Abstract
Assault to DNA that leads to oxidative base damage is repaired by the base excision repair (BER) pathway with specialized enzymes called DNA glycosylases catalyzing the first step of this pathway. These glycosylases can be categorized into two families: the HhH superfamily, which includes endonuclease III (or Nth), and the Fpg/Nei family, which comprises formamidopyrimidine DNA glycosylase (or Fpg) and endonuclease VIII (or Nei). In humans there are three Nei-like (NEIL) glycosylases: NEIL1, 2, and 3. Here we present the first crystal structure of a viral ortholog of the human NEIL2/NEIL3 proteins, Mimivirus Nei2 (MvNei2), determined at 2.04 Å resolution. The C-terminal region of the MvNei2 enzyme comprises two conserved DNA binding motifs: the helix-two-turns-helix (H2TH) motif and a C-H-C-C type zinc-finger similar to that of human NEIL2. The N-terminal region of MvNei2 is most closely related to NEIL3. Like NEIL3, MvNei2 bears a valine at position 2 instead of the usual proline and it lacks two of the three conserved void-filling residues present in other members of the Fpg/Nei family. Mutational analysis of the only conserved void-filling residue methionine 72 to alanine yields an MvNei2 variant with impaired glycosylase activity. Mutation of the adjacent His73 causes the enzyme to be more productive thereby suggesting a plausible role for this residue in the DNA lesion search process.
Keywords: Base excision repair, DNA glycosylase, Mimivirus Nei2, void-filling residue
1. INTRODUCTION
Endogenous DNA damage such as deaminations, depurinations, alkylations, and oxidative damage accounts for ~30,000 lesions per human cell per day [1, 2]. Oxidative damage is predominantly generated by reactive oxygen species, which form as a result of normal cellular processes such as respiration and also originate from various exogenous sources like ionizing radiation and cigarette smoke [3]. If left unrepaired, this type of damage can result in various cell fates including apoptosis, mutagenesis, and eventually cancer. In an effort to maintain genomic stability, cells have developed mechanisms to detect and repair DNA damage. More specifically, oxidized DNA base lesions are recognized and removed by the base excision repair pathway, a highly conserved repair process, the first step of which is catalyzed by a lesion-specific DNA glycosylase [1, 4–6]. Some of these enzymes are bifunctional, i.e., they catalyze the hydrolysis of the N-glycosylic bond linking a base to a deoxyribose (glycosylase activity) and subsequently cleave the DNA 3′ to the newly created apurinic/apyrimidinic site (lyase activity), whereas others are monofunctional and only carry out the glycosylase reaction, generating abasic sites as products. DNA glycosylases that cleave oxidized lesions are grouped into two families, based on sequence homology and structural motifs: the Helix-hairpin-Helix (HhH) family, named after the HhH structural motif involved in DNA binding, and the Fpg/Nei family, named after the prototypical bacterial members formamidopyrimidine DNA glycosylase (Fpg) and endonuclease eight (Nei) [3, 5, 7, 8]. In humans, three Nei-like (NEIL) proteins, NEIL1, NEIL2, and NEIL3 have been characterized [9–15].
Fpg/Nei family members exhibit a distinct two-domain architecture where the N- and C- terminal domains are connected by a flexible hinge region [6, 7, 16, 17]. The N-terminal domain comprises a two-layered β-sandwich flanked by α-helices, while the predominantly α-helical C-terminal domain comprises a conserved helix-two-turn-helix (H2TH) motif, as well as two antiparallel β-strands that form a zinc/zincless-finger motif required for DNA binding. The active site is located in the cleft between the two domains, with two conserved N-terminal residues (generally a proline/valine and a glutamate) important for catalysis. Members of Fpg/Nei family are typically equipped with a “void-filling triad” responsible for filling the void left by the everted lesion and stabilizing the DNA helix. The three residues constituting the void-filling triad are highly conserved and in Escherichia coli Fpg include Met74, Arg109, and Phe111 [18, 19]. Met74 of the β4/5 loop occupies the position of the extruded base; Arg109 of the β7/8 loop stabilizes the orphaned base opposite the lesion, while Phe111 acts as a wedge causing disruption of base stacking between the nucleotide opposite the lesion and the neighboring base, thereby severely kinking the DNA. This non-contiguous arrangement of void-filling residues is observed in most Fpg/Nei enzymes, except for E. coli Nei (EcoNei) where all three residues are located on the same β4/5 loop [7, 20].
Although all Fpg/Nei family members share a similar fold, they differ substantially in their substrate preference. The Fpg proteins preferentially excise oxidized purines, including 8-oxo-7,8-dihydroguanine (8-oxoG) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG), whereas Nei and the NEIL enzymes mainly recognize oxidized pyrimidines, adenine-derived 4,6-diamino- 5-formamidopyrimidine (FapyA), and the further oxidation products of 8-oxoG namely spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh) [7, 14, 17, 21–25].
Two Nei glycosylases from the Acanthameobapolyphaga mimivirus, MvNei1 (L315) and MvNei2 (L720), have recently been cloned and expressed [26, 27]. MvNei1 and MvNei2 have enzymatic properties very similar to their human homologs, NEIL1 and NEIL2/3 [26, 28]. Although MvNei1 and MvNei2 do not recognize 8-oxoG, both enzymes cleave its further oxidation products, Sp and Gh [9, 22, 24, 26]. Single-stranded DNA with the same base lesions, as well as bubble structures, are also substrates for both enzymes [9, 26, 28]. Structural analyses of MvNei1 revealed a zincless-finger β-hairpin motif, first described in human NEIL1, rather than the signature zinc-finger characteristic of the Fpg/Nei family [27, 29]. Sequence alignments of MvNei2, in contrast, predict a C-H-C-C type zinc-finger similar to human NEIL2, for which there is currently no crystal structure [26]. In addition, MvNei2 uses a valine as the active site residue at position 2 as Neil3, instead of the proline found in most Fpg/Nei glycosylases. Furthermore MvNei2 is predicted to have only one of the three conserved void-filling residues, a feature that is also shared with Neil3 [16].
Here we report the first crystal structure of MvNei2, a unique Fpg/Nei family enzyme with a C-H-C-C type zinc finger motif like that seen in the C-terminal domain of human NEIL2, but with an N-terminal domain similar to NEIL3. In addition, mutation of the sole void-filling residue in MvNei2, Met72, to alanine severely diminishes the enzyme's glycosylase activity, underscoring the importance of this residue in recognizing oxidative base damage and stabilizing the DNA upon lesion extrusion.
2. MATERIALS AND METHODS
2.1. Cloning, Overexpression, and Purification
The MvNei2 construct was cloned and purified as described previously [9, 26]. Briefly, the protein was co-expressed in a pETDuet1 (Novagen) vector with the E. coli MAP protein. Expression in Rosetta2 (DE3) pLysS E. coli cells (Novagen) was performed by autoinduction where cells were grown for 48 – 60 hrs at 20 °C [30]. Cells were lysed in a buffer containing 20 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.01% (v/v) NP40, 10% (v/v) glycerol, 2 mM β-ME, and 1 mM PMSF. The lysate obtained was applied onto a fast-flow S-column (GE Healthcare) and the protein was eluted using a gradient from 300 mM - 1 M NaCl. Pooled fractions were then loaded onto a pre-equilibrated nickel affinity column (GE Healthcare) and eluted using an imidazole gradient 10 mM – 500 mM. As a final step, the protein fractions were diluted in half to lower the salt concentration and then loaded onto an SP-FF column as done above. Purified MvNei2 protein was concentrated to about 6 mg/ml, determined by a NanoDrop (NanoDrop Technologies, Wilmington, Delaware) instrument, before flash freezing in LN2 and storage at −80 °C. The QuikChange II XL site-directed mutagenesis kit (Stratagene) was used to introduce the M72A and H73A mutations into MvNei2 in the pETDuet1 (Novagen) expression vector. The resulting variants were completely sequenced to ensure that the appropriate mutations were present.
2.2. DNA Preparation and Complex Formation
The 35-mer oligodeoxynucleotides used for the glycosylase/lyase activity assays and in the multiple-turnover kinetics were purchased from Midland Certified Reagent company (Midland, TX) and purified by urea PAGE. The sequence of the damage-containing strand was 5′-TGTCAATAGCAAG(X)GGAGAAGTCAA TCGTGAGTCT-3′, where X was Tg, OHU, DHU, or uracil (which was used to create an apurinic/apyrimidic site). The complementary oligonucleotide sequence was the following: 5′-AGACTCACGATTGACTTCTCC(Y)CTTGCTATTGACA-3′, in which (Y) denotes the appropriate base (G, A, or C) opposite the damaged base. Gh, Sp1, and 2,6-diamino-4-hydroxy-5N-methyl-formamidopyrimidine (MeFapyG) were synthesized as described previously [31, 32] in the following sequence context: 5′-TGTTCATCATGCGTC(Y)TCGGTATATCCCAT-3′, Y being either Gh or Sp1, and 5′-TCATCATGCGTC(MeFapyG)TCGGTATATCC-3′. The complementary strand for Gh- and Sp1-containing DNA was 5′-ATGGGATATACCGA(C)GACGCATGATGAACA-3′ and for MeFapyG was 5'-GGATATACCGACGACGCATGATGA-3'. 5′-end labeling of substrates was carried out on 1 pmole of each damage-containing strand using T4 polynucleotide kinase (New England Biolabs, Beverly, MA) in the presence of [γ-32P] dATP, for 30 min at 37 °C. The phosphorylation reaction was terminated by addition of 1 mM EDTA and heat inactivation. The end-labeled DNA was separated from the [γ-32P] dATP by ethanol precipitation and diluted in 9 pmoles of the appropriate non-labeled damage-containing oligodeoxynucleotide and 10 pmoles of the complementary oligodeoxynucleotide (for duplex DNA) to a final concentration of 250 nM in 10 mM Tris–HCl (pH 8.0) and 50 mM NaCl. In order to create an AP site, a double-stranded uracil-containing oligodeoxynucleotide was treated with 2 units of uracil DNA glycosylase (UDG, New England Biolabs) for 30 min at 37 °C.
2.3. Glycosylase/Lyase Activity Assays
Glycosylase/lyase assays were performed with 20 nM lesion-containing/AP-site single-stranded (ss) or double-stranded (ds) substrate and increasing concentrations of enzyme (10 – 320 nM) in buffer A: 20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA (pH 8.0) with 100 μg/ml BSA at 37 °C for 30 minutes. The reactions were stopped by adding an equal volume of formamide-containing loading buffer (98% formamide, 5 mM EDTA, 0.1% xylene cyanol and 0.1% bromophenol blue) to measure both glycosylase and lyase activities. The reaction products were separated from the uncleaved substrates with a 12% (w/v) denaturing polyacrylamide gel and quantified with an isotope imaging system (Molecular Imaging System, Bio-Rad).
2.4. Multiple-Turnover Kinetics
Multiple-turnover experiments were performed using buffer A above, with the exception that the substrate was in excess over enzyme. Assays were performed using either 20 nM Sp1:C or ssSp1, incubated with 16 nM total protein. Several concentrations of enzyme from 4 – 18 nM were tested for all enzymes used in this analysis, keeping the substrate concentration constant at 20 nM. The active fraction of the WT enzyme is between ~15–30 %, based on the burst observed under these assay conditions. Therefore a concentration of 16 nM enzyme corresponds to 2–5 nM active enzyme, and the enzyme is fully saturated with 20 nM substrate. The M72A and H73A variants did not exhibit a burst phase and thus an active fraction was not readily obtainable. To comparatively study all three enzymes, we chose an equal enzyme concentration (16 nM) as measured by A280.
Aliquots were removed at various times (15 sec to 60 min) and quenched by the addition of 10 μL of formamide/dye solution. The samples were then run on a 12% denaturing polyacrylamide gel in 1 × TBE at 60 W for 1 hr and imaged using an isotope imaging system (Molecular Imaging System, Bio-Rad). Fitting of the data was performed with GraphPad Prism 6.0a software as described previously [24]. Data from multiple-turnover experiments were fitted with equation (1) where A0 is the amplitude of the burst, kobs is rate constant for the burst phase, and kss is the rate of the linear phase.
| (1) |
2.5. Crystallization and Data Collection
Crystals of unliganded MvNei2 were obtained in sitting-drop vapor diffusion experiments at 18 °C using 20% (w/v) polyethylene glycol 3350, and 0.2 M sodium sulfate decahydrate as the crystallization solution combined in a 1:1 ratio with concentrated protein (4–6 mg/mL) (Peg-Ion HT screen; Hampton Research, Aliso Viejo, CA). Rhombohedral crystals belonging to the R32 space group in the hexagonal setting were obtained after ~3 weeks and grew to ~50 microns in all three dimensions. The crystals were cryoprotected using a solution consisting of the mother liquor supplemented with 20% (w/v) glycerol, mounted onto a cryo-loop and flash cooled into liquid nitrogen. The crystals diffracted in-house using a rotating anode RUH3R X-ray generator (Rigaku, The Woodlands, Texas) and MAR image plate detector (MarResearch, Hamburg, Germany) to 3.2 Å. The same crystal diffracted to 2.04 Å at the Diamond Synchrotron (beamline I04) where a data set was collected at λ= 0.9173 Å. The in-house data were processed using the HKL program suite [33], whereas the synchrotron data were processed with XDS [34] and reduced with SCALA [35].
2.6. Structure Determination and Refinement
The structure of MvNei2 was solved using single isomorphous replacement with anomalous scattering (SIRAS) using a crystal soaked in sodium iodide. The crystal was soaked in 200 mM NaI for 15 minutes and diffracted to 2.25 Å in-house. Three initial iodide sites were located using SHELXD [36–38] and the remaining 5 sites were obtained using Autosol from the PHENIX program suite [39]. The initial model was built using AutoBuild [39, 40]. The rest of the model was built into the electron density map using COOT [41] and refined using Phenix.Refine and CNS [42–44]. Water molecules were picked automatically using AutoBuild and Phenix.Refine [42, 43]. An anomalous difference map calculated using the synchrotron data confirmed the presence of the zinc atom. The final model was found to exhibit good geometry, as determined by Molprobity [45]. Refinement statistics are shown in Table 1. All structure figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.).
TABLE 1.
Data collection, phasing, and refinement statistics for MvNei2
| Native | Iodide | |
|---|---|---|
| Beamlines | Diamond-I04 | Home source |
| Wavelength (Å) | 0.91730 | 1.54 |
| Space group | R32 | R32 |
| Unit-cell parameters (Å, °) | a = b = 121.670, c = 164.180 | a = b = 120.545, c = 164.21 |
| α = β = 90, γ = 120 | α = β = 90, γ = 120 | |
| Molecules per asymmetric unit | 1 | 1 |
|
| ||
| Data collection statistics | ||
| Resolution (Å) | 2.04 | 2.25 |
| Unique reflections | 29962 | 21805 |
| Redundancy | 10.1 (10.5) | 12.3 (6) |
| Rmerge | 0.15 (0.665) | 0.147 (0.636) |
| Overall I/σ | 9.5 (3.3) | 16.37 (2.24) |
| Completeness (%) | 100 (100) | 99.5 (95.3) |
|
| ||
| SIR phasing statistics | ||
| No. of sites from Autosol | 8 | |
| Figure of merit (Resolve) | 0.80 | |
| Refinement statistics | ||
| Rwork (%) | 18.7 | |
| Rfree (%) | 22.7 | |
| r.m.s.d values | ||
| Bond length (Å) | 0.005 | |
| Bond angles (°) | 0.917 | |
| B-factor (Å2) | ||
| Wilson-B | 27.6 | |
| Protein | 34.4 | |
| Water | 38.3 | |
| Zinc | 37.1 | |
| SO42− | 53.2 | |
| Ramachandran plot | ||
| Most favored (%) | 97.5 | |
| Additional allowed (%) | 2.5 | |
| Generously allowed (%) | 0 | |
| Disallowed (%) | 0 | |
Rmerge = Σ|I− 〈I〉 |/ΣI, where 〈I〉 is the average intensity from multiple observations of symmetry-related reflections. Rwork and Rfree = Σ|Fo| − |Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with 10% of the reflections not used in refinement. SIR, single isomorphous replacement. Values for the highest resolution shell are shown in parentheses.
2.7. Protein Data Bank Accession Codes
Atomic coordinates and structure factor amplitudes have been deposited with the Protein data bank (http://www.pdb.org) and are accessible under accession code 4MB7.
3. RESULTS
3.1. Structural features of MvNei2
The viral glycosylase MvNei2 is unique because of its sequence similarity to both human NEIL2 and NEIL3. Structure-based sequence alignments (Figure S1) reveal that MvNei2 shares a C-H-C-C type zinc-finger motif in the C-terminal region similar to that of human NEIL2, while harboring an N-terminal catalytic valine, as seen in Neil3 [16, 25, 26]. In order to study the structural features of this enzyme, we pursued the crystal structure of full-length MvNei2. The structure was determined at a resolution of 2.04 Å. The resulting model was refined to an Rfree of 22.7% and Rwork of 18.7% (Table 1).
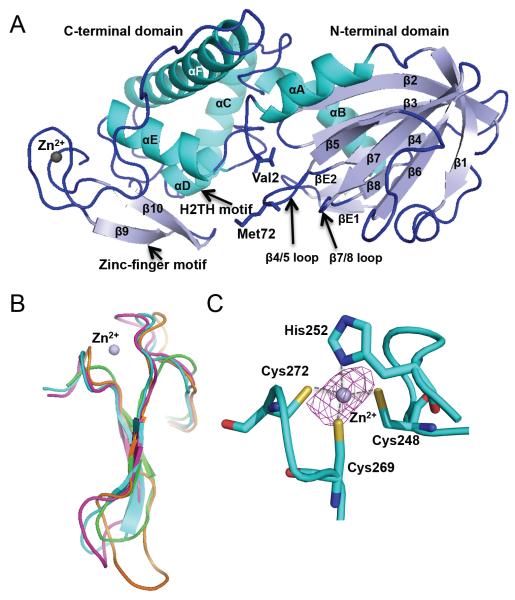
The overall structure of MvNei2 closely resembles that of other members of the Fpg/Nei family and displays a characteristic two-domain architecture (Fig. 1A). The N-terminal domain comprises two α-helices, which flank a two-layered β-sandwich where each layer is composed of four antiparallel β-strands. The C-terminal domain comprises four α-helices, two of which form the conserved H2TH motif (Fig. 1A). The zinc finger motif, which comprises two anti-parallel β-strands, is structurally similar among Fpg/Nei glycosylases, regardless of whether they possess zinc-coordinating residues (as seen in mouse Neil3, MmuNeil3), or a zincless finger motif (in NEIL1 and MvNei1, Fig. 1B). The zinc atom in MvNei2 is coordinated by three cysteines (Cys248, 2.3 Å; Cys269, 2.2 Å; and Cys272, 2.3 Å) and one histidine (His252, 2.0 Å) (Fig. 1C). The corresponding residues in human NEIL2 (Cys291, Cys315, Cys318, and His295) have been shown to play a critical role in the enzyme's function [46]. In addition to the four zinc-coordinating residues, the zinc-finger motif also harbors a highly conserved arginine residue (Arg264) in the loop connecting β strands 9 and 10 (Fig. S1). Mutation of the corresponding arginine in human NEIL1 and NEIL2 adversely affects the enzyme's ability to cleave damaged DNA bases [29, 46].
Figure 1.
Overall structure of Mimivirus Nei2. (A) Ribbon diagram of MvNei2. The secondary structure elements are as follows: αA (3–15), β1 (21–26), β2 (42–48), β3 (51–57), β4 (64–68), β5 (75–79), β6 (89–94), β7 (97–103), β8 (107–110), βE1 (117–119), βE2 (124–126), αB (127–140), αC (148–163), αD (167–173), αE (182–193), αF (205–232), β9 (255–259), and β10 (264–268). Helices are shown in cyan and β-strands in pale blue. The zinc metal ion is shown as a grey sphere. βE1 and 2 are unique to MvNei2 and are located before helix αB. (B) Superposition of the zinc/zincless-finger motifs from MvNei2 (cyan), MmuNeil3 (fuschia, PDB code 3W0F [16]), MvNei1 (orange, PDB code 3A46 [27]) and human NEIL1 (green, PDB code 1TDH [29]). The superposition was performed in COOT [41] using secondary-structure matching (SSM) of all residues in the enzymes. (C) Close-up view of the coordination of the zinc ion by His252, Cys248, Cys262, and Cys272. An anomalous difference Fourier map (magenta mesh) contoured at 3σ is displayed around the zinc ion.
A superposition of MvNei2 with other Fpg/Nei enzymes, NEIL1, MmuNeil3 and MvNei1, indicates that helices αB and αC of MvNei2 superimpose well with those of MmuNeil3, but not those of other glycosylases (Fig. S2, [16]). In both MmuNeil3 and MvNei2, αB and αC are perpendicular with respect to each other, thus creating a more rigid inter-domain segment. In addition, the loop between helix αF and β strand β9 or β10, termed the αF-β9/10 loop or 8-oxoG capping loop, is truncated in MvNei2, as seen in MmuNeil3 [16, 47]. The longer αF-β9/10 loop in bacterial Fpg enzymes wraps around the Hoogsteen face of 8-oxoG and is necessary for cleavage of this lesion but is dispensable for the cleavage of oxidized pyrimidines [17, 47]. The presence of a truncated loop in MvNei2 is consistent with the fact that 8-oxoG is not a preferred substrate for this glycosylase. A structural feature that is so far unique to MvNei2 is a ~16 amino-acid extension (residues 114–127), located between β-strand β8 and helix αB (Figs. S1 and S2). This extension features two antiparallel β-strands (βE1 and βE2) connected by a loop (Fig. 1A).
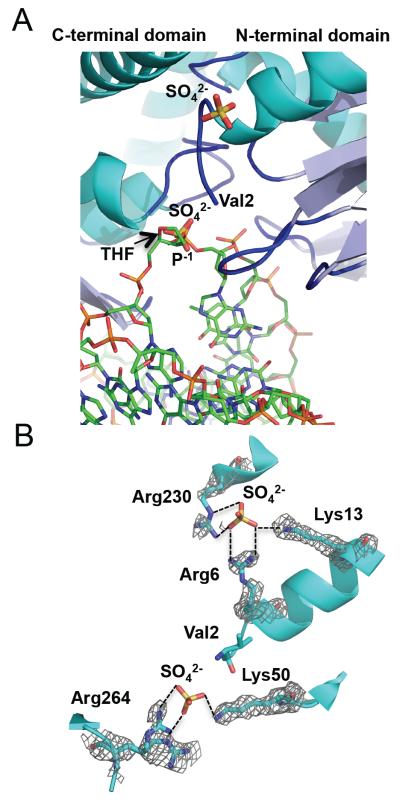
Two sulfate ions were placed in an isomorphous difference map and refined. These anions are likely to be sulfates based on their coordination by positively charged residues, the presence of sulfate in the crystallization conditions, and the crystallographic refinement. The sulfates interact with positively charged residues such as arginines and lysines and thus likely mimic phosphate groups of the DNA backbone, as observed in other DNA or RNA-binding proteins crystallized in the presence of sulfate [48]. A superposition of MvNei2 with the DNA from the MvNei1-DNA complex shows that one of these sulfate ions lies in the same position as the P−1 phosphate immediately following the lesion on the lesion-containing strand (Fig. 2A). The sulfate forms ionic interactions with the highly conserved Arg264 residue located in the loop connecting the two antiparallel β strands of the zinc finger, and Lys50, an absolutely conserved lysine residue critical for glycosylase activity [20] (Fig. 2B).
Figure 2.
The sulfate ions in the unliganded MvNei2 structure likely mimic DNA backbone phosphates. (A) Superposition of unliganded MvNei2 with the DNA from the MvNei1-THF structure (PDB ID 3A46 [27], performed with SSM using all atoms in COOT [41]), indicates that one of the two sulfates found in the MvNei2 structure lies in the same position as a phosphate in the DNA. MvNei2 is colored and positioned as in Fig. 1A. The sulfate ions are colored by atom type, MvNei1-DNA carbon atoms are shown in green and the bases and backbone phosphates are colored by atom type. (B) Coordination of the sulfate ions by basic lysine and arginine residues at the surface of the MvNei2 enzyme. The enzyme is colored in cyan with the side chains and the sulfates colored by atom type. A composite omit map (PHENIX [43]; grey) contoured at 1σ is overlaid on the residues coordinating the sulfate ions.
The second sulfate, which is ~20 Å away from the first one, lies in a basic crevice on the protein surface and makes salt-bridge interactions with Arg230, Arg6, and Lys13 (Fig. 2B). Unlike Lys50 and Arg264, these three residues are not highly conserved across Fpg/Nei enzymes. The binding of a sulfate ion at this position could be fortuitous or it could indicate the path that single-stranded DNA might take (MvNei2 can excise lesions in both ss and dsDNA substrates; see below). An answer to this question will have to await the structure of a Neil2 glycosylase bound to DNA.
3.2. Role of the void-filling residues of MvNei2 in binding to damaged DNA
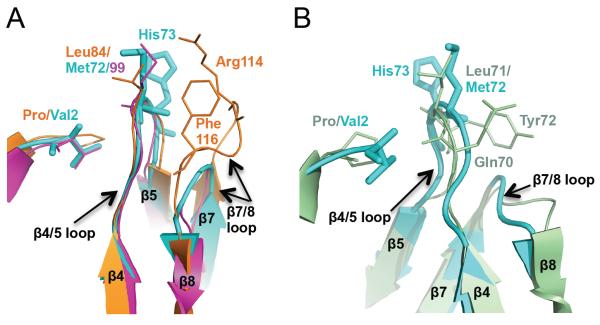
As mentioned previously, MvNei2 bears only one of the three canonical void-filling residues, as seen in Neil3 (Fig. S1). The void-filling Met72 in MvNei2 corresponds to Met99 in MmuNeil3, and Leu84 in MvNei1 (Fig. 3A) and all these residues reside on the β4/5 loop. In the structures of MvNei1 with DNA bound, Leu84 along with Arg114 and Phe116 of the β7/8 loop fill the void created upon extrusion of the damaged base [27, 28]. This non-contiguous arrangement of three void-filling residues is characteristic of many of the Fpg enzymes but is not seen in MvNei2 and MmuNeil3, in which the loop connecting β strands β7 and β8 is much shorter (Fig. 3A) [16]. In EcoNei, the void-filling residues (Gln70, Leu71, and Tyr72) lie on the same β4/5 loop (Fig. 3B) [7, 20]. Superposition of the MvNei2 and EcoNei structures (Fig. 3B) reveals some structural similarity in the β4/5 and β7/8 loops. Previous studies with EcoNei indicated that mutating all three void-filling residues in the β4/5 loop to alanine (i.e. Q70A/L71A/Y72A) or deletion of these three residues resulted in variant with a severely compromised glycosylase activity compared to wild type [49]. In an effort to explore the potential role of Met72 and His73 in base excision, we mutated both residues individually to alanine and measured their glycosylase activity.
Figure 3.
Void-filling residues in MvNei2, MvNei1 and EcoNei (A) Superposition of the β4/5 loop with the void-filling methionine (Met72 in MvNei2, cyan, and Met99 in MmuNeil3, fuchsia, PDB 3W0F [16]) or leucine (Leu84 in MvNei1, orange, PDB 3A46 [27]) and β7/8 loop with void-filling Arg114 and Phe116 of MvNei1 (orange). (B) Superposition of MvNei2 (cyan) with EcoNei (pale green, PDB 1K3W [20]) showing the similarity in the β4/5 and β7/8 loops in these two enzymes. The β4/5 loops contains the void-filling triad in EcoNei, i.e. Gln70, Leu71, and Tyr72. The β7/8 loops are of similar size and shape, and much shorter than that seen in MvNei1 or hNEIL1. Superpositions were performed with residues 67–78 of MvNei2 and the corresponding residues in MmuNeil3, MvNei1, and EcoNei using a least-squares superposition tool (COOT [41]).
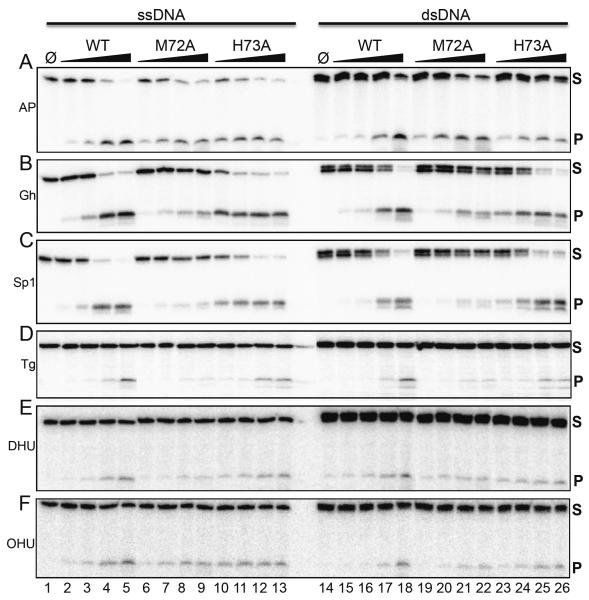
Several lesion-containing substrates (AP site, Gh, Sp, thymine glycol (Tg), dihydrouracil (DHU), and 5-hydroxyuracil (OHU)), which were either single-stranded or in duplex form paired with an appropriate opposite base, were tested with increasing enzyme concentrations and quantified (Fig. 4, Table S1). All three enzymes exhibit a robust lyase activity, as demonstrated by the formation of cleavage products using AP-site containing DNA (Fig. 4A, Table S1). MvNei2 preferentially cleaves its substrates via β-elimination, like mouse Neil3 and human NEIL3 [9, 25, 26]. Of the substrates analyzed, Gh and Sp1 are the preferred substrates for MvNei2 (Fig. 4B, C, Table S1), which is also true for NEIL1, MvNei1, and Neil3 [9, 22, 25, 26]. M72A exhibits a reduced activity for these substrates when compared with WT and H73A. Tg, DHU, and OHU are not preferred substrates for MvNei2, but are still processed by both WT and H73A and to a lesser extent by M72A (Fig. 4 D–F, Table S1). Overall, MvNei2 appears to cleave lesions in ssDNA and dsDNA equally well, in contrast to MmuNeil3, which displays a strong preference for ssDNA [9, 16].
Figure 4.
Glycosylase/lyase activity profile for WT MvNei2 and M72A, H73A variants. Glycosylase assays were performed by incubating 20 nM of single-(lanes 1–13) or double-stranded (14–26) substrates with increasing amounts of enzyme with the following substrate:enzyme ratios: 1:0.5, 1:1, 1:4 and 1:16. “Φ” indicates no enzyme. Panel (A) ssAP-site and AP:C; (B) ssGh and Gh:C; (C) ssSp1 and Sp1:C; (D) ssTg and Tg:A; (E) ssDHU and DHU:A; (F) ssOHU and OHU:G. Assays were performed at 37°C for 30 minutes. S and P indicate substrates and products respectively. Data shown are representative of two repeat experiments. The double bands seen in some dsDNA substrates (e.g., Gh and Sp1) are likely due to the presence of truncated synthesis products.
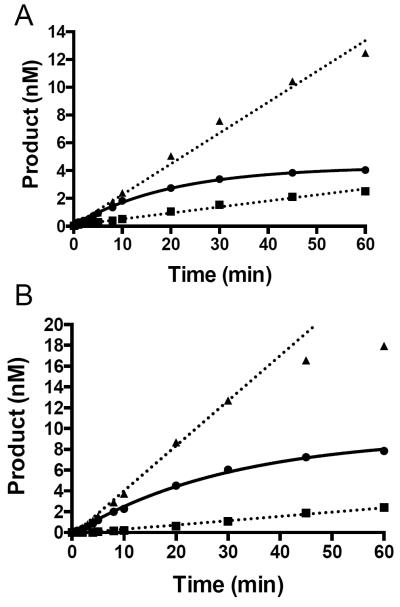
We used an oligodeoxynucleotide substrate containing either Sp1 paired opposite C or ssSp1, under saturating substrate conditions to further evaluate the differences between WT MvNei2 and the M72A and H73A variants. Under these assay conditions, WT MvNei2 displayed a relatively slow burst phase with both Sp1:C and ssSp1 where the values for kobs were 0.05 ± 0.01 min−1 and 0.033 ± 0.003 min−1, respectively (Fig. 5A and B, circles connected by solid line). Mutating the void-filling Met72 to Ala severely diminished this enzyme's ability to cleave the lesion in both Sp1:C and ssSp1 contexts, as compared with WT enzyme (Fig. 5A and B, squares connected by dotted line). These data indicate that, in the absence of the other two void-filling residues, the conserved methionine is a key player in lesion recognition and base extrusion. Mutating Met77 to Ala in a bacterial Fpg glycosylase resulted in diminished efficiency of 8-oxoG repair, implicating this conserved residue in stabilizing the extruded base [19].
Figure 5.
Role of the void-filling Met72 and adjacent His73 in lesion excision. Glycosylase assays with double-stranded Sp1:C (A) and ssSp1 (B) where the DNA substrate (20 nM) was combined with 16 nM of either WT or mutant MvNei2. WT MvNei2 is displayed as circles. M72A and H73A are shown as squares and triangles, respectively. Data plotted represent averages of three independent experiments.
When glycosylase assays were performed with the H73A variant, more substrate was turned over during the course of the experiments when compared to WT, but the burst phase was lost. This behavior was seen for both Sp1:C and ssSp1 (Fig. 5A and B, triangles connected by dotted line). The differences observed between the H73A variant and WT enzyme can be attributed to dissociation and product release: the H73A mutant enzyme appears to no longer be dissociation limited. These data suggest that His73 must interact with the DNA, thereby creating stabilizing interactions, which when absent make the variant more productive. Similar results were observed with the wedge residue variant, Phe111, from E. coli Fpg [50]. In these studies, the diffusive behavior of a F111A variant along DNA in a single-molecule experiment is increased compared to WT, indicating that this mutation makes the enzyme faster and less restricted because it presumably no longer probes DNA for damage [50]. Since MvNei2 lacks the conserved Phe wedge residue, His73 may play an analogous role in the viral enzyme.
3.3 Electrostatic surface potential of MvNei2 compared with other Fpg/Nei glycosylases
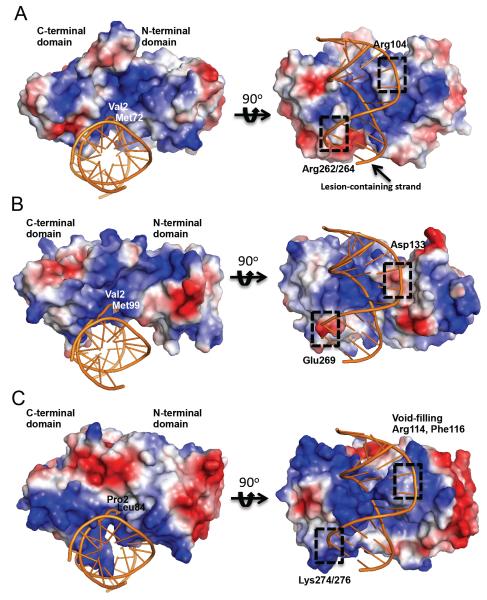
The electrostatic potential of the solvent-accessible surface residues of MvNei2 reveals a positively charged cleft along the long axis of the protein (Fig. 6A) as well as in the active site cleft between the N- and C-terminal domains. This arrangement of positively charged residues along the contour of MvNei2 seems to support the enzyme's ability to bind to duplex as well as single-stranded DNA, which is in agreement with the glycosylase assays conducted with ss and dsDNA substrates. In order to observe the surface potential of MvNei2 juxtaposed with DNA, MvNei2 was superimposed with the DNA from MvNei1 bound to tetrahydrofuran (THF) (PDB code 3A46, [27]) and compared with MmuNeil3, an enzyme with a strong preference for ssDNA, and MvNei1, a glycosylase which binds dsDNA preferentially (Fig. 6) [9, 16]. As reported previously, MmuNeil3 lacks the basic residues required for stabilizing interactions with the DNA. Furthermore, the negatively charged Asp133 residue is predicted to create an unfavorable environment for duplex DNA binding (Fig. 6B) [16, 27]. In addition, Glu269 in the β9/10 loop of MmuNeil3 would similarly discourage dsDNA binding (Fig. 6B). In contrast, in MvNei1 the surface is largely positively charged (Fig. S1). Lys274 and Lys276 of the β9/10 loop, along with the highly conserved Arg277, create a positively charged surface favorable to dsDNA binding (Fig. 6C).
Figure 6.
Surface representation of MvNei2 and comparison with MmuNeil3 and MvNei1. Surface electrostatic potential of (A) MvNei2 superimposed with MvNei1-DNA (COOT [41]) where only the DNA is displayed (PDB ID 3A46, orange); (B) MmuNeil3 superimposed with the above DNA; (C) MvNei1-bound to DNA (PDB ID 3A46). All surfaces were created in PyMOL (vacuum electrostatics option, The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.). The surfaces are colored according to electrostatic potential, where blue and red signify positive and negative charges, respectively. The scale is ±78 kT/e for all surfaces.
A positively charged Arg104 in MvNei2, which corresponds to Asp133 in MmuNeil3, would be conducive to dsDNA binding. Arg262 and Arg264 provide a basic environment to support binding of the opposite strand (Fig. 6A). These key differences between MvNei2 and Neil3 might explain why the former binds ss and dsDNA equally well and the latter has a marked preference for ssDNA.
4. DISCUSSION
4.1. Mimivirus: A giant conundrum
The discovery of the Acanthamoeba polyphaga Mimi (microbe-mimicking) virus in 2003 led to heated debates and discussions of the evolution, origin, and (re)definition of viruses [51–53]. With a genome size of ~1.2-Mb, this virus was the largest described virus. Recently, other close relatives like the Mamavirus, Terra2, and the Marseillevirus were discovered and categorized under double-stranded DNA viruses belonging to the nucleocytoplasmic large DNA virus (NCLDV) superfamily [54–56]. One school of thought suggested that the Mimivirus and other giant protist-associated viruses seem to have evolved from a common ancestor distinct from Bacteria, Archaea, and Eukarya and constitute a new domain of life – the Megavirales [57]. The description of Megavirales as a fourth domain of life distinct from the above three comes from a growing body of evidence that indicates that the genomes of these giant viruses are an amalgamation of genes homologous to eukaryotes, bacteria, and archaea acquired by either gene duplication, gene transfers, orthologous gene displacement, and mobile genetic elements [58]. Others have contested the uniqueness and origin of these giant viruses and describe these viruses as “giant pickpockets”. This theory supports “an escape model” where these viruses are described as chimeras created from fragments of cellular chromosomes, which evolved further by stealing genes from their hosts [59–61].
In the Mimivirus, of the 900 genes present, a majority (~700) are poorly conserved orphan genes, but among the remaining 200, only about half have been suggested to result from lateral gene transfers [58]. Many of these genes encode proteins homologous to those involved in human DNA repair pathways. These include UDG, endonuclease IV (Nfo), O6-methylguanine-DNA-methyltransferase (Mgmt), 1-methyladenine-DNA-dioxygenase (AlkB), UV endonuclease (UvdE), mismatch repair enzyme MutS, and two formamidopyrimidine DNA glycosylases, MvNei1 and MvNei2 [26, 51, 62, 63]. The virus may use these DNA repair proteins during viral DNA replication/synthesis in the nucleus of the host.
4.2. Function of MvNei2 as a DNA glycosylase
MvNei2 is one of the two Fpg/Nei DNA glycosylases from Mimivirus and the combination of structural and biochemical data presented here highlights the unique features of this enzyme. Unlike OGG1 and NTH1 of the Nth family, which remove base damage primarily from duplex DNA, the NEIL enzymes are able to cleave lesions in both ds and ssDNA. Thus, ssDNA intermediates occurring transiently during DNA replication and transcription can be processed efficiently by these enzymes [22, 64–66]. Our data indicate that MvNei2 does not display a strong preference for ssDNA over duplex DNA suggesting that this enzyme can function not only on dsDNA, but also at replication centers and in repair processes where ssDNA intermediates are generated. Despite the presence of only one of the three void-filling residues, this enzyme is able to cleave multiple types of DNA lesions. We present evidence for a role for Met72 and adjacent His73, where the latter may have a function similar to that of the “wedge” residue (typically an aromatic residue) in the other Fpg/Nei enzymes [7, 50].
4.3. Concluding Remarks
We present here the first crystal structure of an Nei enzyme with a C-H-C C-type zinc finger. Another novel structural feature presented here is a unique extension in the N-terminal domain of MvNei2. The function of this segment is currently unknown. The extension could play a role in DNA binding or mediating interactions with other proteins. Whether this extension is unique to the mimivirus glycosylase or not will have to await a crystal structure of a eukaryotic Neil2.
Supplementary Material
Highlights
-
▶
We present the first crystal structure of a viral ortholog of the NEIL2/3 enzymes, MvNei2.
-
▶
MvNei2 is unique because it possesses an N-terminal valine like NEIL3, but a C-terminal C-H-C-C type zinc-finger like NEIL2.
-
▶
MvNei2 lacks two of the three canonical void-filling residues like mouse Neil3.
-
▶
Mutation of the only void-filling methionine in MvNei2 (M72A) renders an enzyme with impaired glycosylase activity.
Acknowledgments
We thank Dr. Cynthia J. Burrows, University of Utah, for providing oligodeoxyribonucleotides containing spiroiminodihydantoin, and guanidinodihydantoin and Dr. Carmelo Rizzo, Vanderbilt University, for the oligodeoxynucleotide containing MeFapyG; Drs. Pierre Aller and Karl Zahn for data collection at the Diamond Light Source, beamline I04, UK; Dr. Mark Rould for helpful crystallographic suggestions; Dr. Robyn Maher, Dr. Jeffrey Bond, and Brittany Carroll for technical advice and support. This work was supported by National Institutes of Health Grant P01CA098993 awarded by the National Cancer Institute. AP was also supported by a J. Walter Juckett post-doctoral fellowship awarded by the Lake Champlain Cancer Research Organization and the Vermont Cancer Center (VCC). Support from the VCC is acknowledged. Financial Support for the Diamond Light Source comes mainly from two shareholders, the UK Government through the Science and Technology Facilities Council (STFC), and the charity, The Wellcome Trust.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
None
References
- [1].Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327:73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Friedberg EC, Walker GC, Seide W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Second Edition ASM Press; Washington, D.C.: 2006. [Google Scholar]
- [3].Duclos SDS, Wallace SS. Consequences and Repair of Oxidative DNA Damage. In: Helmut Greim RA, editor. The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens. The Royal Society of Chemistry; 2012. [Google Scholar]
- [4].David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Fromme JC, Banerjee A, et al. Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14:43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- [6].Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prakash A, Doublié S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog Mol Biol Transl Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair (Amst) 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- [9].Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- [11].Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- [12].Morland I, Rolseth V, Luna L, Rognes T, Bjørås M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- [14].Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- [15].Grin IR, Zharkov DO. Eukaryotic endonuclease VIII-like proteins: new components of the base excision DNA repair system. Biochemistry (Mosc) 2011;76:80–93. doi: 10.1134/s000629791101010x. [DOI] [PubMed] [Google Scholar]
- [16].Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural Characterization of a Mouse Ortholog of Human NEIL3 with a Marked Preference for Single-Stranded DNA. Structure. 2013;21:247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace SS, Doublié S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012;11:714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- [19].Sung RJ, Zhang M, Qi Y, Verdine GL. Structural and biochemical analysis of DNA helix-invasion by the bacterial 8-oxoguanine DNA glycosylase MutM. J Biol Chem. 2013 doi: 10.1074/jbc.M112.415612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc Natl Acad Sci U S A. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjørås M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013 doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst) 2007;6:1629–1641. doi: 10.1016/j.dnarep.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Imamura K, Wallace SS, Doublié S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Imamura K, Averill A, Wallace SS, Doublié S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J Biol Chem. 2012;287:4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- [31].Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, Rizzo CJ. Site-specific synthesis and characterization of oligonucleotides containing an N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylfor mamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem Res Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- [33].Otwinowski ZM, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- [34].Kabsch W. Xds, Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- [36].Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- [37].Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- [38].Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner RD, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2010;66:1153–1163. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- [45].Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Das A, Rajagopalan L, Mathura VS, Rigby SJ, Mitra S, Hazra TK. Identification of a zinc finger domain in the human NEIL2 (Nei-like-2) protein. J Biol Chem. 2004;279:47132–47138. doi: 10.1074/jbc.M406224200. [DOI] [PubMed] [Google Scholar]
- [47].Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462:762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Keppetipola N, Shuman S. Mechanism of the phosphatase component of Clostridium thermocellum polynucleotide kinase-phosphatase. RNA. 2006;12:73–82. doi: 10.1261/rna.2196406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006;45:12039–12049. doi: 10.1021/bi060663e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- [52].Claverie JM, Ogata H, Audic S, Abergel C, Suhre K, Fournier PE. Mimivirus and the emerging concept of “giant” virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- [53].Claverie JM, Abergel C. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet. 2010;26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- [54].Filee J, Chandler M. Convergent mechanisms of genome evolution of large and giant DNA viruses. Res Microbiol. 2008;159:325–331. doi: 10.1016/j.resmic.2008.04.012. [DOI] [PubMed] [Google Scholar]
- [55].Filee J, Pouget N, Chandler M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by Nucleocytoplasmic large DNA viruses. BMC Evol Biol. 2008;8:320. doi: 10.1186/1471-2148-8-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Colson P, Yutin N, Shabalina SA, Robert C, Fournous G, La Scola B, Raoult D, Koonin EV. Viruses with more than 1,000 genes: Mamavirus, a new Acanthamoeba polyphaga mimivirus strain, and reannotation of Mimivirus genes. Genome Biol Evol. 2011;3:737–742. doi: 10.1093/gbe/evr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Colson P, de Lamballerie X, Fournous G, Raoult D. Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales. Intervirology. 2012;55:321–332. doi: 10.1159/000336562. [DOI] [PubMed] [Google Scholar]
- [58].Filee J, Chandler M. Gene exchange and the origin of giant viruses. Intervirology. 2010;53:354–361. doi: 10.1159/000312920. [DOI] [PubMed] [Google Scholar]
- [59].Forterre P. Giant viruses: conflicts in revisiting the virus concept. Intervirology. 2010;53:362–378. doi: 10.1159/000312921. [DOI] [PubMed] [Google Scholar]
- [60].Moreira D, Lopez-Garcia P. Comment on “The 1.2-megabase genome sequence of Mimivirus”. Science. 2005;308:1114. doi: 10.1126/science.1111195. author reply 1114. [DOI] [PubMed] [Google Scholar]
- [61].Moreira D, Lopez-Garcia P. Ten reasons to exclude viruses from the tree of life. Nat Rev Microbiol. 2009;7:306–311. doi: 10.1038/nrmicro2108. [DOI] [PubMed] [Google Scholar]
- [62].Chen R, Wang H, Mansky LM. Roles of uracil-DNA glycosylase and dUTPase in virus replication. J Gen Virol. 2002;83:2339–2345. doi: 10.1099/0022-1317-83-10-2339. [DOI] [PubMed] [Google Scholar]
- [63].Suzan-Monti M, La Scola B, Raoult D. Genomic and evolutionary aspects of Mimivirus. Virus Res. 2006;117:145–155. doi: 10.1016/j.virusres.2005.07.011. [DOI] [PubMed] [Google Scholar]
- [64].Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.