Abstract
This study presents a novel method to visualize protein aggregate and particle formation data to rapidly evaluate the effect of solution and stress conditions on the physical stability of an IgG1 monoclonal antibody (mAb). Radar chart arrays were designed so that hundreds of Microflow Digital Imaging (MFI) solution measurements, evaluating different mAb formulations under varying stresses, could be presented in a single figure with minimal loss of data resolution. These MFI radar charts show measured changes in subvisible particle number, size and morphology distribution as a change in the shape of polygons. Radar charts were also created to visualize mAb aggregate and particle formation across a wide size range by combining data sets from size exclusion chromatography (SEC), Archimedes resonant mass measurements, and MFI. We found that the environmental/mechanical stress condition (e.g., heat vs. agitation) was the most important factor in influencing the particle size and morphology distribution with this IgG1 mAb. Additionally, the presence of NaCl exhibited a pH and stress dependent behavior resulting in promotion or inhibition mAb particle formation. This data visualization technique provides a comprehensive analysis of the aggregation tendencies of this IgG1 mAb in different formulations with varying stresses as measured by different analytical techniques.
Keywords: particle size, protein aggregation, data visualization, morphology, microflow imaging, Archimedes, monoclonal antibody, formulation, stability
Introduction
Improving our basic understanding of protein aggregate and particle formation pathways during the manufacturing and storage of therapeutic protein candidates is of increasing interest to protein formulation scientists and regulators alike. This interest primarily arises from the potential ability of these particles to induce an unwanted, humoral immune response resulting in the production of anti-drug antibodies that reduce the efficacy of the administered therapy.1 Previous investigators have shown that particles generated by different stresses vary in average size, shape, concentration, transparency, and chemical modifications, however, the physicochemical properties of a protein particle that potentially elicit an immune response remain elusive.2–4 The immunogenic potential of particle subpopulations is difficult to study because separating and isolating species from a highly heterogeneous population, while maintaining their structural integrity, is technically challenging.4 Additionally, sample handling must be minimized during particle characterization to preserve the integrity of the measured particle populations.5,6 Because of these challenges, formulation development strategies for protein therapeutics are increasingly focused on reducing the number of submicron and subvisible particle degradants that form during manufacturing and storage.7 The formation of proteinaceous particles can be reduced by increasing both the conformational and colloidal stability of the protein in solution as well as by decreasing the number of nucleating species.8 Non-native aggregation is often initiated by partially unfolded or misfolded monomers that associate if there is a favorable change in the free energy of the system.9–14 Certain anions (e.g., chloride) preferentially accumulate on the surface of proteins and can influence both the conformational and colloidal stability of proteins in solution.15,16
Multiple analytical instruments are needed to measure the full range of aggregates and particles in solution because no single technique is currently capable of providing an accurate, quantitative description of the entire aggregation profile of a protein solution.17–20 Subvisible and submicron are terms used to classify particles that are 1–100 µm and 0.1 to <1 µm, respectively.7 Microflow digital imaging (MFI) has become a popular technique to characterize subvisible particles and has been used to study particle formation in monoclonal antibody formulations.21–24 MFI captures digital images of particles from a solution passing through a flow cell and uses algorithms to measure a wide variety of parameters including, but not limited to, equivalent circular diameter, concentration, counts, circularity, aspect ratio, and intensity. These parameters provide an advantage over other subvisible particle counting techniques because morphological filters can be used to discriminate between proteinaceous particles and non-proteinaceous silicone oil droplets or air bubbles.25 A limitation of MFI, however, is the possible undercounting of particles that have a refractive index similar to that of the solvent.26 This experimental artifact can be partially corrected through the proper use of the optimize illumination function.22,26,27 The Archimedes Particle Metrology System uses the principle of resonant mass measurement (RMM) to measure the buoyant mass of particles from a solution passing through a suspended microchannel resonator (SMR). When equipped with a Hi-Q microsensor, the Archimedes is able to measure submicron particles from 200 nm to subvisible particles <5 µm in equivalent spherical diameter. Additionally, RMM has been used to study submicron particle as low as ~104 particles/ml in a monoclonal antibody formulation.28 Weinbuch et al. describes a comparative analysis of MFI and RMM measurements and includes more discussion of the measurement principles and limitations of both instruments.29 Both RMM and MFI provide high resolution particle size and concentration data by counting individual particles, but the large quantity of data produced makes it challenging to compare trends across multiple data sets.
Data visualization methods have been developed to aid in the formulation development of protein-based therapeutic drugs and vaccines. The empirical phase diagram (EPD) visualizes macromolecule structural changes as a shift in colors as a function of environmental stresses (e.g., temperature and solution pH). The EPD visually summarizes biophysical stability data from multiple techniques that have been reduced in dimensionality through principle components analysis.30,31 There are several reports in which the EPD has been used to visualize changes in mAb conformational stability and dynamics.32–34 Additional data visualization methods have more recently been developed to examine protein stability data including radar chart analysis and comparative signature diagrams.35,36 Radar charts have the advantage of displaying multidimensional data without truncating the data or using statistical methods such as principle components analysis.
In this report, we apply radar chart array analysis to compare multiple, large data sets from different analytical methods to determine the effect of formulation variables on the nature of IgG1 mAb aggregates and particles generated through different stress conditions. First, we assess the effect of solution pH and NaCl on the concentration, size, and formation kinetics of subvisible particles, measured by MFI, created due to stirring and shaking of the mAb solutions. Additionally, we use radar chart array analysis to evaluate how IgG1 mAb particle morphology, i.e., the particle’s aspect ratio (a measure of elongation) and intensity (a measure of particle transparency), are affected by formulation conditions. Lastly, we examine the potential of radar chart analysis to visualize formation of the particles and aggregates across the entire size profile, from subvisible and submicron sized particles to small soluble aggregates as measured by MFI, RMM, and size exclusion high performance liquid chromatography (SE-HPLC), respectively.
Experimental Procedures
Materials and Sample Preparation
An IgG1 mAb (pI ~9) was supplied by Janssen Biotech Inc. (Horsham, PA) at ~40 mg/ml. Working mAb solutions were made by diluting the stock solution into 20 mM citrate-phosphate buffer to a final protein concentration of 1.0 mg/ml. Individual samples were prepared at pH 4.0, 6.0 and 8.0 and NaCl concentrations of 0, 0.15, and 1.0 M. These working solutions (3 mL) were prepared in glass vials (37.7×16.75 mm) from Schott (Lebanon, PA) and capped with 20 mm lyophilization vial stoppers (West Pharmaceutical Services, Exton, PA).
Generations of Aggregates
Heat stressed aggregates were generated by incubating samples at 65°C for 10, 120, and 240 min. Stirred stressed aggregates were generated by stirring samples with teflon coated micro spinbars (Fisher) for 10, 120, and 240 min. The stirring speed was adjusted to setting 5 on a Pierce Reacti-Therm III from Thermo Scientific (West Palm Beach, FL). Shaking stressed protein aggregates were made by shaking samples side to side at 300 RPM for 10, 120, and 240 min using a HS 260 shaker from IKA Works Inc. (Wilmington, NC). Stress conditions were selected based on separate studies examining the effect of environmental stresses on the extent of aggregate and particle formation with same IgG1 mAb (Telikepalli et al, submitted).
Differential Scanning Calorimetry
DSC was performed with 1.0 mg/ml protein samples, prior to stress exposure, using a high throughput capillary VP-DSC (MicroCal, Piscataway, NJ). Thermograms were obtained by scanning from 10° to 90°C at a rate of 60°C/hr as described previously.37 Tm values were obtained by iteratively fitting the thermogram to a non-2-state model for unfolding. To values were obtained by determining the point at which a specific interpolated tangent line intersects the horizontal axis.
Extrinsic Fluorescence Spectroscopy
IgG1 mAb samples were diluted to 0.1 mg/ml in their corresponding buffers and a 20 molar excess of 1-anilinonaphthalene-8-sulfonate (ANS, Sigma) was added. The fluorescence emission was measured using a QM-40 spectrofluorometer from Photon Technology International (Birmingham, NJ) as described previously.21
Size-exclusion High Performance Liquid Chromatography
A Tosoh Bioscience TSKgel G3000SWx1 stainless steel column (San Francisco, CA) along with the corresponding guard column (TSKgel SWxl guard column) operated at 30°C was used in all experiments. A Shimadzu Prominence UFLC HPLC system (Kyoto, Japan) connected to a diode array detector was used. Twenty five µL of each sample were injected into a column that was equilibrated with at least 10 column volumes of 200 mM sodium phosphate buffer (pH 6.8). The flow rate was 0.7 mL/min and the measurement time was 30 minutes. LC Solution software was used for data analysis and peak integration. Percent total area loss (compared to time=0), percent monomer loss, and percent soluble aggregate in solution were reported as described previously in Bond et al., which also details the column conditioning procedure and column performance/recovery for characterizing protein aggregates.38
Resonant Mass Measurement
An Archimedes Particle Metrology System (Affinity Biosensors, Santa Barbara, CA) was used to quantify submicron and small subvisible particles from 0.275 to 1.85 µm. A Hi-Q micro sensor, calibrated using 1.03 µm polystyrene beads (Affinity Biosensors), was used for all measurements. Reference solutions of 1:20 D2O:H2O or pure H2O were used depending upon the solution density of the sample being measured. Prior to daily measurements, the accuracy of the Hi-Q microsensor was tested using the 1.03 µm bead standard. Before each measurement, the sensor and the micro tubing, was rinsed with 2% PCC-54 detergent, by applying high pressure on the sample and reference vials to clean the bypass channels. In addition to the high pressure applied to the sample and reference vials, low pressure was also applied to the sample waste vial to induce a flow to rinse the sensor with 2% PCC-54 detergent. This process was applied until a clean frequency trace was achieved. The sensor was then loaded with particle free water and two “sneeze” operations were performed. The sample was loaded for 30 s, and a stop trigger of 200 particles or 30 min measurement time was used. The limit of detection was empirically determined (0.035 Hz) and used throughout the study. The density value for the mAb was estimated at 1.41g/ml based on protein molecular weight.39
Microflow digital imaging
A DPA-4200 flow microscope (Protein Simple, Santa Clara, CA) was used to capture digital images of subvisible particles with ECD from 2–70 µm. Before each measurement, the flow cell was primed in the following manner: particle free water was flushed through the flow cell at the maximum flow rate until the flow cell was observed to be particle free for at least 30 s. The optimize illumination function was then used to ensure the proper level of solvent illumination. Samples underwent the minimal required amount of sample handling prior to measurement and were degassed by exposure to a vacuum chamber for 10 min. To prevent clogging of the flow cell, samples containing approximately 50 or more visible particles (observed through visual inspection) were set aside for 30 min to allow the largest particles to settle. Samples with particle concentrations over the limit of quantitation (900,000 counts/ml)22 were diluted and the results were corrected for the dilution factor (samples subjected to 240 min of stirring were diluted 1:3, and samples stressed for ≥120 min by heat were diluted 1:10). Particle statistics were generated using MFI View Analysis Suite (MVAS) version 1.3 (Protein Simple, Santa Clara, CA). Each MFI measurement was separated into five subpopulations based upon particle equivalent circular diameter (ECD); 2≤x<5, 5≤x<10, 10≤x<25, 25≤x<50, and 50≤x<70 µm.
Radar Chart Construction
Radar charts were generated using software developed in our laboratory (MiddaughSuite) as described previously36. This report is, however, the first time that radar chart arrays have been applied to protein aggregation and particle formation data sets from MFI, RMM and SEC. To create the MFI particle number and size range radar charts, particle concentration data, obtained from analysis using MVAS 1.3 software described above, were pre-processed by calculating the mean, standard deviation, and mean minus standard deviation (m−SD) for the five particle size populations listed in the previous section. The mean and m−SD values were then uploaded to MiddaughSuite software. The radar charts were set to a log scale and each axis was defined to have a minimum of 10 and a maximum of 1,000,000 particles. Radar charts were generated for both the mean and the m−SD values and then precisely superimposed using pixel alignment in Adobe Photoshop CS6 (Adobe Systems, San Jose, CA). The purpose of superimposing the two radar charts was to include an indication of error for n=3 replicate experiments because particle measurements are often highly variable. The m−SD was chosen because m+SD values would be hard to visualize because positive error bars appear smaller on a log scale.
Particle morphology radar charts were created using the average mean intensity and aspect ratio values for each of the five particle size populations (obtained through the filter manager tool in MVAS 1.3). These data were pre-processed and the mean and m+SD values were uploaded to MiddaughSuite. The axes of this radar chart were set to an inverted linear scale and each axis was defined to have a minimum value of 0.35 and a maximum of 0.85. Axes with an insufficient number of particles (empirically determined to be < 25 counted) were highlighted with a light red background color. The mean and the m+SD particle morphology radar charts were also superimposed using Adobe Photoshop to indicate variability between n=3 measurements. The m+SD were used to maintain the consistency of radar chart interpretation because the morphology radar chart arrays use an inverted linear axis.
To create the “multiple instruments” radar chart, data from SEC, RMM, and MFI were preprocessed to obtain the mean and the m−SD for each condition. The data were then uploaded to MiddaughSuite and each axis was highlighted with a background color according its scale. Axes 1 and 2, corresponding to total area loss and monomer loss were set to an inverted linear scale between 0 and 100%. Axis 3, the soluble aggregate content, was set to a linear scale between 0 and 25%. Axes 4–6, representing submicron and small subvisible particle concentrations between 0.275 and 1.85 µm were set to a log scale with a minimum value of 104 and a maximum value of 109 particles (approximate limit of quantitation for the Archimedes). Lastly, axes 7–9, representing subvisible particles from 2–70 µm, were set to a log scale with a minimum of 100 and a maximum of 107 particles. The mean and m−SD radar charts were generated and superimposed using Adobe Photoshop to indicate error between the n=3 measurements.
Results
Conformational stability of IgG1 mAb vs. solution pH and NaCl concentration
Prior to examining the effect of formulation variables on particle formation, differential scanning calorimetry was used to study the relative conformational stability of the IgG1 mAb in solutions with varying pH and NaCl concentrations. For example, at pH 4, three thermal melting events were observed at 56.1, 70.7, and 72.6 °C, which are referred to as Tm1, Tm2, and Tm3, respectively (See Supplemental Figure S1). The effects of solution pH and NaCl on the observed Tm values for the mAb are summarized in Table 1. mAb solutions at pH 4 had the lowest thermal melting value (and onset temperatures) while the addition of 0.15 and 1 M NaCl to the pH 4 solutions caused further reductions in melting and onset temperatures (Table 1). At pH 6 and 8 with no NaCl, only Tm2 and Tm3 were reproducibly detected. Addition of 0.15 M NaCl appeared to slightly lower the melting and onset temperatures, and the addition of 1 M NaCl resulted in detection of Tm1. In addition, fluorescence measurements using ANS as an extrinsic probe were used to determine if solution pH and NaCl concentration influenced ANS interactions with hydrophobic surfaces within the mAb (or potentially between mAbs). No major differences in ANS fluorescence intensity were observed in the various mAb solutions described above suggesting no major alterations of mAb’s overall tertiary structure (or self-associations) across the pH range of 4–8 and NaCl concentrations of 0 to 1M (data not shown).
Table 1.
Effect of solution pH and NaCl concentration on thermal onset (Tonset) and thermal melting (Tm1, Tm2, Tm3) temperatures of an IgG1 mAb as determined by DSC. See methods section for experimental details. SD is standard deviation from triplicate (n=3) experiments. ND indicates a Tm value could not be determined from experimental data.
| pH | [NaCl] (M) | Tonset | SD | Tm1 | SD | Tm2 | SD | Tm3 | SD |
|---|---|---|---|---|---|---|---|---|---|
| 4.0 | 0.00 | 48.1 | 0.9 | 56.1 | 0.1 | 70.7 | 0.2 | 72.6 | 0.1 |
| 0.15 | 46.6 | 0.4 | 53.0 | 0.1 | 68.3 | 0.1 | 70.8 | 0.1 | |
| 1.00 | 44.7 | 0.2 | 50.9 | 0.1 | 67.3 | 0.1 | 68.5 | 0.1 | |
| 6.0 | 0.00 | 66.1 | 0.1 | ND | ND | 73.0 | <0.1 | 83.4 | 0.3 |
| 0.15 | 65.4 | 0.2 | ND | ND | 72.7 | <0.1 | 83.3 | <0.1 | |
| 1.00 | 62.6 | 0.5 | 69.9 | 0.1 | 74.3 | <0.1 | 84.1 | <0.1 | |
| 8.0 | 0.00 | 66.0 | 0.2 | ND | ND | 72.3 | <0.1 | 82.7 | <0.1 |
| 0.15 | 66.0 | 0.3 | ND | ND | 71.9 | <0.1 | 82.7 | <0.1 | |
| 1.00 | 64.5 | 0.1 | 71.0 | <0.1 | 73.6 | <0.1 | 83.6 | <0.1 |
Radar chart analysis of effects of solution pH, NaCl concentration, and stress on IgG1 mAb particle formation as measured by MFI
Particle number and size
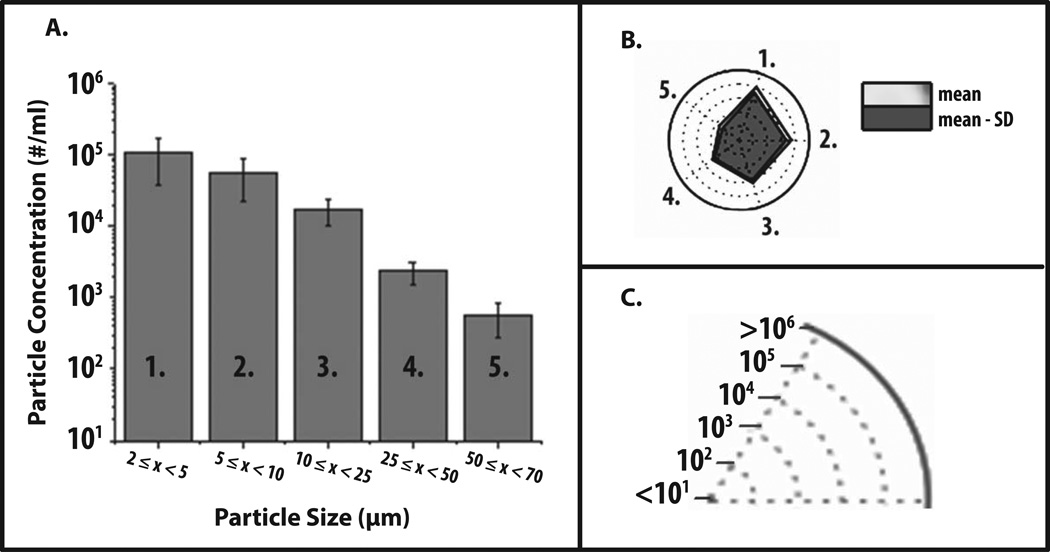
Radar Charts were initially constructed to visualize various data sets of subvisible particle formation as obtained by MFI with an IgG1 mAb under different formulation conditions. First, we present some generic MFI data to demonstrate how to interpret a radar chart. Figure 1A shows a commonly used graphical representation of generic MFI data from one experimental condition, i.e., a histogram of particle concentration (log scale) as function of particle size (grouped together in different size bins). Figure 1B shows the same MFI data in the form of a radar chart. The numbered axes of the radar chart correspond to the same numbered size bins on the histogram (e.g., number one refers to particle size bin 2–5 microns in both Figures 1A and 1B). The axes of radar charts are linearly connected so that the data set can be visualized as a polygon. In addition, particle concentration is displayed in this radar chart using concentric rings positioned from the center to the edge of the circle. For example, as shown in Figure 1C, each ring shows a ten-fold increase in particle concentration.
Figure 1. Application of radar chart array analysis to evaluate subvisible particle concentration and size distribution data measured by MFI.
(a) Histogram (n=3) of representative MFI data plotting particle concentration versus particle size (ECD) for an IgG1 mAb solution containing 20 mM citrate-phosphate buffer at pH 8.0 and 1 M NaCl subjected to 4 hours of shaking. (b) Radar chart presentation of the same data set; each axis is labeled with a number and is the same size range as the corresponding numbered bin size in the histogram. The values along the each axis of the radar chart are connected linearly to visualize the data set as a polygon. The perimeter of the outermost polygon represents the mean particle concentration, the polygon labeled ‘mean - SD’ represents the mean minus one standard deviation, and the distance between the perimeters of the two polygons (along an axis) is equivalent to the lower error bar in the equivalent bin of the histogram (one standard deviation). (c) Illustration of the particle concentration (log scale) used in the radar charts in this work to display experimentally measured micro flow imaging subvisible particle concentrations.
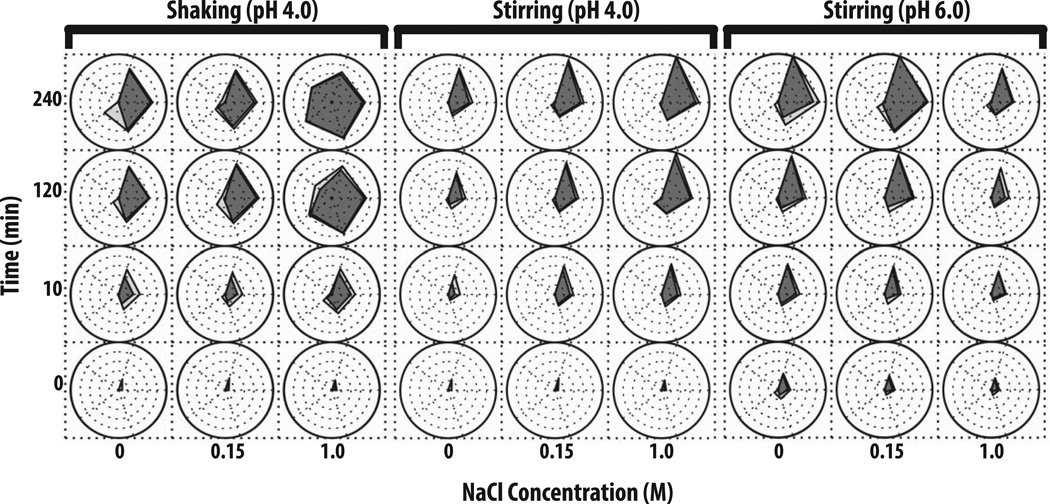
In Figure 2, radar chart analysis was used to elucidate the effect of varying solution pH, NaCl concentration, and stress conditions on the kinetics of subvisible particle formation (number and the size distribution as measured by MFI) for an IgG1 mAb. Prior to stress, a low number (i.e., 100 to 3,000) subvisible particles were detected in the various protein samples with samples at pH 4 having fewer particles than pH 6 regardless of the NaCl concentration. After 10 min of shaking stress (left panel of Figure 2), more than 5,000 particles (2–25 µm) had formed at pH 4 with no added NaCl. Samples containing 0.15 M NaCl formed particles at a similar rate, however, ~50 particles from 25–50 µm had also formed. Samples in the presence of 1 M NaCl had the highest particle concentration in size range of 2–25 µm as well as >100 particles/ml from 25–50 µm. The concentration of subvisible particles, generated through shaking, increased over the time of shaking with increased values noted at 120 min. Particle formation decelerated after 120 min (no longer forming at a logarithmic rate) with the 240 min shaking sample showing similar particle concentrations and size distributions across the pH and NaCl conditions examined. The presence of 1 M NaCl had a more pronounced effect on IgG1 mAb samples at pH 4 that were shaken for more than 2 h since an average of 10,000 particles from 25–50 µm and 1,000 particles from 50–70 µm had formed. In general, the presence of 1 M NaCl at pH 4, 6 or 8 caused larger subvisible particles to form when these protein samples were shaken (Figure 2, left panel and Supplemental Figure S1).
Figure 2. Radar chart array analysis of subvisible particle concentration and size data for IgG1 mAb solutions as measured by MFI.
IgG1 mAb solutions of varying pH values and NaCl concentrations were exposed to different stress types over time as indicated in the figure (n=3), and resulting formation of subvisible particles was measured by MFI. The Y-axis represents the amount of time the formulations were stressed (in minutes), the X-axis represents NaCl concentrations, and each radar chart panel (left, middle, right) signifies a change in stress or pH. See methods for experimental details of MFI measurements from each of these accelerated stress studies. Refer to Figure 1 for an explanation of MFI particle concentration and size range scales/units shown in individual radar charts.
The IgG1 mAb samples stirred at pH 4 resulted in an entirely different subvisible particle distribution, compared to shaking under the same solution conditions (Figure 2, middle panel). For example, particles generated from stirring stress tended to be primarily 2–25 µm in size. After 10 min of stirring, samples with 0 M NaCl had the least amount of subvisible particles, 0.15 M NaCl had ~104 particles/ml, and 1 M NaCl had >104 particles. The concentration of small particles noticeably increased between 2 and 4 h of stirring. After 4 h of stirring, for IgG1 mAb samples with 1 M NaCl, the most subvisible particles (> 106 particles/ml from 2–5 µm) were formed, while samples with 0.15 M NaCl formed ~300,000 particles/ml from 2–5 µm and samples with no NaCl formed <50,000 particles/ml (Figure 2, middle panel).
Samples stirred at pH 6 (Figure 2, right panel) and pH 8 (Supplemental Figure S2, right panel) exhibited very different behaviors in terms of the effect of NaCl on particle formation compared to pH 4. Samples at pH 6 in the absence of NaCl had the highest concentration of subvisible particles over 4 h (particle concentrations >106 particles/ml). In contrast, the presence 1 M NaCl greatly reduced the concentration of subvisible particles to ~50,000/ml. At pH 8, the presence of either 0.15 or 1M NaCl greatly reduced the concentration of subvisible particles for IgG1 mAb samples stirred for 2 and 4 h (Supplemental Figure S2).
Particle Morphology
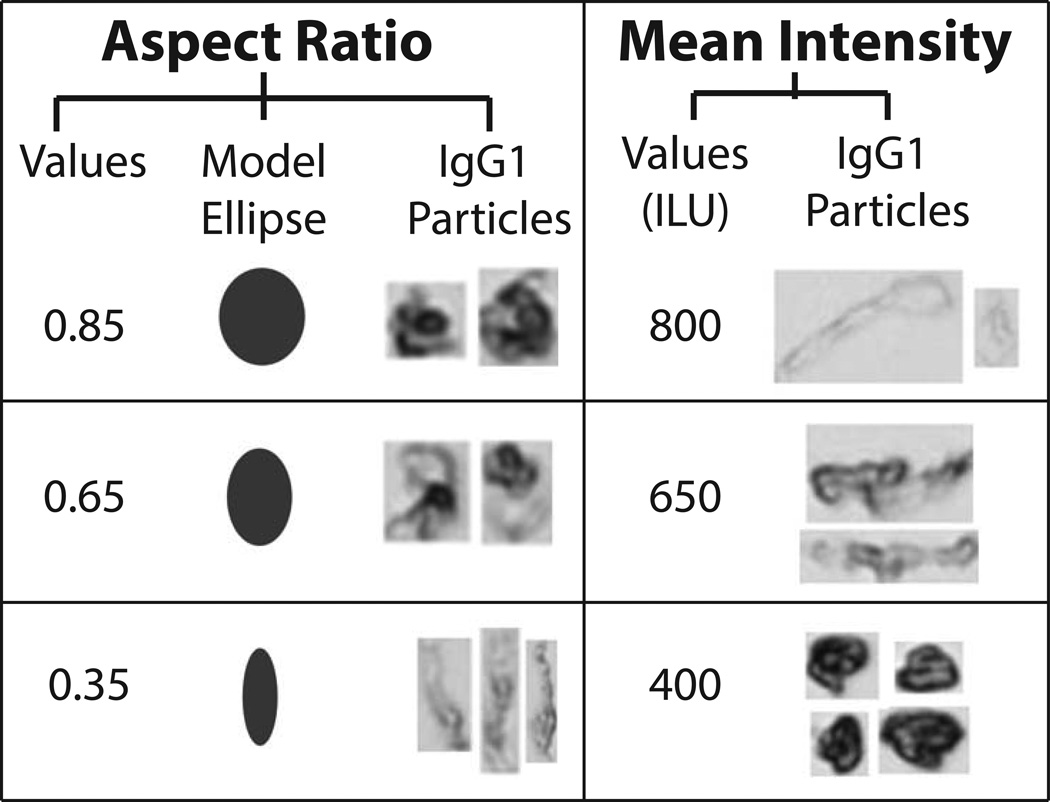
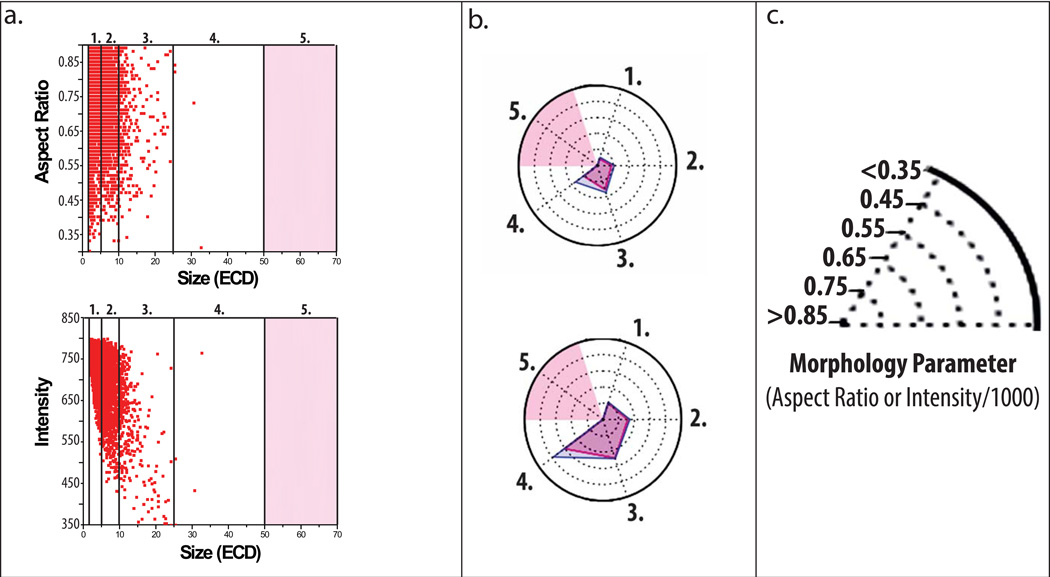
Representative particle images captured by MFI demonstrate formation of low, medium, and high aspect ratio/intensity valued protein particles in the IgG1 mAb solutions depending on the formulation and stress (Figure 3). Model ellipses that correspond to particle aspect ratio values are also presented as a simplified method to visualize particle elongation (i.e., aspect ratio). We present some MFI data to demonstrate how to interpret a radar chart for particle morphology. In Figure 4A, these data are presented in a manner similar to other reports with published MFI morphological data.40 Figure 4A shows scatter plots (for the first 3,000 particles counted by MFI, n=1) of particle size vs. a morphological parameter, one plot for particle aspect ratio and the other for particle intensity. Morphological data from additional particles cannot be added to these scatter plots since resolution would be lost as additional data points merge into one continuous color (not shown). In contrast, the aspect ratio and intensity values from the same experiment, but this time using the entire, complete data set (>200,000 particles/ml, n =3) are presented as an radar chart (Figure 4B) showing particle size vs. morphological parameter, using a scale for the morphology parameters as shown in Figure 4C (i.e., aspect ratio or intensity/1000). Regions shaded in light red in the radar charts in Figure 4B denote insufficient particles were present in the samples (empirically defined as < 25 particles as measured by MFI) to effectively compare the effect NaCl, pH, and stress on particle morphology. One reason this was done was so that the few subvisible particles present at time zero did not affect the radar chart analysis of the morphological properties of particles formed from stress and solution conditions over time.
Figure 3. Representative morphological parameters for protein particles from MFI analysis.
Examples of three different aspect ratios and intensity values (low, moderate, and high) are presented along with corresponding IgG1 mAb particle images from MFI. Model ellipses of equivalent aspect ratios are presented to help visualize changes in particle shape. For ease of presentation, protein particle images from MFI are not presented to scale.
Figure 4. Application of radar chart array analysis to evaluate subvisible protein particle morphological parameters measured by MFI.
(a) Scatter plots of a subset of particle data displaying aspect ratio (top) and intensity (bottom) versus ECD (first 3,000 from n=1 experiments) counted within an IgG1 mAb solution containing 20 mM citrate buffer at pH 8.0 and 0.15 M NaCl subjected to 4 h of shaking. Light red shading represents size bins with insufficient data as defined by x < 25 particles counted. (b) Radar chart presentations of the same experiment showing average aspect ratio (top) or intensity/1000 (bottom) per size range except with the complete data set (~200,000 particles, n=3); each numerical axis in the radar chart is an equivalent size range to the corresponding numbered region in the scatter plots. The perimeter of the outermost polygon displays the mean morphology parameter, the perimeter of the polygon labeled ‘mean + SD’ displays the mean plus one standard deviation, and the distance between the perimeters of the two polygons (along an axis) is one standard deviation. (c) Illustration of the scale/units used in the radar charts to display experimentally measured (MFI) subvisible particle aspect ratio and intensity/1000 values.
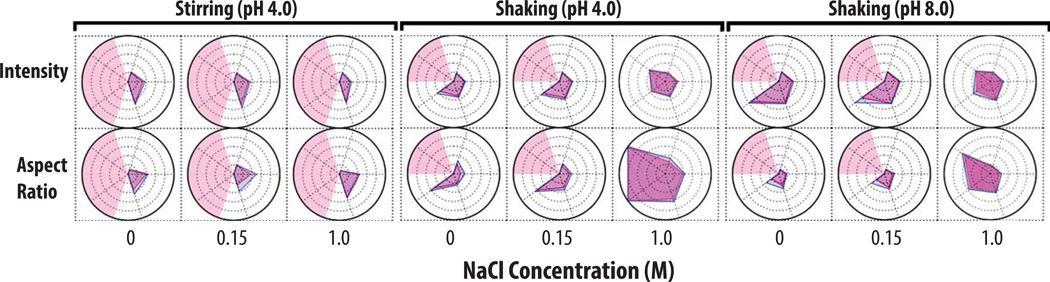
As an example, radar chart analysis of MFI particle morphological parameters are shown for IgG1 mAb samples stressed for 4 h with varying pH, NaCl, and stress conditions (Figure 5). As shown on the left panel, the average particle aspect ratio and intensity values did not change with NaCl concentration when samples were stirred for 4 h at pH 4. As another example, the addition of 1 M NaCl had a profound effect on particle morphology when the samples were subjected to 4 h of shaking at pH 4 and 8. When the mAb samples were shaken at pH 4 (Figure 5, middle panel), the presence 1 M NaCl caused the average aspect ratio of all size ranges in the particle distribution to decrease (i.e., the particle elongation increases). Particles larger than 25 µm had an average aspect ratio of less than 0.35 (refer to the ellipse models presented in Figure 3). Additionally, the intensity of subvisible particles in the size range from 25–50 µm increased (became more transparent) in the presence of 1M NaCl when compared with both 0 M and 0.15 M NaCl. Samples of IgG1 mAb subjected to the same shaking stress but now at pH 8 (Figure 5, right panel), in the presence of 0 M and 0.15M NaCl, contained particles with low intensity (i.e., were not transparent) and had a high aspect ratio (i.e., more globular). Interestingly, the presence of 1 M NaCl at pH 8 caused a large increase in average intensity and a substantial reduction in the aspect ratios for particles in the size range over 10–50 µm.
Figure 5. Radar chart array analysis of protein particle morphological parameters versus particle size for IgG1 mAb solutions as measured by MFI.
IgG1 mAb solutions (at indicated pH and salt concentration) were stressed to generate protein particles (n=3), and solutions were analyzed by MFI. See methods section for experimental details of each accelerated stress study. The Y-axis is labeled with the morphological parameter, the X-axis represents the NaCl concentration, and each panel (left, middle, right) represents a change in stress or pH as indicated. The numerical value/units of each concentric circle in this radar chart is shown in Figure 4C. Light red shading represents a region where an insufficient number of particles (x < 25) were counted. Refer to Figure 4 for an explanation of particle morphological parameters shown in individual radar charts.
Radar charts incorporating protein aggregate and particle formation data from multiple analytical techniques
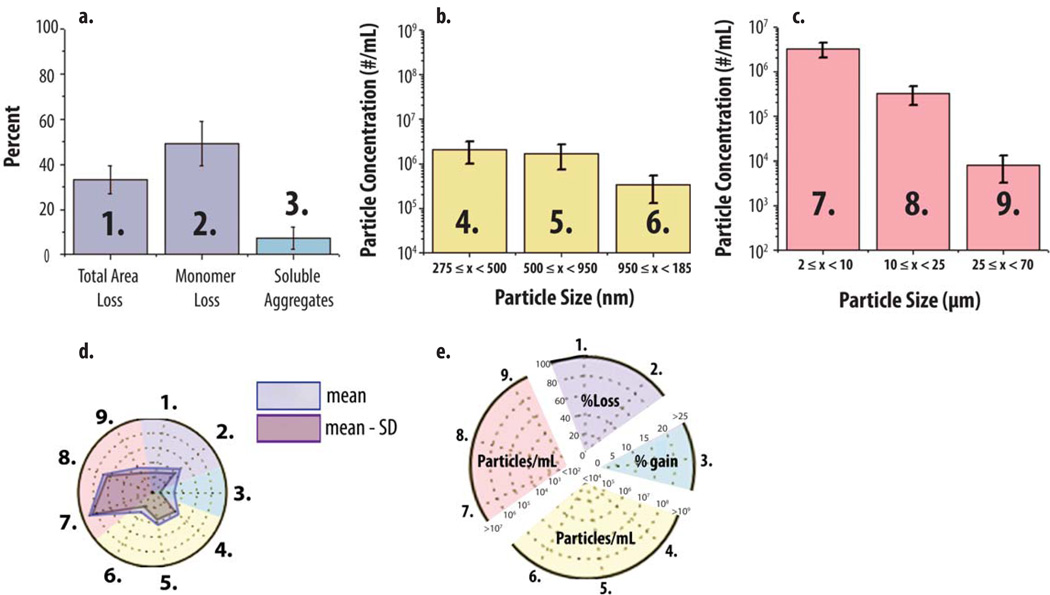
Radar chart array analysis was then applied to data sets from multiple analytical techniques, which measured mAb particle and aggregate formulation over different size ranges. The ‘multiple-instruments radar chart’ was designed to provide mechanistic information relating to the potential interrelationships of soluble aggregate and particle formation. A representative example of the multiple-instrument radar chart is shown in Figure 6. For SEC, Figure 6A shows a typical bar graph of SEC data for pH 6, 0.15 M NaCl mAb samples that were heated for 2 h. SEC data includes the percent of monomer loss, soluble aggregates formed and total area loss (i.e., loss of protein content compared to t=0 as an indirect measure of insoluble aggregate formation). The same SEC data are also presented in radar chart form in Figure 6D (axes 1–2 in the purple region, and axis 3 in the blue region, of the radar chart in Figure 6D correspond to bars 1–3 in Figure 6A). Similarly for submicron particle data, Figure 6B is a histogram of RMM data, for the same heat stressed mAb sample, providing particle size data in the submicron range up to particles with an equivalent spherical radius of 1.85 µm. The numbered size bins (4–6) in Figure 6B correspond to axes 4–6 in the yellow region of the corresponding radar chart (Figure 6D). Finally, Figure 6C is a histogram of MFI particle size and concentration data for the heat stressed mAb sample for subvisible particles of size 2–70 µm. The particle size bins numbered 7–9 in the histogram (Figure 6C) correspond to axes 7–9 in the red region of the radar chart (Figure 6D). A more detailed key showing the labeling of the multiple-instruments radar chart in terms of scale and measurements, are provided in Figure 6E.
Figure 6. Application of radar chart array analysis to protein aggregate and particle formation data from multiple analytical techniques.
An IgG1 mAb solution containing 20 mM citrate phosphate buffer (pH 6.0) and 0.15 M NaCl was heated at 65°C for 120 min and the levels of soluble aggregates, submicron and subvisible particles were measured using SEC, RMM and MFI, respectively. (a, b, c) Bar graph (n=3) of SEC, RMM particle concentrations (0.275–1.85 µm) and MFI particle concentrations (2–70 µm). (d) Radar chart presentation of the same data sets shown bar graphs (a,b,c). The SEC, RMM and MFI data denoted 1–3, 4–6 and 7–9 in the bar charts (a, b, c, respectively) corresponds to axes 1–3, 4–6 and 7–9 in the corresponding radar chart (d). The perimeter of the outermost polygon in the radar chart displays the mean value (particle concentration or area gain/loss), the perimeter of the polygon labeled ‘mean - SD’ represents the mean value minus one standard deviation, and the distance between perimeters of the two polygons (along an axis) is one standard deviation. (e) Illustration of the numerical scales and units for the radar chart axes with different scales being highlighted with distinct background colors for results from SEC (1,2,3), RMM (4,5,6) and MFI (7,8,9), respectively.
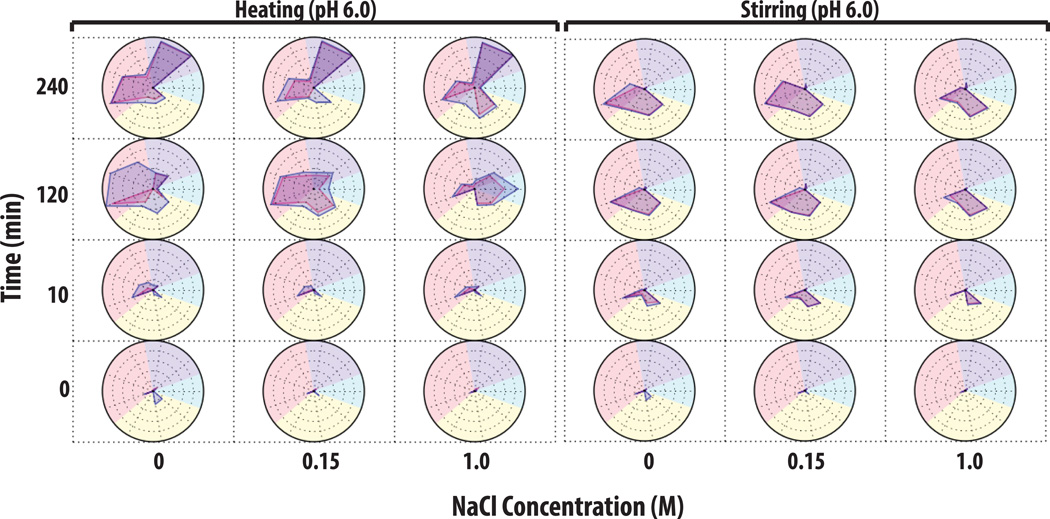
An example of utilizing a ‘multiple-instruments radar chart’ to characterize aggregate and particle formation is shown for heat and stirring stresses of an IgG1 mAb solution at pH 6 with varying NaCl concentrations (Figure 7). At time zero (at all NaCl concentrations), the mAb samples contained <1,000 subvisible particles, >105 submicron particles, and were > 99% monomer with <0.2% soluble aggregates. After 10 min of heat stress at 65°C, mAb samples without NaCl showed a ~10% total peak area loss as well as a ~10% monomer loss as measured by SEC, and after 2 h of heating, the samples had 35% total area loss, 40% monomer loss, and 5% soluble aggregates. In addition, an extremely high concentration of subvisible particles (>107 particles/ml) was now observed. Interestingly, under the same conditions, the addition of 0.15 M NaCl increased soluble aggregate levels to 10% based on SEC with a concomitant decrease the total concentration of subvisible particles by ~10 fold as measured by MFI (and did not substantially change the amount of submicron particles). The presence of 1 M NaCl in the mAb solution greatly increased the levels of soluble aggregates to ~22% while the measured total area loss was approximately 20%, and the monomer loss was ~50% as measured by SEC. In addition, the 1M NaCl containing mAb samples showed a greatly reduced concentration of subvisible particles (<50,000 measured by MFI), as well as a reduced concentration of submicron particles by more than 10× as measured by RMM. After 4 h of heating, there was essentially complete aggregation and the monomer species was virtually undetectable by SEC (left panel of Figure 7).
Figure 7. Radar chart analysis of protein aggregate and particle formation data from multiple analytical techniques to visualize the extent of IgG1 physical instability.
IgG1 mAb solutions containing 20 mM citrate phosphate buffer (pH 6.0) and varying NaCl concentrations were stressed by either heat at 65°C or stirring over time (n=3). Samples were evaluated by a combination of SEC, RMM and MFI. See methods section for experimental details of each accelerated stress study. The Y-axis represents the amount of time the samples was stressed (in minutes), the X-axis represents NaCl concentration, and each radar chart panel (left and right) denotes heating and stirring, respectively. Refer to Figure 6 for an explanation of units and scales used in the individual radar charts for each analytical method (SEC, RMM and MFI data).
The effect of stirring stress on IgG1 mAb solutions is shown in the right panel of Figure 7. In contrast to heating stress, no major changes in soluble aggregate levels with less than 10% total area loss were observed by SEC in the majority of samples subjected to stirring stress. This result in itself suggests different aggregation mechanisms are dominant for heating and stirring stresses. After 10 min of stirring, ~106 submicron sized particles formed in all of the samples, and the mAb sample without NaCl had formed the most subvisible particles ranging from 2–10 µm. After 2 and 4 h of stirring, the particle number and size distributions in the mAb samples with 0 and 0.15 M NaCl appeared similar, but samples with 1 M NaCl showed a ~50 fold reduction of subvisible particles along with a substantially increased number of submicron particles (~108 particles/ml) as shown in right panel of Figure 7. Finally, mAb samples were shaken at pH 4 and displayed a third dominant aggregation mechanism (See Supplemental Figure 2): no soluble aggregates were detected by SEC, while submicron particle formation leveled off after 10 min as measured by Archimedes. The subvisible sized particle populations continued increasing over time as measured by MFI. Under these conditions, the addition of 1 M NaCl caused increased formation of subvisible particles in the 25–70 µm size range.
Discussion
Utility of radar chart arrays to summarize and analyze protein aggregate and particle formation data sets from different instruments
The ability of radar charts to better visualize protein aggregation and particle formation results for an IgG1 mAb in different formulations, as measured by multiple analytical instruments, was investigated in this report. We first used radar charts to examine the effect of pH, NaCl concentration, stress type, and length of stress exposure on the size and concentration of the sub-visible particles that were formed as measured by MFI. A broader application of radar charts incorporating data from multiple analytical instruments was also investigated. Radar chart array analysis was shown to be an effective tool for characterizing the effect of individual formulation composition and environmental stress variables on protein growth kinetics and particle size distribution. For example, the pH dependent behavior of particle formation (number and size distribution) as a function of NaCl and stress (i.e., shaking and stirring) was established for an IgG1 mAb. It is important to emphasize the utility of radar charts to plot large data sets of MFI data and compare trends across formulation conditions. For example, the MFI data in Figure 2 represents 36 different experimental conditions (e.g., time, stress type, formulation composition) performed in triplicate resulting in a total of 108 experiments. If histograms were to be used as shown in Figure 1A, many plots would be needed to summarize the results and compare the effects of time, stress type, and formulation composition on mAb particle formation (including particle concentration and size distribution) in solution. This makes analysis of trends in particle size, concentration and morphology more challenging. Using radar chart array analysis, however, all of these results are displayed, and trends across the 36 conditions are summarized in a single figure (Figure 2). The radar chart arrays in this paper were modified to display statistical variability: the m−SD was included in the design of the concentration vs. size radar chart to represent a lower error bar. m+SD values were used in the morphology vs. size radar chart to maintain consistency of interpretation because it was designed with an inverted linear axis. The ability to display the error makes these modified radar-chart arrays a more powerful tool to study protein aggregation than other data visualization methods that have previously been applied to study protein physical stability such as the empirical phase diagram. It is critical for protein particle data visualization methods to include a measure of statistical variability because protein particle measurements often have run-to-run as well as vial-to-vial variability.6,21,23,24
Another application of radar chart analysis is to analyze the effect of formulation composition and environmental stresses on the average morphology of any subvisible particle size range. MFI is capable of measuring multiple particle morphological parameters such as intensity, feret diameter, aspect ratio, circularity, etc. Unfortunately, these data remain largely underutilized, especially in comparison to particle concentration and size range measurements. This is largely due to unfamiliarity with the parameters and the difficulty in presenting such large data sets in an organized fashion. Several reports have recently presented MFI morphological data using scatter plots and line graphs.27,40 These graphical methods are effective at presenting a few morphological data sets, however, scatter plots and line graphs become cumbersome to interpret as the quantity of particles and/or the number of data sets (e.g., formulation conditions and time points) increase. Other data visualization methods used for displaying biophysical stability data, such as the EPD, would also be inefficient at visualizing particle morphological trends because principle components analysis would complicate data interpretation since all of morphological parameters correlate to varying degrees. Radar chart analysis was successful in elucidating the effect of buffer composition (pH and NaCl concentration) and stress conditions (shaking and stirring) on the morphology of sub-visible particles (aspect ratio and intensity) formed from IgG1 mAb solutions (Figure 5). Radar-chart-array analysis could be applied in the future to elucidate the effect of similar formulation variables on other particle parameters including particle mass, reversibility (defined as a decrease in particle size when diluted to stable pH conditions),3 density, and surface irregularity (fractal dimension); such work is currently underway in our laboratory.
Radar chart arrays were also used to visualize results from multiple analytical instruments that monitor protein aggregate and particle formation across size ranges from mAb soluble aggregates (SEC), to submicron particles (RMM), and to subvisible particles (MFI). To our knowledge, there are no other data visualization methods available which provide such a comprehensive picture of protein aggregation data across a wide size range. Although radar chart array analysis provided qualitative comparisons of the effect of formulation variables on mAb aggregation, one limitation of this approach is that SEC, RMM and MFI use different measurement methods and provide information on different numerical scales. For example, SEC has known limitations to monitor protein aggregates including irreversible binding of aggregates to the column matrix. In this work, an optimized SEC procedure was utilized to minimize such issues and maximize recovery.38 In addition to loss of monomer and increase in soluble aggregate levels, we also monitored loss of total area of samples compared to time zero (an indirect measure of insoluble aggregate formation). RMM measures a frequency shift caused by a particle passing through a resonator and calculates the buoyant mass based on the shift and the sensitivity of the resonator. An equivalent spherical diameter is calculated using the appropriate density value and the buoyant mass (refer to Burg et al. for further explanation).41 The density of pure protein reported in the literature is variable with values used ranging from 1.32 g/ml29 to 1.43 g/ml42. We have chosen to use a density value of 1.41 g/ml as suggested by Fischer et al. based on the molecular weight of IgGs.39 It is also important to mention that the Archimedes software uses extrapolation to determine the particle concentration per ml. MFI measures the 2-dimensional area of particles and, through image analysis, calculates the equivalent circular diameter (ECD) of the particle.43 ECD is not directly related to spherical diameter, and assuming such may result in an overestimation of particle volume. Another analytical limitation in this work is the presence of a detection gap to accurately measure very small particles from ~50 nm to 275 nm. The application of nanoparticle tracking analysis (NTA) or dynamic light scattering would be of interest in future studies to address a more complete size range. NTA requires, however, a high minimum concentration of submicron particles (>106/ml) and would likely be applied to select samples. Nonetheless, radar chart analysis was an effective tool for probing complex protein growth mechanisms monitored by multiple analytical techniques (Figures 6 and 7) and, in general, offered the ability to more easily follow trends in the data, thus providing some additional insights as described below.
Effects of pH, NaCl and stress on IgG1 mAb aggregate and particle formation and morphology
The focus of this work was on the development of radar chart data visualization techniques for MFI data on subvisible particle formation (concentration, size range, and morphology) due to different stresses (shaking, stirring, heating). Once developed, however, interesting trends in the physical instability of this model IgG1 mAb were readily apparent. For example, the pH and stress-type dependence of the NaCl effect on the aggregation profile of this IgG1 mAb was striking. During stirring and heating, but not shaking, the presence of 1 M NaCl promoted particle growth at pH 4, but inhibited particle growth at pH 6 and 8 (refer to Figure 7 and Supplemental Figure S3). In contrast, during shaking stress, the presence of NaCl increased the size and concentration of particles present at pH 4, 6, and 8 (refer to Supplemental Figure S4). This pH-dependent effect of NaCl on the physical instability of this IgG1 mAb was also observed by separate turbidity measurements (data not shown).
Studies have previously shown that anion binding promotes aggregation of an IgG mAb through charge shielding and by affecting conformational stability.16 In this work, the presence of NaCl (> 0.15 M) destabilized the conformational of the mAb at pH 4 as measured by DSC in terms of Tonset and three Tm values (See Table 1). NaCl had a much less pronounced effect on Tonset and Tm values for this IgG1 mAb at pH 6 and 8 (See Table 1). The observed decrease in conformational stability provides some insight into why the presence of NaCl destabilizes this mAb at pH 4 during stress. It does not, however, necessarily address the observed effects of NaCl on slowing particle formation during heating or stirring at pH 6 or 8. Kroetsch et al. showed that soluble aggregates of alpha-chymotrypsinogen A rapidly coalesce to form insoluble aggregates once a critical salt concentration was reached.13 Our results suggest that addition of 1 M NaCl at pH 6 actually increases the colloidal stability of this mAb since the particles formed during heating and stirring are on average much smaller in size compared to samples with or without 0.15 M NaCl. Accurate particle mass (or density) measurements/calculations, however, are needed in addition to particle size, to more conclusively understand the effect of NaCl on the colloidal stability of this mAb. Such measurements and calculations, combined with radar chart array analysis of the results, are currently underway.
Another interesting observation is that the average morphology of subvisible particles formed varies with solution pH and NaCl for shaking, but not stirring stress. The particles created through stirring appear to form by a similar mechanism since they have similar size and morphology across the pH values and NaCl concentrations as measured by MFI. In contrast, subvisible particles created through shaking have a diverse morphology (i.e., aspect ratio, intensity) that is sensitive to both the pH and NaCl concentration. This result suggests that in addition to changing the conformational and colloidal stability of a mAb, the formulation composition can also influence the mechanism of aggregation under certain stresses, potentially by influencing the nature of the interactions between the associating molecules.
Since formation of aggregates and particulates during long-term and accelerated stability studies affect formulation composition choices and decisions about storage conditions and shelf life, radar chart analysis could assist in these important comparisons during formulation development. Stability data are also a key part of comparability assessments, since changes in mAb drug product formulations or primary containers may affect the protein’s physical stability.44,45 Radar charts could potentially assist in these types of comparisons as well. For example, the effect of formulation impurities from prefilled syringes such as silicon oil46,47 and tungsten48 on protein particle formation could be evaluated by radar chart analysis using appropriate filters of the MFI data to separate silicon oil droplets from protein particles.25,29
Conclusions
When conducting high throughput screening studies of protein instability under various formulation conditions, data visualization methods are crucial to quickly and effectively interpret large quantities of physical stability data.49,50 Radar chart analysis offers an exciting opportunity to improve the throughput and effectiveness of analyzing protein aggregation and particle formation studies. Additionally, there is potential to combine particle size/concentration data sets with more accurate measurements/approximations of protein particle volume, partial specific volume, and mass (in development). Radar chart analysis should also provide an improved understanding of the effect of formulation composition and environmental stresses on the mechanisms of mAb particle formation.
Supplementary Material
Acknowledgments
The authors wish to thank and acknowledge Janssen R&D for providing an IgG1 mAb for this study as well as financial support from the Higuchi Predoctoral Fellowship, the Kansas Bioscience Authority and CK support from NIH biotechnology training grant 5-T32-GM008359.
References
- 1.Carpenter JF, Randolph TW, Jiskoot W, Crommelin DJ, Middaugh CR, Winter G, Fan YX, Kirshner S, Verthelyi D, Kozlowski S, Clouse KA, Swann PG, Rosenberg A, Cherney B. Overlooking subvisible particles in therapeutic protein products: gaps that may compromise product quality. Journal of pharmaceutical sciences. 2009;98(4):1201–1205. doi: 10.1002/jps.21530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo Q, Joubert MK, Stevenson R, Ketchem RR, Narhi LO, Wypych J. Chemical modifications in therapeutic protein aggregates generated under different stress conditions. The Journal of biological chemistry. 2011;286(28):25134–25144. doi: 10.1074/jbc.M110.160440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joubert MK, Luo Q, Nashed-Samuel Y, Wypych J, Narhi LO. Classification and characterization of therapeutic antibody aggregates. The Journal of biological chemistry. 2011;286(28):25118–25133. doi: 10.1074/jbc.M110.160457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filipe V, Jiskoot W, Basmeleh AH, Halim A, Schellekens H, Filipe V. Immunogenicity of different stressed IgG monoclonal antibody formulations in immune tolerant transgenic mice. mAbs. 2012;4(6) doi: 10.4161/mabs.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherer TM, Leung S, Owyang L, Shire SJ. Issues and challenges of subvisible and submicron particulate analysis in protein solutions. The AAPS journal. 2012;14(2):236–243. doi: 10.1208/s12248-012-9335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu L, Randolph TW, Carpenter JF. Particles shed from syringe filters and their effects on agitation-induced protein aggregation. Journal of pharmaceutical sciences. 2012;101(8):2952–2959. doi: 10.1002/jps.23225. [DOI] [PubMed] [Google Scholar]
- 7.Ripple DC, Dimitrova MN. Protein Particles: What We Know and What We Do Not Know. Journal of pharmaceutical sciences. 2012 doi: 10.1002/jps.23242. [DOI] [PubMed] [Google Scholar]
- 8.Chi EY, Krishnan S, Randolph TW, Carpenter JF. Physical stability of proteins in aqueous solution: mechanism and driving forces in nonnative protein aggregation. Pharmaceutical research. 2003;20(9):1325–1336. doi: 10.1023/a:1025771421906. [DOI] [PubMed] [Google Scholar]
- 9.Philo JS, Arakawa T. Mechanisms of protein aggregation. Current pharmaceutical biotechnology. 2009;10(4):348–351. doi: 10.2174/138920109788488932. [DOI] [PubMed] [Google Scholar]
- 10.Neudecker P, Robustelli P, Cavalli A, Walsh P, Lundstrom P, Zarrine-Afsar A, Sharpe S, Vendruscolo M, Kay LE. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336(6079):362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 11.Brummitt RK, Nesta DP, Chang L, Chase SF, Laue TM, Roberts CJ. Nonnative aggregation of an IgG1 antibody in acidic conditions: part 1. Unfolding, colloidal interactions, and formation of high-molecular-weight aggregates. Journal of pharmaceutical sciences. 2011;100(6):2087–2103. doi: 10.1002/jps.22448. [DOI] [PubMed] [Google Scholar]
- 12.Arosio P, Rima S, Morbidelli M. Aggregation mechanism of an IgG2 and two IgG1 monoclonal antibodies at low pH: from oligomers to larger aggregates. Pharmaceutical research. 2013;30(3):641–654. doi: 10.1007/s11095-012-0885-3. [DOI] [PubMed] [Google Scholar]
- 13.Kroetsch AM, Sahin E, Wang HY, Krizman S, Roberts CJ. Relating particle formation to salt- and pH-dependent phase separation of non-native aggregates of alpha-chymotrypsinogen A. Journal of pharmaceutical sciences. 2012;101(10):3651–3660. doi: 10.1002/jps.23264. [DOI] [PubMed] [Google Scholar]
- 14.Kim N, Remmele RL, Jr, Liu D, Razinkov VI, Fernandez EJ, Roberts CJ. Aggregation of anti-streptavidin immunoglobulin gamma-1 involves Fab unfolding and competing growth pathways mediated by pH and salt concentration. Biophysical chemistry. 2013;172:26–36. doi: 10.1016/j.bpc.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Gokarn YR, Fesinmeyer RM, Saluja A, Razinkov V, Chase SF, Laue TM, Brems DN. Effective charge measurements reveal selective and preferential accumulation of anions, but not cations, at the protein surface in dilute salt solutions. Protein science : a publication of the Protein Society. 2011;20(3):580–587. doi: 10.1002/pro.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fesinmeyer RM, Hogan S, Saluja A, Brych SR, Kras E, Narhi LO, Brems DN, Gokarn YR. Effect of ions on agitation- and temperature-induced aggregation reactions of antibodies. Pharmaceutical research. 2009;26(4):903–913. doi: 10.1007/s11095-008-9792-z. [DOI] [PubMed] [Google Scholar]
- 17.Das TK. Protein particulate detection issues in biotherapeutics development--current status. AAPS PharmSciTech. 2012;13(2):732–746. doi: 10.1208/s12249-012-9793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Narhi LO, Jiang Y, Cao S, Benedek K, Shnek D. A critical review of analytical methods for subvisible and visible particles. Current pharmaceutical biotechnology. 2009;10(4):373–381. doi: 10.2174/138920109788488905. [DOI] [PubMed] [Google Scholar]
- 19.Demeule B, Messick S, Shire SJ, Liu J. Characterization of particles in protein solutions: reaching the limits of current technologies. The AAPS journal. 2010;12(4):708–715. doi: 10.1208/s12248-010-9233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolls S, Tantipolphan R, Wiggenhorn M, Winter G, Jiskoot W, Friess W, Hawe A. Particles in therapeutic protein formulations, Part 1: overview of analytical methods. Journal of pharmaceutical sciences. 2012;101(3):914–935. doi: 10.1002/jps.23001. [DOI] [PubMed] [Google Scholar]
- 21.Kumru OS, Liu J, Ji JA, Cheng W, Wang YJ, Wang T, Joshi SB, Middaugh CR, Volkin DB. Compatibility, physical stability, and characterization of an IgG4 monoclonal antibody after dilution into different intravenous administration bags. Journal of pharmaceutical sciences. 2012;101(10):3636–3650. doi: 10.1002/jps.23224. [DOI] [PubMed] [Google Scholar]
- 22.Sharma DK, King D, Oma P, Merchant C. Micro-flow imaging: flow microscopy applied to sub-visible particulate analysis in protein formulations. The AAPS journal. 2010;12(3):455–464. doi: 10.1208/s12248-010-9205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnard JG, Singh S, Randolph TW, Carpenter JF. Subvisible particle counting provides a sensitive method of detecting and quantifying aggregation of monoclonal antibody caused by freeze-thawing: insights into the roles of particles in the protein aggregation pathway. Journal of pharmaceutical sciences. 2011;100(2):492–503. doi: 10.1002/jps.22305. [DOI] [PubMed] [Google Scholar]
- 24.Wuchner K, Buchler J, Spycher R, Dalmonte P, Volkin DB. Development of a microflow digital imaging assay to characterize protein particulates during storage of a high concentration IgG1 monoclonal antibody formulation. Journal of pharmaceutical sciences. 2010;99(8):3343–3361. doi: 10.1002/jps.22123. [DOI] [PubMed] [Google Scholar]
- 25.Strehl R, Rombach-Riegraf V, Diez M, Egodage K, Bluemel M, Jeschke M, Koulov A. Discrimination Between Silicone Oil Droplets and Protein Aggregates in Biopharmaceuticals: A Novel Multiparametric Image Filter for Sub-visible Particles in Microflow Imaging Analysis. J Pharm Res. 2011 doi: 10.1007/s11095-011-0590-7. [DOI] [PubMed] [Google Scholar]
- 26.Zolls S, Gregoritza M, Tantipolphan R, Wiggenhorn M, Winter G, Friess W, Hawe A. How subvisible particles become invisible-relevance of the refractive index for protein particle analysis. Journal of pharmaceutical sciences. 2013 doi: 10.1002/jps.23479. [DOI] [PubMed] [Google Scholar]
- 27.Sharma DK, Oma P, Pollo MJ, Sukumar M. Quantification and characterization of subvisible proteinaceous particles in opalescent mAb formulations using micro-flow imaging. Journal of pharmaceutical sciences. 2010;99(6):2628–2642. doi: 10.1002/jps.22046. [DOI] [PubMed] [Google Scholar]
- 28.Patel AR, Lau D, Liu J. Quantification and characterization of micrometer and submicrometer subvisible particles in protein therapeutics by use of a suspended microchannel resonator. Analytical chemistry. 2012;84(15):6833–6840. doi: 10.1021/ac300976g. [DOI] [PubMed] [Google Scholar]
- 29.Weinbuch D, Zolls S, Wiggenhorn M, Friess W, Winter G, Jiskoot W, Hawe A. Micro-flow imaging and resonant mass measurement (archimedes) - complementary methods to quantitatively differentiate protein particles and silicone oil droplets. Journal of pharmaceutical sciences. 2013;102(7):2152–2165. doi: 10.1002/jps.23552. [DOI] [PubMed] [Google Scholar]
- 30.Maddux NR, Joshi SB, Volkin DB, Ralston JP, Middaugh CR. Multidimensional methods for the formulation of biopharmaceuticals and vaccines. Journal of pharmaceutical sciences. 2011 doi: 10.1002/jps.22618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark NR, Ma'ayan A. Introduction to statistical methods to analyze large data sets: principal components analysis. Science signaling. 2011;4(190):tr3. doi: 10.1126/scisignal.2001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng W, Joshi SB, He F, Brems DN, He B, Kerwin BA, Volkin DB, Middaugh CR. Comparison of high-throughput biophysical methods to identify stabilizing excipients for a model IgG2 monoclonal antibody: conformational stability and kinetic aggregation measurements. Journal of pharmaceutical sciences. 2012;101(5):1701–1720. doi: 10.1002/jps.23076. [DOI] [PubMed] [Google Scholar]
- 33.Bhambhani A, Kissmann JM, Joshi SB, Volkin DB, Kashi RS, Middaugh CR. Formulation design and high-throughput excipient selection based on structural integrity and conformational stability of dilute and highly concentrated IgG1 monoclonal antibody solutions. Journal of pharmaceutical sciences. 2012;101(3):1120–1135. doi: 10.1002/jps.23008. [DOI] [PubMed] [Google Scholar]
- 34.Thakkar SV, Joshi SB, Jones ME, Sathish HA, Bishop SM, Volkin DB, Middaugh CR. Excipients differentially influence the conformational stability and pretransition dynamics of two IgG1 monoclonal antibodies. Journal of pharmaceutical sciences. 2012;101(9):3062–3077. doi: 10.1002/jps.23187. [DOI] [PubMed] [Google Scholar]
- 35.Iyer V, Maddux N, Hu L, Cheng W, Youssef AK, Winter G, Joshi SB, Volkin DB, Middaugh CR. Comparative signature diagrams to evaluate biophysical data for differences in protein structure across various formulations. Journal of pharmaceutical sciences. 2013;102(1):43–51. doi: 10.1002/jps.23367. [DOI] [PubMed] [Google Scholar]
- 36.Kim JH, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein science : a publication of the Protein Society. 2012;21(10):1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manikwar P, Majumdar R, Hickey JM, Thakkar SV, Samra HS, Sathish HA, Bishop SM, Middaugh CR, Weis DD, Volkin DB. Correlating excipient effects on conformational and storage stability of an IgG1 monoclonal antibody with local dynamics as measured by hydrogen/deuterium-exchange mass spectrometry. Journal of pharmaceutical sciences. 2013;102(7):2136–2151. doi: 10.1002/jps.23543. [DOI] [PubMed] [Google Scholar]
- 38.Bond MD, Panek ME, Zhang Z, Wang D, Mehndiratta P, Zhao H, Gunton K, Ni A, Nedved ML, Burman S, Volkin DB. Evaluation of a dual-wavelength size exclusion HPLC method with improved sensitivity to detect protein aggregates and its use to better characterize degradation pathways of an IgG1 monoclonal antibody. Journal of pharmaceutical sciences. 2010;99(6):2582–2597. doi: 10.1002/jps.22034. [DOI] [PubMed] [Google Scholar]
- 39.Fischer H, Polikarpov I, Craievich AF. Average protein density is a molecular-weight-dependent function. Protein science : a publication of the Protein Society. 2004;13(10):2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simler BR, Hui G, Dahl JE, Perez-Ramirez B. Mechanistic complexity of subvisible particle formation: links to protein aggregation are highly specific. Journal of pharmaceutical sciences. 2012;101(11):4140–4154. doi: 10.1002/jps.23299. [DOI] [PubMed] [Google Scholar]
- 41.Burg TP, Godin M, Knudsen SM, Shen W, Carlson G, Foster JS, Babcock K, Manalis SR. Weighing of biomolecules, single cells and single nanoparticles in fluid. Nature. 2007;446(7139):1066–1069. doi: 10.1038/nature05741. [DOI] [PubMed] [Google Scholar]
- 42.Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta crystallographica Section D, Biological crystallography. 2000;56(Pt 7):791–794. doi: 10.1107/s090744490000679x. [DOI] [PubMed] [Google Scholar]
- 43.Wilson GA, Manning MC. Flow imaging: moving toward best practices for subvisible particle quantitation in protein products. Journal of pharmaceutical sciences. 2013;102(3):1133–1134. doi: 10.1002/jps.23445. [DOI] [PubMed] [Google Scholar]
- 44.Lubiniecki A, Volkin DB, Federici M, Bond MD, Nedved ML, Hendricks L, Mehndiratta P, Bruner M, Burman S, Dalmonte P, Kline J, Ni A, Panek ME, Pikounis B, Powers G, Vafa O, Siegel R. Comparability assessments of process and product changes made during development of two different monoclonal antibodies. Biologicals : journal of the International Association of Biological Standardization. 2011;39(1):9–22. doi: 10.1016/j.biologicals.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 45.Federici M, Lubiniecki A, Manikwar P, Volkin DB. Analytical lessons learned from selected therapeutic protein drug comparability studies. Biologicals : journal of the International Association of Biological Standardization. 2013;41(3):131–147. doi: 10.1016/j.biologicals.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Gerhardt A, Bonam K, Bee JS, Carpenter JF, Randolph TW. Ionic strength affects tertiary structure and aggregation propensity of a monoclonal antibody adsorbed to silicone oil-water interfaces. Journal of pharmaceutical sciences. 2013;102(2):429–440. doi: 10.1002/jps.23408. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Pinnamaneni S, Quan Y, Jaiswal A, Andersson FI, Zhang X. Mechanistic understanding of protein-silicone oil interactions. Pharmaceutical research. 2012;29(6):1689–1697. doi: 10.1007/s11095-012-0696-6. [DOI] [PubMed] [Google Scholar]
- 48.Seidl A, Hainzl O, Richter M, Fischer R, Bohm S, Deutel B, Hartinger M, Windisch J, Casadevall N, London GM, Macdougall I. Tungsten-induced denaturation and aggregation of epoetin alfa during primary packaging as a cause of immunogenicity. Pharmaceutical research. 2012;29(6):1454–1467. doi: 10.1007/s11095-011-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Razinkov VI, Treuheit MJ, Becker GW. Methods of high throughput biophysical characterization in biopharmaceutical development. Current drug discovery technologies. 2013;10(1):59–70. doi: 10.2174/157016313804998915. [DOI] [PubMed] [Google Scholar]
- 50.Samra HS, He F. Advancements in high throughput biophysical technologies: applications for characterization and screening during early formulation development of monoclonal antibodies. Molecular pharmaceutics. 2012;9(4):696–707. doi: 10.1021/mp200404c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.