Abstract
Oxidative stress in humans causes damage to biomolecules by generating reactive oxygen species (ROS). DNA can be oxidatively damaged by ROS, which may lead to carcinogenesis. Here we report a microfluidic electrochemical array designed to rapidly detect oxidation in intact DNA in replicate measurements. Sensor arrays were fabricated by wet-chemistry patterning of gold compact discs. The 8-sensor array is incorporated into a 60 µL microfluidic channel connected to a pump and sample valve. The array features 7 nm thick osmium bipyridyl poly(vinylpyridine) chloride [Os(bpy)2(PVP)10Cl]+ films assembled layer-by-layer with polyions onto the gold sensors. 7,8-dihydro-8-oxoguanine (8-oxodG) in ds-DNA is selectively oxidized by [Os(bpy)2(PVP)10Cl]+ in intact ds-DNA to provide catalytic square wave voltammograms (SWV). The device is easy-to-use, fast, inexpensive, reusable and can detect one 8-oxodG per 6600 nucleobases. The mass detection limit is 150-fold lower than a previously reported dip-and-read voltammetric sensor for oxidized DNA. Fast assays (<1 min) and moderate sample consumption (15 pmol DNA) suggest potential for research and clinical applications. Practical use is illustrated by detecting DNA oxidation from cigarette smoke and ash extracts in dispersions with NADPH and Cu2+.
Introduction
Oxidative stress in living organisms is an imbalance of the oxidant/antioxidant ratio in favor of oxidants. This condition can lead to oxidative damage to biomolecules like DNA, lipids, and proteins.1 Oxidative stress is implicated in cancer, aging, inflammation, and other conditions. 2-4 DNA oxidation can be facilitated by exposure to pollutant and drug metabolites. In particular, quinone moieties can undergo redox cycling induced by NADPH and metal ions in the body. Such processes generate reactive oxygen species (ROS) including singlet oxygen, superoxide, and hydroxyl radicals1,5,6 that oxidize DNA, 7,8 in a process that may be coupled with DNA adduct formation.9 NADPH in some tissues is estimated at 100–200 µM,10 and CuII is present in significant amounts as nuclear histone complexes and bound to N7 positions of guanines on DNA.11 DNA oxidation by such pathways may exceed the capacity of human repair mechanisms and facilitate disease.12 Thus, measurement of oxidized DNA can be used to screen disease development and environmental exposure.13-15
Guanine is the most easily oxidized DNA base.16,17 Oxidation product 8-hydoxy-7,8-hydro-2'-deoxyguanosine (8-oxodG),8,18 is an important biomarker for human oxidative DNA damage. A number of analytical methods for 8-oxodG have been reported, but many require hydrolysis of the DNA.19 We previously developed electrochemical and electrochemiluminescent biosensors that detect 8-oxodG in intact ds-DNA.20-22 These sensors measure oxidized DNA in solution in dip-and-read mode or in DNA films. They employ Os(bpy)2Cl+-polyvinylpyridine (OsPVPCl) metallopolymer films on pyrolytic graphite electrodes that selectively oxidize 8-oxodG in DNA to provide catalytic voltammetric peaks or electrochemiluminescence (ECL). Selectivity for 8-oxodG is high because OsPVPCl with redox potential 0.4 V vs. Ag/AgCl (pH 5.5) can electrocatalytically oxidize 8-oxodG, but does not oxidize the natural nucleobases that have much more positive oxidation potentials.20,23
8-OxodG has been measured after DNA hydrolysis by using high performance liquid chromatography with electrochemical detection (HPLC-ECD),24,25 gas chromatography-mass spectrometry (GC-MS) 26, and HPLC-MS/MS. 27, 28 However, 8-oxodG may form during sample pretreatment in some protocols,19,29,30 and further oxidation may destroy 8-oxodG.31,32 Oxidative DNA lesions can be measured without hydrolysis by using specific antibodies along with Comet assays or enzyme-linked immunosorbent assay (ELISA)33-36. Comet assays are relatively nonspecific, and do not directly yield concentrations of 8-oxodG. In ELISA, antibodies may be cross-reactive, and this has been cited as the cause of differences in results between ELISA and HPLC- ECD.37-39
In the present paper, we describe an inexpensive, wet etched gold CD electrochemical array40 coated with OsPVPCl in a microfluidic channel41 to measure 8-oxodG directly in samples of intact DNA in a single-step assay. This device is faster, provides 150-fold better mass detection limit in replicate readings, and requires less sample than our previous dip-and-read voltammetric sensor. Calibration was achieved using oxidized DNA standards with 8-oxodG concentrations measured by LC-MS/MS. Application of the sensor array is illustrated by measuring oxidative DNA damage caused by cigarette smoke and ash extracts combined with NADPH and Cu2+.
Experimental Section
Chemicals and Materials
Polyguanylic acid potassium salt (polyG), deoxyguanosine (dG), 8-oxoguanosine (8-oxodG) and salmon testes (ST) ds-DNA (2000 base pairs avg., 41.2 % G/C), poly(diallyldimethylammonium chloride) (PDDA), poly(sodium 4-styrenesulfonate) (PSS), CuCl2, MgCl2, catechol, 1,2-naphthoquinone (1,2-NPQ), K2OsCl6, D-glucose 6-phosphate sodium salt (G6P), β-nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP+), glucose-6- phosphate dehydrogenase (G6PDH) were from Sigma. Water was treated with a Hydro Nanopure system to specific resistance > 16 mΩ-cm. Synthesis and characterization of [Os(bpy)2(PVP)10Cl]+ (bpy = 2,2'-bipyridyl; PVP = poly(4-vinylpyridine)) followed literature procedures.42 Film voltammetry and UV spectrum of this materials were consistent with [Os(bpy)2(PVP)10Cl]+ (Figures S1, and S2)
Sensor Array Fabrication
The gold CD microwell array was fabricated by wet chemistry etching and printed insulation as reported previously.40 The 8 gold sensor elements reside in the bottoms of 1 µL microwells formed by heat transferring a computer laserjet-printed pattern. The microwells facilitate modification of the sensor surfaces for specific assays, as previously described for immunosensing.40 To detect oxidized DNA, sensor elements were coated with a monolayer of 3-mercaptopropionic acid (MPA), and [Os(bpy)2(PVP)10Cl]+ was immobilized on the MPA monolayers between layers of PSS by layer-by-layer electrostatic self-assembly, as reported previously.20,21 Films are denoted in the order of layer deposition as PDDA/PSS/OsPVPCl/PSS. The microfluidic detection chamber consists of a molded poly(dimethoxy)silane (PDMS) slab with 60 µL rectangular microchannel sandwiched between two hard poly(methylmethacrylate) (PMMA) plates. 43 An Ag/AgCl wire reference and a Pt wire counter electrode are inserted in the top plate running along the full channel length. A syringe pump (Harvard Apparatus) and injection valve with 100 µL sample loop are connected in series to the inlet of the detection chamber.
Film deposition was monitored at formation of each layer by using a Quartz Crystal Microbalance (QCM, USI Japan) with 9 MHz QCM quartz resonators (AT-cut, International Crystal Mfg. Co.) The gold coated resonator (0.16 ± 0.01 cm2) was treated with 0.5 mM 3- mercaptopropionic acid (MPA) in ethanol to create a negatively charged monolayer on the surface of the gold. The same films used for the sensor electrodes were assembled on the gold resonators, and frequency change was measured at room temperature after drying to monitor layer mass. 44
Voltammetric Analysis
Square wave voltammetry (SWV) was done at 4 mV step, 25 mV pulse, and 15 Hz frequency. 10 mM pH 5.5 ammonium acetate buffer with 50 mM NaCl was first pumped into the microfluidic chamber at 240 µL min-1. Then, the oxidized DNA sample (containing 50 mM NaCl) was loaded into the 100 µL loop in the injector valve and injected at the same flow rate. At the time when the sample solution completely filled the detection microchannel, estimated previously with dyes, flow was stopped and SWV was measured simultaneously on all sensors by using an 8-electrode CHI 1040b electrochemical workstation.43
Oxidation of deoxyguanosine and DNA
PolyG (0.04 mg/mL) and DNA (0.2 mg/mL) in 10 mM pH 5.5 acetate buffer with 50 mM NaCl was oxidized by Fenton's reagent containing 1.4 mM FeSO4 and 40 mM H2O2 at ambient temperature (22 °C) with constant stirring. Then, 1 mL aliquots were taken every few seconds from the reaction mixture and frozen immediately in liquid nitrogen, followed by freeze drying. For electrochemical analysis, samples were reconstituted in 1 mL of 10 mM pH 5.5 acetate buffer with 50 mM NaCl.
HPLC-MS/MS
The freeze-dried oxidized DNA samples were purified by ethanol precipitation to eliminate the inorganic ions, 45and then were reconstituted in 1 mL Tris buffer, pH 7.4. The DNA was later hydrolyzed by incubating with deoxyribonuclease I (400 unit mg-1 of DNA) for 3 h, followed by adding phosphodiesterase I (0.2 unit mg-1 of DNA) and alkaline phosphatase (1.2 unit mg-1 of DNA) for 12 h in microcentrifuge tubes at 37 °C. Hydrolyzed DNA samples were vacuum filtered using a 96-well filtration plate (3KDa MW cutoff).46 Samples were analyzed using a Waters 2690 high-performance liquid chromatograph (HPLC) interfaced to an Applied Biosystems 4000 Q TRAP® triple quadrupole linear ion trap mass spectrometer equipped with an electrospray ionization source. An Agilent Zorbax SB-C18 (4.6 mm × 150 mm, id, 5 µm particle size) reversed phase analytical column was used with 10 mm C18 guard column. The mobile phase was 10 mM ammonia acetate buffer, pH 4.0 (solvent A) and methanol + 0.1% formic acid (solvent B). Gradient was isocratic at 10% B for 40 min at 0.8 mL/min, and the ionization source was positive electrospray. The method was validated by spiking 0.04 mg/mL deoxyguanosine with 8-oxodG (stock solution 10 µM), followed by HPLC-MS/MS detection by multiple reaction monitoring (MRM).28
Oxidation of DNA by cigarette smoke and ash
Cigarette smoke from a full cigarette was trapped in a cotton plug in a syringe by pulling smoke into the syringe from the lit cigarette. The cotton plug was then extracted with 1 mL dimethyl sulfoxide (DMSO). Cigarette particulate ash after combustion was dispersed in 1 mL DMSO and filtered. 47 DNA (0.4 mg/mL) was incubated with catechol, 1,2-naphthoquinone, cigarette smoke extract or cigarette ash after adding 100 µM Cu2+ and an NADPH regeneration system consisting of 1 U/mL G6PDH enzyme, 2.5mM G6P, 0.5 mM NADP+, 1 mM Mg2+, in 50mM pH 7.4 Tris buffer at 37 °C. 48
Results
Film Characterization by Quartz Crystal Microbalance
Adsorbed mass/area (M/A) of each deposited, dried polyion layer during fabrication of PDDA/PSS/OsPVPCl/PSS films was obtained from the frequency change (ΔF) on a QCM resonator using the Sauerbrey equation: 49
| (1) |
Nominal thickness (d) for films deposited on one side of the resonator was estimated using an expression confirmed previously by high-resolution electron microscopy assuming average polymer density 1.3 g cm-3: 50
| (2) |
Linear relationships of -ΔF versus layer number indicated reproducible increases in mass (Figure S3) with small standard deviations indicating that the metallopolymer was successfully immobilized with good reproducibility. 49 Mass and thickness of layers of PDDA, PSS and OsPVPCl calculated from QCM frequency differences (ΔF) are in the supporting information file, Table S1. Average OsPVPCl in the films was 1.95±0.14 µg cm-2 and nominal film thickness was 14.4±1.6 nm.
Stability
Figure 1a shows 40 repetitive multiple-scan cyclic voltammograms (CVs) of a PDDA/PSS/OsPVPCl/PSS film on the gold-CD array with excellent reproducibility. We tested long-term stability by incubating the thin film up to 8 hours in DNA solutions without Fenton's reagent, and measuring final to initial current ratios. The oxidation-reduction peak current decreased to 95.3 % of initial response after 8 hr incubation. When arrays were stored dry at 4 °C for 2 weeks, responses were within 85 % of initial responses. Arrays used in this study were stored for less than 1 week.
Figure 1.

Stability and reproducibility of OsPVPCl films and oxidized DNA assays: (a) Forty segments of CV at 50 mV s-1 of PDDA/PSS/OsPVPCl/PSS film on one sensor in the array in pH 5.5 buffer containing 50 mM NaCl, (b) Difference SWVs of 8 sensors in the array coated with PDDA/PSS/OsPVPCl/PSS films after introducing an oxidized DNA sample made by reacting DNA with Fenton's reagent for 60 s. (Baselines are offset to facilitate labeling)
Oxidation-reduction peak separation of CVs, ΔEp (= Epc - Epa), was 36 ± 2 mV, and peak current ratio (Ipa/Ipc) was 1.05 ± 0.02 (Epc = cathodic peak potential, Epa = anodic peak potential, Ipc = cathodic peak current, Ipa = anodic peak current). The midpoint potential of PDDA/PSS/OsPVPCl/PSS film is 0.17 V vs. Ag/AgCl (0.05 M NaCl). This value is converted to 0.23 V vs. SCE by using the Nernst Equation, which is close to the value of 0.34 V reported for OsPVP films on glassy carbon electrodes in acetonitrile.42b We found a formal potential of 0.24 V for PSS/OsPVP/PSS films on a pyrolytic graphite electrode vs. a true SCE electrode (Figure S2, SI) Voltammetric characteristics are consistent with non-ideal quasireversible surface voltammetry.51 Figure 1b shows SWV responses of all 8 sensors in the array to a sample of oxidized DNA. The catalytic peaks had relative standard deviation of ±4.4%.
Measurements on oxidized polyG
Array performance was first investigated by using poly(guanosine) (polyG) oxidized by Fenton's reagent to form 8-oxodG in polyG strands (see Experimental Section). Oxidized polyG solutions were injected into the detection chamber, flow was stopped when the chamber was full, and the SWV was obtained at 0.22 V vs. Ag/AgCl (50 mM NaCl). Increases in peak current over the control were found due to the electrochemical oxidation of OsIIPVPCl to OsIIIPVPCl in the sensor film (Figure 2a), followed by selective catalytic oxidation of 8-oxodG in oxidized polyG by OsIIIPVPCl.20 Longer periods of oxidation of polyG produced increases in peak current suggesting increased amounts of 8-oxodG (Figure 2b). Controls showed that peak currents are the same in buffer alone and after exposure to control polyG solutions that had been treated with only Fe2+ or only H2O2. Relatively small error bars demonstrate good reproducibility.
Figure 2.

Difference SWV at pH 5.5 in the microfluidic array for 0.04 mg mL-1 polyG incubated with Fenton's reagent. Controls are 0.04 mg mL-1 polyG treated with Fenton's reagent lacking Fe2+: (a) sensor responses; (b) ratio of final to initial SWV peak currents in solution of polyG after oxidation with Fenton's reagent. (Error bars represent standard deviations, n=8.)
Detection of Oxidized DNA
Array performance was then tested for oxidized DNA made by incubating DNA with Fenton's reagent for specific times. Controls were made by incubating DNA with Fe2+ and buffer without H2O2, or with H2O2 but without Fe2+. As with oxidized polyG, oxidized DNA samples gave peak current increases with oxidation time. Peak heights for these 0.2 mg mL-1 DNA samples were similar to those of the oxidized 0.04 mg mL-1 polyG (Figure 1), consistent with the 20.6% of dG originally in the DNA, giving dG concentration equivalent to that in 0.04 mg mL-1 polyG. No significant response over a zero DNA sample was found for controls. The detection limit of the array was 150 8-oxoG's per 106 nucleobases, estimated as 3 standard deviations above the average SWV peak current of control samples.
HPLC-MS/MS
Amounts of 8-oxodG in DNA oxidized by Fenton's reagent for specific times were measured using HPLC-MS/MS in order to validate the array. Oxidized DNA was purified by ethanol precipitation, enzymatically hydrolyzed, and filtered through a 96-well filtration plate (3KDa molar weight). In LC-MS/MS analysis using MRM with m/z 268→152, a large peak eluted at 6 min corresponding to dG, while the peak at 8 min with m/z 284→168 represents oxidation product 8-oxodG (Figure 4a). Figure 4b shows the ratio of 8-oxodG to the total amount of dG found in the LC-MS/MS assay that increased with oxidation time. These data were used to obtain relative amounts of 8-oxodG for each oxidation time, which were used to construct a calibration graph for the sensor peak current data (Figure 4c). The sensor peak ratio vs. relative 8-oxodG concentration from LC-MS/MS was linear and can be used with the array to quantitate 8-oxodG in unknown DNA samples. The LC-MS/MS limit of detection of 8-oxodG was 0.044 pmol 8-oxodG per 15.2 pmol of DNA (one 8-oxodG per 106 nucleobases.). In unoxidized DNA, 57 8-oxodG per 106 nucleobases were found by HPLC-MS/MS. These data allow the relative number of oxidized nucleobases to be estimated from the array results (Table 2).
Figure 4.

HPLC-MS/MS results for array calibration: (a) HPLC-MS/MS chromatogram in MRM mode obtained for the ions at m/z 268→152 transition (dG) and 284→168 transition (8-oxodG, inset) for 2 mg mL-1 DNA incubated with FeSO4 and H2O2 for 45 s. (b) Ratio of [8-oxoguanine]/{[8-oxoguanine] + [guanine]} for DNA reacted with Fenton's reagent for different time, then hydrolyzed. (c) Calibration curve for sensor current ratio vs. relative amount of 8-oxodG. (Error bars represent standard deviations for three measurements.)
Table 2.
Correlation of SWV current peak ratio and 8-oxodG concentration.
| Sensor current Ratio | 8-oxodG moieties per 106 nucleobases |
|---|---|
| 1.001 | 57 |
| 1.330 | 315 |
| 1.446 | 401 |
| 1.629 | 503 |
| 1.721 | 560 |
| 1.903 | 636 |
| 2.137 | 795 |
| 2.236 | 872 |
Reusability
Sensor response was measured repetitively for oxidized DNA and buffer, with an intermediate buffer wash for 1 min at 240 µL min-1 (4 x channel vol.). Response for the oxidized DNA and the blank remained relatively constant (Supporting Information, Figure S4). The mean response of the 10th oxidized DNA injection was 97.3% of the 1st oxidized DNA response.
Detection in complex sample matrices
Since extraction of DNA from living systems may result in impurities in the samples, we investigated several potential interferences. Oxidized DNA was measuring in pH 5.5 acetate buffer containing 50 mM NaCl, fetal calf serum (1% v/v in buffered saline), human serum (1% v/v in buffered saline), fat-free milk (diluted to 1% with buffered saline and used within 30 min). Solutions were spiked with 200 μL of a solution containing 40 µg oxidized DNA and 50 mM NaCl. Control SWVs is the same media were made by adding 200 µL pH 5.5 buffer + 50 mM NaCl. 52,53Figure 5 shows the relative 8-oxodG concentration obtained from sensor SWV currents using the calibration curve (Figure 4c). Solid bars represents the relative 8-oxodG concentration in each solution spiked with the same amount of oxidized DNA. Reported values represent the mean and standard deviations of measurements from three independent sensors. Recovery expressed as the number of (8-oxodG/106 bases measured in each sample)/ (the number of 8-oxodG/106 bases measured in buffer) × 100% was 99.7% for milk, 97.4% in calf serum and 96.2% in human serum.
Figure 5.

Determination of 8-oxodG in 1 mL pH 5.5 buffer containing 1% fat-free milk, 1% fetal calf serum and 1% human serum spiked with 40 μg of oxidized DNA reacted with Fenton's reagent for 45 s. Control SWVs were measured in the same media without oxidized DNA. The solid bars represent the relative 8-oxodG concentration in each solution spiked with the same amount oxidized DNA. The dashed bars represents controls without spiked oxidized DNA. (Standard deviations are from three independent sensors)
Detection of DNA oxidation induced by cigarette smoke using sensor arrays
Quinones can generate ROS via redox cycling involving NADPH and Cu2+ (Scheme 2).54 Cigarette smoke and ash contain quinones 5 and smoking can lead to oxidative stress and the formation of 8-oxodG.48,55 To evaluate the array to explore this source of DNA damage, catechol and 1,2-naphthoquinone (300 µM) were incubated with DNA in solutions with Cu2+ and an NADPH regeneration system. Sensor array responses indicated an increase in amounts of 8-oxodG with incubation time (Figure 6a). We thus prepared samples by incubating 1 mL 0.4 mg mL-1 DNA solutions in pH 7.4 buffer with 100 µL cigarette smoke extract in DMSO, and 100 µL of cigarette particulate extract from ash in DMSO (see Experimental for details), after adding 100 µM Cu2+ and an NADPH regeneration system. 46 Sensor array results after overnight incubations are consistent with significant 8-oxodG formation from both cigarette smoke and ash in these systems (Figure 6b). The signals for the quinone samples decreased at longer incubation times. This may be due to further oxidation of the 8-oxodG.31 For controls, the catechol, 1, 2-naphthoquinone (1,2-NPQ), cigarette smoke or ash extracts were replaced with DMSO.
Scheme 2.

Possible quinone redox cycling pathway for oxidative DNA damage induced by catechol, NADPH and Cu2+.54
Figure 6.
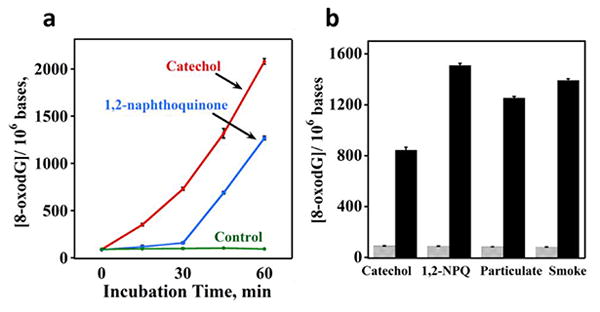
DNA oxidation induced by quinones, cigarette smoke and ash: (a) 8-oxodG per 106 nucleobases for 0.4 mg mL-1 DNA incubated with 300 mM catechol, 1,2-NPQ, or control buffer with 100 mM Cu2+ and NADPH regeneration system at 37°. (b) 8-oxodG formation in DNA solutions incubated with catechol, 1, 2-NPQ, cigarette ash and smoke extracts (black bars) that included 100 mM Cu2+ and NADPH regeneration system at 37°C overnight. In controls (grey bars), quinones and cigarette extracts were replaced with DMSO.
Discussion
Results described above demonstrate the successful fabrication and validation of an electrochemical microwell array to measure 8-oxodG on intact DNA in solution. OsPVPCl films provide catalysis of electrochemical oxidation of 8-oxodG in microwells containing array sensors. The wells eliminate cross-contamination of film-forming solutions and help control uniformity during film deposition.40 The 7 nm thick OsPVPCl films selectively oxidize 8-oxodG20,23 to provide a catalytic SWV response proportional to the amount of 8-oxodG in the DNA.
Peak heights for 0.2 mg mL-1 DNA containing ~ 0.04 mg mL-1 dG were similar to those for oxidized 0.04 mg mL-1 polyG (Figures 2 and 3) for equivalent DNA oxidation times, consistent with selective catalytic oxidation of the 8-oxodG in those samples by OsPVPCl. The films are relatively stable (Figure 1), the array is reusable (Figure S4), and results were reasonably unaffected by several types of interferences (Figure 6). Calibration was effected by making oxidized DNA standards using Fenton's reagent for fixed reaction times and verifying 8-oxodG content using LC-MS/MS (Figure 4).
Figure 3.

Difference SWV peaks for 0.2 mg mL-1 DNA at pH 5.5 incubated in Fenton's reagent. Control is 0.2 mg mL-1 DNA treated with Fenton's reagent lacking Fe2+: (a) sensor responses; (b) ratios of final to initial SWV peak currents for DNA incubated with Fenton's reagent (Error bars represent standard deviations for n=8).
The detection limit of the array for 8-oxodG in oxidized DNA was 150 8-oxodG sites in 106 nucleobases. Thus, the array can detect one 8-oxodG per 6600 intact nucleobases. This is marginally improved over our previously reported single electrode sensor, (1 8-oxodG per 6000 nucleobases).20 However, the new array requires only 20 ug DNA, 150-fold less that the single electrode sensor that required 3 mg DNA samples, and thus the mass detection limit is 150-fold less than the previous sensor. The native DNA used was analyzed by HPLC-MS/MS and contained 57 8-oxodG per 106 nucleobases, consistent with a low level of oxidized bases in natural DNA,56 but below the current detection limit of the sensor array.
In clinical or research applications, amounts of DNA may be limited (e.g., µg DNA extracted from 0.5 cm3 tissue),57 so minimization of sample size is crucial. In HPLC assays, the typical sample size is 50-100 ug.28,58 In the array, about 20 µg DNA (15.2 pmol) is enough to provide 8 replicate measurements. The microchannel housing volume of the detection unit is 60 µL, and could be decreased further by redesign and sensor miniaturization. Unlike most competing methods, the oxidized DNA can be analyzed directly by the array with minimal sample workup, although with tissue samples, simple DNA isolation and purification may be necessary. In addition, the 8-sensor configuration adds versatility to include companion assays in the array, e.g. detection of other types of DNA damage.59
As a practical application, the array was used to measure oxidized DNA induced by extracts of cigarette smoke and ash. These materials contain polycyclic aromatic hydrocarbons (PAHs) and their oxidized derivatives that are suspected carcinogens.48,60 Cigarette smoke extract has been shown to induce oxidative stress in vitro and in vivo.47 Catechols, quinones, and other PAH derivatives in the smoke and ash are suspected to contribute to oxidative DNA damage.61 A general pathway related to cigarette smoke-mediated DNA oxidation is shown in Scheme 2. The quinones in cigarette smoke lead to ROS formation, which is a predominate cause of 8-oxodG formation.
Pure catechol and 1, 2-naphthoquinone (1, 2-NPQ) were used to demonstrate that catecholquinone redox cycling caused oxidation of DNA (Figure 6a), consistent with previous reports.62 Cigarette smoke and particulate ash extracts incubated with DNA in the presence of Cu2+ and the NADPH regeneration system also resulted in the formation of 8-oxoG in the DNA samples (Figure 6b). Results are consistent with oxidative damage of DNA caused by cigarette smoke and ash.
In summary, this paper describes an inexpensive, easily fabricated 8-electrode gold-CD chip in a simple microfluidic array capable of replicate measurements of 8-oxodG, a biomarker for oxidative stress, in intact DNA. Features include low sample consumption, low cost, reusability, and fast analyses (~ 20 s per 8 determinations). These features suggest future applications for toxicity screening as well as clinical and research measurements. Additional sensitivity, which we are currently pursuing, would be an asset in these applications.
Supplementary Material
Scheme 1.

Configuration of the microfluidic voltammetric array. The 8-sensor microwell detection chip is incorporated into a 60 µL PDMS channel (not shown) integrating reference (blue line) and counter (red line) wire electrodes along the channel.
Acknowledgments
This work was supported financially by US PHS grant No. ES03154 from the National Institute of Environmental Health Sciences (NIEHS), NIH, USA.
Footnotes
Supporting Information: The file contains 5 additional figures and one table documenting RuPVPCl characterization and additional control experiments for the oxidized DNA array. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kohen R, Nyska A. Toxicol Pathol. 2002;30:620–650. doi: 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- 2.Hussain SP, Hofseth LJ, Harris CC. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 3.Henchcliffe C, Beal MF. Nat Clin Pract Neuro. 2008;4:600–609. doi: 10.1038/ncpneuro0924. [DOI] [PubMed] [Google Scholar]
- 4.Krohn K, Maier J, Paschke R. Nat Clin Pract Endocrinol & Metabol. 2007;3:713–720. doi: 10.1038/ncpendmet0621. [DOI] [PubMed] [Google Scholar]
- 5.Trachootham D, Alexandre J, Huang P. Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 6.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Free Radical Biol Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oikawa S. Environ Health & Prev Med. 2005;10:65–71. doi: 10.1007/BF02897995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valavanidis A, Vlachogianni T, Fiotakis C. J Environ Sci & Health, Part C. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 9.Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Chem Res Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 10.Malaisse WJ, Hutton JC, Kawazu S, Herchuelz A, Valverde I, Sener A. Diabetologia. 1979;16:331–341. doi: 10.1007/BF01223623. [DOI] [PubMed] [Google Scholar]
- 11.Bryan SE, Vizard DL, Beary DA, LaBiche R, Hardy K. J Nucleic Acids Res. 1981;9:5811–5823. doi: 10.1093/nar/9.21.5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benz CC, Yau C. Nat Rev Cancer. 2008;8:875–879. doi: 10.1038/nrc2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ames BN, Gold LS. Mutat Res-Fund Mol M. 1991;250:3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 14.Ames BN, Gold LS, Willett WC. Proc Nat Acad Sci. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Møller P, Cooke MS, Collins A, Olinski R, Rozalski R, Loft S. Free Radical Res. 2012;46:541–553. doi: 10.3109/10715762.2011.644241. [DOI] [PubMed] [Google Scholar]
- 16.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 17.(a) Yanagawa H, Ogawa Y, Ueno MJ. Biol Chem. 1992;267:13320–13326. [PubMed] [Google Scholar]; (b) Shigenaga MK, Park JW, Cundy KC, Gimeno CJ, Ames BN. Methods Enzymol. 1990;186:521–530. doi: 10.1016/0076-6879(90)86146-m. [DOI] [PubMed] [Google Scholar]; (c) Goyal RN, Jain N, Garg DK. Bioelectrochem Bioenerg. 1997;43:105–114. [Google Scholar]; (d) Duarte V, Muller JG, Burrows CJ. Nucleic Acids Res. 1999;27:496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Shigenaga MK, Ames BN. Free Radical Biol Med. 1991;10:211–216. doi: 10.1016/0891-5849(91)90078-h. [DOI] [PubMed] [Google Scholar]; (b) Lunec J, Holloway KA, Cooke MS, Faux S, Griffiths HR, Evans MD. Free Radical Biol Med. 2002;33:875–885. doi: 10.1016/s0891-5849(02)00882-1. [DOI] [PubMed] [Google Scholar]; (c) Kasai H. Mutat Res. 1997;387:147–163. doi: 10.1016/s1383-5742(97)00035-5. [DOI] [PubMed] [Google Scholar]; (d) Halliwell B. Free Radical Biol Med. 2002;32:968–974. doi: 10.1016/s0891-5849(02)00808-0. [DOI] [PubMed] [Google Scholar]; (e) Gedik CM, Boyle SP, Wood SG, Vaughan NJ, Collins AR. Carcinogenesis. 2002;23:1441–1446. doi: 10.1093/carcin/23.9.1441. [DOI] [PubMed] [Google Scholar]
- 19.Cadet J, Douki T, Ravanat JL, Wagner JR. Bioanal Rev. 2012;4:55–74. [Google Scholar]
- 20.(a) Mugweru A, Rusling JF. Electrochem Commun. 2001;3:406–409. [Google Scholar]; (b) Mugweru A, Wang B, Rusling JF. Anal Chem. 2004;76:5557–5563. doi: 10.1021/ac049375j. [DOI] [PubMed] [Google Scholar]; (c) Rusling JF. In: Electrochemistry of nucleic acids and proteins. Palecek E, Scheller F, Wang J, editors. Elsevier; 2005. pp. 433–450. [Google Scholar]
- 21.Mugweru A, Rusling J. Electroanalysis. 2006;18:327–332. [Google Scholar]
- 22.Dennany L, Forster RJ, White B, Smyth M, Rusling JF. J Am Chem Soc. 2004;126:8835–8841. doi: 10.1021/ja048615+. [DOI] [PubMed] [Google Scholar]
- 23.Ropp PA, Thorp HH. Chem Biol. 1999;6:599–605. doi: 10.1016/s1074-5521(99)80111-2. [DOI] [PubMed] [Google Scholar]
- 24.Lagadu S, Pottier I, Sichel F, Laurent C, Lefaix JL, Prevost V. Biomarkers. 2010;15:707–714. doi: 10.3109/1354750X.2010.511269. [DOI] [PubMed] [Google Scholar]
- 25.Kasai H. J Radiat Res. 2003;44:185–189. doi: 10.1269/jrr.44.185. [DOI] [PubMed] [Google Scholar]
- 26.Cadet J, Douki T, Ravanat JL. Mutat Res-Fund Mol M. 2011;711:3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 27.Serrano J, Palmeira CM, Wallace KB, Kuehl DW. Rapid Commun Mass Sp. 1996;10:1789–1791. doi: 10.1002/(SICI)1097-0231(199611)10:14<1789::AID-RCM752>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 28.Taghizadeh K, McFaline JL, Pang B, Sullivan M, Dong M, Plummer E, Dedon PC. Nat Protocols. 2008;3:1287–1298. doi: 10.1038/nprot.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ravanat JL, Turesky RJ, Gremaud E, Trudel LJ, Stadler RH. Chem Res Toxicol. 1995;8:1039–1045. doi: 10.1021/tx00050a007. [DOI] [PubMed] [Google Scholar]
- 30.Hamberg M, Zhang LY. Anal Biochem. 1995;229:336–344. doi: 10.1006/abio.1995.1422. [DOI] [PubMed] [Google Scholar]
- 31.White B, Smyth MR, Stuart JD, Rusling JF. J Am Chem Soc. 2003;125:6604–6605. doi: 10.1021/ja0343252. [DOI] [PubMed] [Google Scholar]
- 32.White B, Tarun MC, Gathergood N, Rusling JF, Smyth MR. Molec BioSys. 2005;1:373–381. doi: 10.1039/b511756a. [DOI] [PubMed] [Google Scholar]
- 33.Yin B, Whyatt RM, Perera FP, Randall MC, Cooper TB, Regina M, Santella RM. Free Radical Bio Med. 1995;18:1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- 34.Garratt LW, Mistry V, Singh R, Sandhu JK, Sheil B, Cooke MS, Sly PD. Free Radical Bio Med. 2010;48:1460–1464. doi: 10.1016/j.freeradbiomed.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 35.Kain J, Karlsson HL, Möller L. Mutagenesis. 2012;27:491–500. doi: 10.1093/mutage/ges010. [DOI] [PubMed] [Google Scholar]
- 36.Hartwig A, Dally H, Schlepegrell R. Toxicol Lett. 1996;88:85–90. doi: 10.1016/0378-4274(96)03722-8. [DOI] [PubMed] [Google Scholar]
- 37.Mitsumoto H, Santella RM, Liu X, Bogdanov M, Zipprich J, Wu HC, Mahata J, Kilty M, Bednarz K, Bell D, Gordon PH, Hornig M, Mehrazin M, Naini A, Flint Beal M, Factor-Litvak P. Amyotrophic Lateral Sclerosis. 2008;9:177–183. doi: 10.1080/17482960801933942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooke MS, Barregard L, Mistry V, Potdar N, Rozalski R, Gackowski D, Siomek A, Foksinski M, Svoboda P, Kasai H, Konje JC, Sallsten G, Evans MD, Olinski R. Biomarkers. 2009;14:103–110. doi: 10.1080/13547500802706012. [DOI] [PubMed] [Google Scholar]
- 39.Weimann A, Broedbaek K, Henriksen T, Stovgaard ES, Poulsen HE. Free Radical Res. 2012;46:531–540. doi: 10.3109/10715762.2011.647693. [DOI] [PubMed] [Google Scholar]
- 40.Tang CK, Vaze A, Rusling JF. Lab on a Chip. 2012;12:281–286. doi: 10.1039/c1lc20833k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chikkaveeraiah BV, Liu H, Mani V, Papadimitrakopoulos F, Rusling JF. Electrochem Commun. 2009;11:819–822. doi: 10.1016/j.elecom.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.(a) Buckingham DA, Dwyer FP, Goodwin HA, Sargeson AM. Aust J Chem. 1964;17:325–336. [Google Scholar]; (b) Forster RJ, Vos JG. Macromolecules. 1990;23:4372–4377. [Google Scholar]
- 43.Chikkaveeraiah BV, Mani V, Patel V, Gutkind JS, Rusling JF. Biosens Bioelectron. 2011;26:4477–4483. doi: 10.1016/j.bios.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munge B, Estavillo C, Schenkman JB, Rusling JF. ChemBioChem. 2003;4:101–108. doi: 10.1002/cbic.200390018. [DOI] [PubMed] [Google Scholar]
- 45.Allen GC, Flores-Vergara MA, Krasnyanski S, Kumar S, Thompson WF. Nat Protocols. 2006;1:2320–2325. doi: 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 46.Zhao L, Schenkman JB, Rusling JF. Anal Chem. 2010;82:10172–10178. doi: 10.1021/ac102317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mani V, Kadimisetty K, Malla S, Joshi AA, Rusling JF. Environ Sci Technol. 2013;47:1937–1944. doi: 10.1021/es304426j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vadhanam MV, Thaiparambil J, Gairola CG, Gupta RC. Chem Res Toxicol. 2012;25:2499–2504. doi: 10.1021/tx300312f. [DOI] [PubMed] [Google Scholar]
- 49.Lvov YM, Lu Z, Schenkman JB, Zu X, Rusling JF. J Am Chem Soc. 1998;120:4073–4080. [Google Scholar]
- 50.Lvov Y, Aviga K, Ichinose I, Kunitake T. J Am Chem Soc. 1995;22:6117–6123. [Google Scholar]
- 51.Rusling JF, Zhang Z. Designing Functional Biomolecular Films on Electrodes. In: Rusling JF, editor. Biomolecular Films: Design, Function and Applications. Marcel Dekker; N.Y.: 2003. pp. 1–64. [Google Scholar]
- 52.Lubin AA, Lai RY, Baker BR, Heeger AJ, Plaxco KW. Anal Chem. 2006;78:5671–5677. doi: 10.1021/ac0601819. [DOI] [PubMed] [Google Scholar]
- 53.Du J, Liu M, Lou X, Zhao T, Wang Z, Xue Y, Zhao J, Xu Y. Anal Chem. 2012;84:8060–8066. doi: 10.1021/ac301954j. [DOI] [PubMed] [Google Scholar]
- 54.Oikawa S, Hirosawa I, Hirakawa K, Kawanishi S. Carcinogenesis. 2001;22:1239–1245. doi: 10.1093/carcin/22.8.1239. [DOI] [PubMed] [Google Scholar]
- 55.Gupta RC, Arif JM. Chem Res Toxicol. 2001;14:951–957. doi: 10.1021/tx000131d. [DOI] [PubMed] [Google Scholar]
- 56.Lim KS, Cui L, Taghizadeh K, Wishnok JS, Chan W, DeMott MS, Babu IR, Tannenbaum SR, Dedon PC. J Am Chem Soc. 2012;134:18053–18064. doi: 10.1021/ja307525h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Embrechts J, Lemière F, Van Dongen W, Esmans EL, Buytaert P, Van Marck E, Kockx M, Makar A. J Am Soc Mass Spec. 2003;14:482–491. doi: 10.1016/S1044-0305(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 58.Cui L, Ye W, Prestwich E, Wishnok JS, Taghizadeh K, Dedon PC, Tannenbaum SR. Chem Res Toxicol. 2013;26:195–202. doi: 10.1021/tx300294d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wasalathanthri DP, Faria RC, Malla S, Joshi AA, Schenkman JB, Rusling JF. Analyst. 2013;138:171–178. doi: 10.1039/c2an35993f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffmann D, Hoffmann I, El-Bayoumy K. Chem Res Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 61.Valavanidis A, Vlachogianni T, Fiotakis C. J Environ Sci Heal C. 2009;27:120–139. doi: 10.1080/10590500902885684. [DOI] [PubMed] [Google Scholar]
- 62.Park JH, Gelhaus S, Vedantam S, Oliva AL, Batra A, Blair IA, Troxel AB, Field J, Penning TM. Chem Res Toxicol. 2008;21:1039–1049. doi: 10.1021/tx700404a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


