemblies of 454/Roche reads at 13-18.1 fold coverages, using Arachne49. Scaffolds were validated using Mekano interface (Genoscope). Primer walking, PCRs and in-vitro transposition were used for finishing. The assembled consensus sequences were validated using Illumina reads at 45-105 fold coverages and consed functionalities, and by mapping of termini-sequences from bacterial artificial chromosome libraries50. High quality, contiguous genome sequences of 4420 kb (STB-L, 9 contigs), 4432 kb (STB-D, 12 contigs), 4524 kb (STB-J, 11 contigs), and 4525 kb (STB-K, 9 contigs) were generated. Remaining gaps estimated not to exceed 2 kb correspond to GC-rich and repetitive regions coding for PE_PGRS proteins, and/or the pks5 region (STB-J). For STB-A, a fully finished, contiguous sequence of 4,482,059 bp was obtained by using ~ 80,000 shotgun Sanger reads, Illumina-generated reads and finishing12,28. WGS data from STB strains E, G, H, and I were generated using Illumina HiSseq technology and single lanes. Resulting reads that covered the genomes of these STB up to 900x were assembled using the Velvet software51 and contigs were ordered using M. canettii CIPT 140010059 (STB-A) and M. tuberculosis H37Rv as reference genomes.
Annotation and comparative genomics
Annotation and genome comparisons were performed with the Microscope platform52, Artemis and Artemis comparison tool (ACT)53. When applicable, annotations were transferred from those of M. tuberculosis orthologs in the TubercuList/Mycobrowser database, using BLAST matches of > 90% protein sequence identity, an alignable region of > 80% of the shortest protein length in pairwise comparisons and visual inspection of the gene synteny. Pairwise average nucleotide identities were calculated using JSpecies54. The core/accessory genomes of STB and M. tuberculosis were determined as described16.
SNP and indel analysis
SNiPer pipeline (Genoscope) based on the SSAHA2 package55 was used to map Illumina reads and detect SNPs and indels of STB strains against a corrected version56 of the M. tuberculosis H37Rv reference sequence (NC_000962)12. After exclusion of ambiguous maps on repeat regions, an average of 4.7 million split paired-end reads of 36 bp (STB-A, -D, -L, - J) or trimmed at 50 bp (STB-K) were mapped at a resulting genome coverage > 40x. SNPs with base coverage < 10, base quality < 25, or heterozygosity > 0.2 were removed. ACT53 comparison files were created by using MUMmer and NUCmer softwares57 to visualize the SNP distribution in local genome regions.
Calculation of dN/dS
dN/dS ratios were calculated on orthologs conserved in all STB and M. tuberculosis H37Rv, as identified by bidirectional best hits, alignable region of >80% and sequence identity >=30%. Pairwise, concatenated codon alignments between M. tuberculosis H37Rv and each STB strain were generated using PAL2NAL58, after respective protein alignments obtained with MUSCLE59. Synonymous and non-synonymous substitutions were defined using Nei-Gojobori method-based SNAP35 or maximum likelihood-based PAML36. STB T-cell antigen, essential and non-essential gene categories, as well as T-cell epitope codon concatenates were constructed as described30.
Recombination
The genomes of M. tuberculosis H37Rv, M. bovis AF2122/97 and the five STB strains were aligned using progressive Mauve60. Given a pair ij of aligned genomes, the number of SNPs xij observed between i and j within a region of length l follows a binomial distribution B(l, pij), where pij is the expected proportion of recombination-free nucleotide differences between taxa i and j. Regions containing at least one pair of sequences ij with an unexpectedly large or low number xij of SNPs, i.e. min [Pr (X ≥ xij ), Pr (X ≤ xij )] < 0.05 where X ~ B(l, pij), were identified by using a 200 character-long sliding window along the conserved (core) portions of the multiple genome alignment. The value pij inside each window was estimated as the proportion of SNPs between i and j within the 10000 aligned characters flanking the sliding window on both sides. To obtain a reference phylogeny, all regions of length ≥ 500 characters (excluding gaps) that did not contain an unexpected number of SNPs were concatenated. The derived supermatrix was used to infer a phylogenetic tree by using the Neighbor-Joining algorithm on the pairwise nucleotide p-distances61. All regions of length ≥ 500 characters with a significantly high or low number of SNPs were inspected visually for detection of concentration of homoplasic characters using ACT53, leading to similarities between strains incongruent with the phylogenetic tree. The proportion of protein coding sequences within the core genome likely affected by inter-strain recombination was assessed with the Pairwise Homoplasy Index62, the Maximum x2 test63, and the Neighbour Similarity Score64.
Bacterial growth assays
Growth rates of STB and reference MTBC strains in liquid media were measured by using a BACTEC 460 system (Beckton-Dickinson) as recommended by the manufacturer.
Mouse infection experiments , histopathological and cell analyses
Mice were maintained according to the Institut Pasteur de Lille and Paris guidelines for laboratory animal husbandry. Animal experiments were approved by the Nord-Pas-De-Calais ethical committee (CEEA 15/2009) and the Institut Pasteur Hygiene Committee (authorization number 75-1469), in accordance with European and French guidelines (Directive 86/609/CEE and Decree 87–848). Eight-week-old female BALB/c mice were infected by the intranasal route with 103 CFUs of either STB or M. tuberculosis H37Rv strains, respectively. At indicated times, 4 mice per group were sacrificed, and colony counting was performed from homogenized individual lungs and spleens as described65. For histopathological evaluation, whole lungs were harvested from 3 BALB/c mice per group 128 days post-infection, fixed in 4% formalin, and embedded in paraffin. Four mm-thick sections were stained with hematoxylin-eosin. Virulence and cell-analysis-based immunological assays using C57BL/6 and/or SCID mice were performed as described66,67. Adaptive immune cells from infected mice were prepared, incubated with conjugated mAbs (Beckton-Dickinson), fixed, and analyzed using a CyAn system and Summit (Beckman Coulter) and FlowJo (Treestar) softwares.
Supplementary Material
Acknowledgements
We are grateful to Stewart Cole for help in initiating the M. canettii CIPT 140010059 genome sequencing project and advice, to Thierry Garnier and Adamandia Kapopoulou for help in data management and to Zoe Rouy for help with sequence deposal. The work was supported in part by the Institut Pasteur (PTR 314, PTR 383), the European Community’s FP7 program Grant no. 260872, the Wellcome trust grant 098051, and a Genoscope collaborative grant no. 114.
Footnotes
URLs Magnifying Genome (MaGe) server, https://www.genoscope.cns.fr/agc/microscope/about/collabprojects.php?P_id=44#ancreLogin; MycoBrowser database, http://mycobrowser.epfl.ch/; The Mycobacteriophage database, http://phagesdb.org/.
Accession Codes The complete genome sequence for strain STB-A (CIPT 140010059) was deposited under Accession No. HE572590. Genome sequences of strains STB-D (CIPT 140060008), STB-J (CIPT 140070017), STB-K (CIPT 140070010), and STB-L (CIPT 140070008) were deposited in the EMBL database under accession numbers PRJEB94 - BN44 - FO203507, PRJEB93 - BN43 - FO203508, PRJEB92 - BN42 - FO203509, and PRJEB95 - BN45 - FO203510, respectively. Illumina-derived WGS sequences for strains STB-E (CIPT 140070002), STB-G (CIPT 140070005), STB-H (CIPT 140070013), and STB-I (CIPT 140070007) were deposited in the EMBL WGS repository under project numbers PRJEB584, PRJEB585, PRJEB586, and PRJEB587, respectively.
References
- 1.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–861. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 2.Sreevatsan S, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wirth T, et al. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosch R, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A. 2002;99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Supply P, et al. Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol Microbiol. 2003;47:529–538. doi: 10.1046/j.1365-2958.2003.03315.x. [DOI] [PubMed] [Google Scholar]
- 6.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A. 2004;101:4871–4876. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Soolingen D, et al. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 8.Fabre M, et al. High genetic diversity revealed by variable-number tandem repeat genotyping and analysis of hsp65 gene polymorphism in a large collection of “Mycobacterium canettii” strains indicates that the M. tuberculosis complex is a recently emerged clone of “M. canettii”. J Clin Microbiol. 2004;42:3248–3255. doi: 10.1128/JCM.42.7.3248-3255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutierrez MC, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabre M, et al. Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect Genet Evol. 2010;10:1165–1173. doi: 10.1016/j.meegid.2010.07.016. [DOI] [PubMed] [Google Scholar]
- 11.Koeck JL, et al. Clinical characteristics of the smooth tubercle bacilli ‘Mycobacterium canettii’ infection suggest the existence of an environmental reservoir. Clin Microbiol Infect. 2011;17:1013–1019. doi: 10.1111/j.1469-0691.2010.03347.x. [DOI] [PubMed] [Google Scholar]
- 12.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Garnier T, et al. The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci U S A. 2003;100:7877–7882. doi: 10.1073/pnas.1130426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Springer B, Stockman L, Teschner K, Roberts GD, Bottger EC. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 16.Stinear TP, et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008;18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veyrier FJ, Dufort A, Behr MA. The rise and fall of the Mycobacterium tuberculosis genome. Trends Microbiol. 2011;19:156–161. doi: 10.1016/j.tim.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 18.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 19.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becq J, et al. Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol. 2007;24:1861–1871. doi: 10.1093/molbev/msm111. [DOI] [PubMed] [Google Scholar]
- 22.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: all the world’s a phage. Proc Natl Acad Sci U S A. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon SV, Bottai D, Simeone R, Stinear TP, Brosch R. Pathogenicity in the tubercle bacillus: molecular and evolutionary determinants. Bioessays. 2009;31:378–388. doi: 10.1002/bies.200800191. [DOI] [PubMed] [Google Scholar]
- 25.Deshayes C, et al. Detecting the molecular scars of evolution in the Mycobacterium tuberculosis complex by analyzing interrupted coding sequences. BMC Evol Biol. 2008;8:78. doi: 10.1186/1471-2148-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV. Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2009;7:537–544. doi: 10.1038/nrmicro2165. [DOI] [PubMed] [Google Scholar]
- 27.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature. 2004;431:84–87. doi: 10.1038/nature02837. [DOI] [PubMed] [Google Scholar]
- 28.Bentley SD, et al. The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS Negl Trop Dis. 2012;6:e1552. doi: 10.1371/journal.pntd.0001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brosch R, et al. Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci U S A. 2007;104:5596–5601. doi: 10.1073/pnas.0700869104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comas I, et al. Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet. 2010;42:498–503. doi: 10.1038/ng.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hershberg R, et al. High functional diversity in M. tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocha EP, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 33.Castillo-Ramirez S, et al. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 2011;7:e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korber B. In: Computational Analysis of HIV Molecular Sequences. Rodrigo AG, Learn GH, editors. Kluwer Academic Publishers; 2000. pp. 55–72. Ch. 4. [Google Scholar]
- 36.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 37.Simeone R, et al. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012;8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalscheuer R, Weinrick B, Veeraraghavan U, Besra GS, Jacobs WR., Jr. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Achtman M. Insights from genomic comparisons of genetically monomorphic bacterial pathogens. Philos Trans R Soc Lond B Biol Sci. 2012;367:860–867. doi: 10.1098/rstb.2011.0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namouchi A, Didelot X, Schock U, Gicquel B, Rocha EP. After the bottleneck: Genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 2012;22:721–734. doi: 10.1101/gr.129544.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mba Medie F, Ben Salah I, Henrissat B, Raoult D, Drancourt M. Mycobacterium tuberculosis complex mycobacteria as amoeba-resistant organisms. PLoS One. 2011;6:e20499. doi: 10.1371/journal.pone.0020499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen KT, Piastro K, Gray TA, Derbyshire KM. Mycobacterial biofilms facilitate horizontal DNA transfer between strains of Mycobacterium smegmatis. J Bacteriol. 2010;192:5134–5142. doi: 10.1128/JB.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Medzhitov R, Janeway CA. Innate Immunity cecognition and control of adaptive immune responses. Semin Immunol. 1998;10:351–353. doi: 10.1006/smim.1998.0136. [DOI] [PubMed] [Google Scholar]
- 45.Aagaard C, et al. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat Med. 2011;17:189–194. doi: 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- 46.Cadieux N, et al. Induction of cell death after localization to the host cell mitochondria by the Mycobacterium tuberculosis PE_PGRS33 protein. Microbiology. 2011;157:793–804. doi: 10.1099/mic.0.041996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiden MC, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 49.Batzoglou S, et al. ARACHNE: a whole-genome shotgun assembler. Genome Res. 2002;12:177–189. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brosch R, et al. Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics. Infect Immun. 1998;66:2221–2229. doi: 10.1128/iai.66.5.2221-2229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallenet D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database (Oxford) 2009 doi: 10.1093/database/bap021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carver T, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niemann S, et al. Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS One. 2009;4:e7407. doi: 10.1371/journal.pone.0007407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 2002;30:2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suyama M, Torrents D, Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 62.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- 64.Jakobsen IB, Easteal S. A program for calculating and displaying compatibility matrices as an aid in determining reticulate evolution in molecular sequences. Comput Appl Biosci. 1996;12:291–295. doi: 10.1093/bioinformatics/12.4.291. [DOI] [PubMed] [Google Scholar]
- 65.Pethe K, et al. The heparin-binding haemagglutinin of M. tuberculosis is required for extrapulmonary dissemination. Nature. 2001;412:190–194. doi: 10.1038/35084083. [DOI] [PubMed] [Google Scholar]
- 66.Majlessi L, et al. Influence of ESAT-6 secretion system 1 (RD1) of Mycobacterium tuberculosis on the interaction between mycobacteria and the host immune system. J Immunol. 2005;174:3570–3579. doi: 10.4049/jimmunol.174.6.3570. [DOI] [PubMed] [Google Scholar]
- 67.Bottai D, et al. ESAT-6 secretion-independent impact of ESX-1 genes espF and espG1 on virulence of Mycobacterium tuberculosis. J Infect Dis. 2011;203:1155–1164. doi: 10.1093/infdis/jiq089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.