Summary
Diffuse large B-cell lymphoma (DLBCL) is the most common form of non-Hodgkin's lymphoma (NHL) with the greatest challenge for improving patient survival being the management of chemo-refractory disease upon relapse. Epigenetic dysregulation has been correlated with more aggressive malignancies and chemo-resistance. In this issue of Cancer Discovery, Clozel and colleagues demonstrate the potential for low-dose DNA methyltransferase inhibitors (DNMTIs) as both a rational and effective neo-adjuvant approach for chemo-sensitization in DLBCL.
Diffuse large B-cell lymphoma (DLBCL) is an aggressive and heterogeneous disease generally treated with the anthracycline based regimen, R-CHOP. Chemo-resistance remains the primary obstacle for the eradication of DLBCL, which may be conferred through either intrinsic mechanisms related to enhanced drug inactivation/transport or acquired resistance through altered gene expression that prevents cell death. Epigenetic mechanisms have been explored in the context of acquired drug resistance as epigenetic modification can result in altered gene expression without altering the DNA sequence itself. Epigenetics can be defined as the regulation of DNA architecture and the accessibility of chromosomal DNA to transcription factors, which is influenced by how compactly (heterochromatin) or relaxed (euchromatin) the DNA is coiled around the nucleosome. DNA methylation may influence gene expression by either directly interfering with transcription factor binding to gene promoters or by encouraging the recruitment of HDACs by methyl-CpG binding proteins, thus remodeling the chromatin structure.
In this issue of Cancer Discovery, Clozel and colleagues expand upon previous studies to establish DNA methylation as a critical component of epigenetic dysregulation during DLBCL lymphomagenesis as well as a rational epigenetic mechanism to target in chemo-refractory DLBCL; much like acetylation and methylation of his tones have previously been shown (1-3). The authors utilized gene expression and DNA methylation profiling to compare eight doxorubicin-resistant cell lines, which demonstrated no significant signs of intrinsic drug resistance, to six doxorubicin-sensitive cell lines. They hypothesized that the acquired resistance was mediated through epigenetic silencing of genes that contribute to doxorubicin resistance. Furthermore, Clozel and colleagues conducted a phase I clinical study evaluating the capacity for 5-aza-2′-deoxycytidine, decitabine (DAC) to prime potentially chemo-refractory patients for doxorubicin treatment in a personalized manner. Their hypothesis was founded on previous studies that have demonstrated both recurrent aberrant epigenetic mechanisms in DLBCL as well as potential efficacy of epigenetic-based therapies. As a related matter, both his tone acetylation and methylation in the context of DLBCL will be briefly discussed as these epigenetic events are physically and functionally related to DNA methylation and exemplify personalized epigenetic therapies in DLBCL (2-4).
Acetylation of his tones on conserved lysine residues in the N-terminal tail or on the core of the nucleosome removes a positive charge, generally resulting in relaxed, transcriptionally active DNA.HDACs catalyze the removal of acetyl groups back to coenzyme A, thus repressing gene expression. Methyl-CpG-binding proteins, MBD2 and MeCP2, are able to recruit HDACs to the site of CpG island methylation, which can result in enhanced gene suppression (5). Nearly 70% of DLBCLs overexpress BCL6, a corepressor of numerous genes including EP300, a putative tumor suppressor (2).The differentiation of germinal center (GC) B-cells can be inhibited by BCL6 proteins, which regulate and coordinate plasmacytic differentiation in conjunction with HDACs and transcription factors such as BLIMP1, PAX4 and XBP1. The Melnick group previously identified the BCL6-EP300 axis through ChIP-on-chip experiments and has subsequently demonstrated the efficacy of HDAC inhibitors inducing cell death in GCB-DLBCLs(2). Although HDACIs have yet to be explored as chemo-sensitizers in DLBCL, they have demonstrated potential as potent chemo- and radio-sensitizers in cell lines from lung, breast, ovary, esophageal, gastric, colon, thyroid, prostate and pancreatic cancers and appear to be rational therapies for DLBCLs of GC origin (6).
Histone methylation, especially H3K9 and H3K27, is generally associated with transcriptional repression and often concurrent with deacetylation. EZH2 is the catalytic component of the polycomb repressive complex 2 (PRC2) responsible for the methylation of H3K27 and repression of select genes. EZH2 mutations on residues Y641 and A677were identified in 22% of DLBCL and 10% of follicular lymphoma (FL) patient samples resulting in altered substrate preference and enhanced di- and trimethylation of H3K27 (3). A recent study demonstrated a correlation between H3K27me3 mediated silencing through EZH2 overexpression, inactivation of pathways such as TGF-β and chemo-resistance in serous ovarian cancer (7).McCabe and colleagues recently employed the SAM-competitive small molecular inhibitor, GSK126, and demonstrated its selectivity for EZH2 activity in GCB-DLBC Land its ability to reactivate PRC2 suppressed genes.Pharmacological inhibition of EZH2 has yet to be explored in the context of chemo-sensitization in DLBCL but studies suggest that combinatorial epigenetic or neoadjuvant approaches may be rational for some hematological malignancies.
DNA hypermethylation on CpG islands located in the promoter of tumor suppressors can be aberrantly methylated by DNMT1, DNMT3A and DNMT3B. A replication-coupled passive DNA demethylation process has been described but the mechanism behind active DNA demethylation remains elusive. While no enzyme has been attributed to the catalysis of DNA demethylation, some research suggest that hydroxylmethylation of 5-methylcytosine (5mC) to 5hmC may be an intermediate for the removal of methylated cytosine (8).TET proteins are capable of promoting demethylation by catalyzing 5mC to 5hmC and a recent study employing genome-wide profiling identified DNA hypermethylation signatures associated with DLBCLs harboring inactivating mutations in TET2 (9).
Clozel and colleagues utilized the pyrimidine nucleoside analogs DAC to demonstrate a mechanism for chemo-sensitization of DLBCL (1). DAC and 5-azacytine were initially utilized because of their intrinsic cytotoxic properties but are now known to be DNA hypomethylating agents through their irreversible inhibition of DNMTs. During cellular replication, inhibition of efficient incorporation of methyl groups into newly synthesized DNA results in hypomethylation and global reactivation of genes. Clozel and colleagues exploited this event in rapidly dividing tumor cells believing they would be more susceptible to this augmented replication-coupled passive DNA demethylation. Their hypothesis was that low dose DAC would result in the reactivation of tumor suppressor genes and pathways necessary for chemo-sensitization. Previous studies by the Melnick group have demonstrated that DNMT1, which primarily maintains DNA methylation due to its preference for hemimethylated DNA, is highly expressed in normal GC-B cells (10). In addition to the genetic heterogeneity induced by AICDA mediated mutagenesis during somatic hypermutation, GC B-cells are characterized by DNA methylation heterogeneity. This diverse repertoire of GC B-cells is under considerable pressure to maintain homogeneous DNA methylation patterning in all clonagenic progeny during rapid proliferation and clonal expansion, which requires high expression of DNMT1. Clonal populations may acquire aberrant DNA methylation, resulting in the inactivation of tumor suppressor genes and pathways, thus conferring a malignant phenotype.
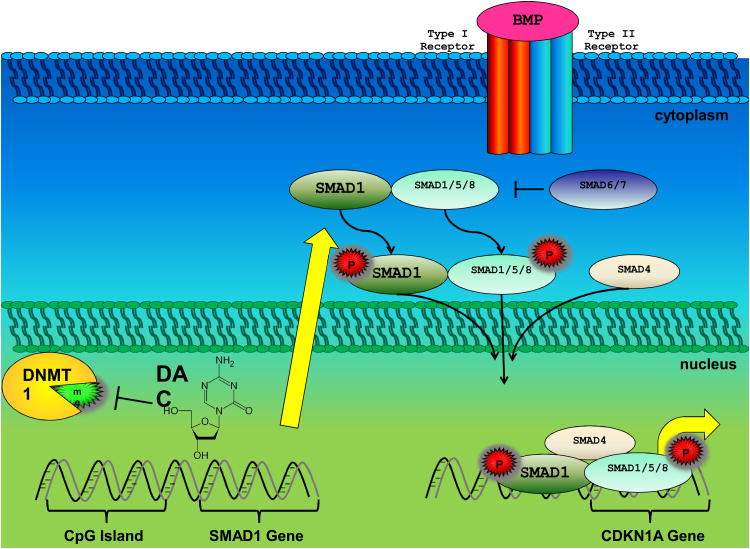
In their study, Clozel and colleagues identified SMAD1 inactivation mediated through aberrant DNA methylation and as the primary culprit conferring chemo-resistance in DLBCL (1).Interestingly, this finding agrees with previous observations that disruption of TGF-β related pathway through epigenetic mechanisms results in more stem-like tumors, resistant to chemotherapies (7).Conversely, activation of TGF-β signaling has been established as a critical mediator of epithelial to mesenchymal transition (EMT) in numerous solid tumors, resulting in a more metastatic and chemo-refractory phenotype (11). This TGF-β paradox has been attributed the ability for specific SMAD trimers to associate with distinct transcription factors and DNA sequences, thus allowing each SMAD to regulate a unique repertoire of genes. SMAD1, along with SMAD5, mediates TGF-β related BMP-dependent pathways of which tumor suppressor CDKN1A is a target gene (12). With the exception of indolent FL, SMAD1 activation upon TGF-β stimulation rarely occurs in B-cell malignancies and overexpression of SMAD1 in DLBCL results in anti-proliferative effects (13). Furthermore, Clozel and colleagues observed the induction of the tumor specific senescence with incomplete growth arrest (SWING) program upon SMAD1 induction and subsequent activation of DNA damage response gene CDKN1A (1,14). Depicted in Figure 1 is a model Clozel et al. proposed for the mechanism-based epigenetic chemosensitization of diffuse large B cell lymphoma.
Figure 1.
A depiction of the model Clozel et al. proposed for the mechanism-based epigenetic chemosensitization of diffuse large B cell lymphoma upon low dose 5-aza-2-deoxycytidine, decitibine (DAC) treatment (1). Acquired resistance to anthracyclines may occur through DNMT1 methylation of CpG islands located in the promoter of genes such as SMAD1. Hypomethylation and reactivation of genes occurs after inhibition of DNMT1 by DAC. This hypomethylation, which is augmented in rapidly dividing cells, results in the reactivation of genes (e.g. SMAD1). SMAD1 confers chemosensitization upon its phosphorylation, trimerization, translocation into the nucleus and transcription of target genes including CDKN1A.
While many clinical trials employing the maximally tolerated dose are demonstrating a low therapeutic index for epigenetic therapies, low dose administration of these agents is showing great promise in their ability to “prime” tumors for subsequent chemotherapy. In addition, the combination of HDACIs with demethylating agents has become a very attractive approach due to the physical and functional relationship between his tones and DNA (15).This collective body of work demonstrates the utility of epigenetic agents in combination with current regimens in the treatment of refractory diseases such as DLBCL and these advances may serve as an overture to the modified regimens that may be pursued more extensively in the near future. We eagerly anticipate the results of ongoing trials in chemo-refractory DLBCLs incorporating these epigenetic-based neo-adjuvant approaches.
References
- 1.Clozel T, Yang SN, Elstrom RL, Tam W, Martin P, Kormaksson M, et al. Mechanism-Based Epigenetic Chemosensitization Therapy of Diffuse Large B Cell Lymphoma. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010 doi: 10.1172/JCI42869. doi: pii: 42869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;7427:108–12. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 4.Zhu WG, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anticancer Agents. 2003;3:187–99. doi: 10.2174/1568011033482440. Review. [DOI] [PubMed] [Google Scholar]
- 5.Müller-Tidow C, Kügler K, Diederichs S, Klümpen S, Möller M, Vogt U, et al. Loss of expression of HDAC-recruiting methyl-CpG-binding domain proteins in human cancer. Br J Cancer. 2001;8:1168–74. doi: 10.1054/bjoc.2001.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis L, Pili R. Histone Deacetylase Inhibitors: Advancing Therapeutic Strategies in Hematological and Solid Malignancies. Pharmaceuticals (Basel) 2010;8:2411–69. doi: 10.3390/ph3082441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman-Rothe N, Curry E, Zeller C, Liber D, Stronach E, Gabra H, et al. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene. 2012 doi: 10.1038/onc.2012.477. [DOI] [PubMed] [Google Scholar]
- 8.Guibert S, Weber M. Functions of DNA methylation and hydroxymethylation in mammalian development. Curr Top Dev Biol. 2013;104:47–83. doi: 10.1016/B978-0-12-416027-9.00002-4. [DOI] [PubMed] [Google Scholar]
- 9.Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, Nielsen Ab, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013 doi: 10.3324/haematol.2013.088740. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood. 2011;(13):3559–69. doi: 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;34:4741–51. doi: 10.1038/onc.2010.215. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamato K, Hashimoto S, Imamura T, Uchida H, Okahashi N, Koseki T, et al. Activation of the p21(CIP1/WAF1) promoter by bone morphogenetic protein-2 in mouse B lineage cells. Oncogene. 2001;32:4383–92. doi: 10.1038/sj.onc.1204572. [DOI] [PubMed] [Google Scholar]
- 13.Munoz O, Fend F, de Beaumont R, Husson H, Astier A, Freedman AS. TGFβ-mediated activation of Smad1 in B-cell non-Hodgkin's lymphoma and effect on cell proliferation. Leukemia. 2004;12:2015–25. doi: 10.1038/sj.leu.2403485. [DOI] [PubMed] [Google Scholar]
- 14.Sherman MY, Meng L, Stampfer M, Gabai VL, Yaglom JA. Oncogenes induce senescence with incomplete growth arrest and suppress the DNA damage response in immortalized cells. Aging Cell. 2011;6:949–61. doi: 10.1111/j.1474-9726.2011.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu WG, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anticancer Agents. 2003;3:187–99. doi: 10.2174/1568011033482440. Review. [DOI] [PubMed] [Google Scholar]