Abstract
The ruthenium (II) polypyridyl complexes (RPCs) Δ-[(phen)2Ru(tatpp)]Cl2 (Δ-[3]Cl2) and ΔΔ-[(phen)2Ru(tatpp)Ru(phen)2]Cl4 (ΔΔ-[4]Cl4) are a new generation of metal-based anti-tumor agents. These RPCs bind DNA via intercalation of the tatpp ligand which itself is redox-active and easily reduced at biologically relevant potentials. We have previously shown that RPC 44+ cleaves DNA when reduced by glutathione to a radical species, and that this DNA cleavage is potentiated under hypoxic conditions in vitro. Here we show that 32+ also exhibits free-radical mediated DNA cleavage in vitro, and that 32+ and 44+ both exhibit selective cytotoxicity towards cultured malignant cell lines, and marked inhibition of tumor growth in vivo. The murine acute toxicity of RPCs 32+ and 44+ (maximum tolerable doses (MTD’s) ~ 65 µmol/kg) is comparable with that for cisplatin (LD50 ~57 µmol/kg) but unlike cisplatin, RPC’s are generally cleared from the body unchanged via renal excretion without appreciable metabolism or nephrotoxic side effects. RPCs 32+ and 44+ are demonstrated to suppress growth of human non-small cell lung carcinoma (~83%), show potentiated cytotoxicity in vitro under hypoxic conditions, and induce apoptosis through both intrinsic and extrinsic pathways. The novel hypoxia-enhanced DNA cleavage activity and biological activity suggest a promising new anti-cancer pharmacophore based on metal complexes with aromatic ligands that are easily reduced at biologically accessible potentials.
Keywords: Ruthenium, lung cancer, hypoxia, tumor xenografts
INTRODUCTION
The success of cisplatin (cis-Pt(NH3)2Cl2) as a chemotherapeutic agent (1) has led to the development of other complexes of platinum, as well as complexes of ruthenium, which have similar substitution kinetics, but differ in biological reactivity and toxicities in vivo. (2–4) To date, there are no ruthenium-based anti-cancer drugs in routine clinical use, however imidazolium [trans-imidazoledimethylsulfoxide-tetrachlororuthenate (III)] (NAMI-A) and indazolium [trans-tetrachlorobis(1H-indazole) ruthenate(III)] (KP1109) have advanced to preclinical and/or clinical trials.(5–8) Most of the success to date with ruthenium based compounds has been with complexes bearing one or more labile ligands which, like cisplatin, allows the metal to directly bind with biological targets, particularly DNA. NAMI-A, KP1109, and the anti-tumor agents [cis-bis(acetonitrile)-1,10-phenanthroline-2-phenylpyridineruthenium(II)] hexafluorophosphate (RDC11) and ruthenium-aryl-X complexes(9, 10) all contain at least one labile ligand and it is postulated that these complexes directly bind biological targets upon loss of this ligand in vivo.
The biological activity of coordinately saturated ruthenium (II) polypyridyl complexes (RPC’s), such as the trisphenanthroline complex (12+) (Fig. 1), was extensively studied by Dwyer and Schulman(11) in the 1950’s and 60’s and even prior to that by Beccari in the late 1930’s.(12) These complexes differ from cisplatin and ruthenium complexes such as NAMI-A and KP1109 in that the ruthenium (II) ion lacks labile ligands and cannot directly form bonds with biological targets. Instead, early studies with radiolabeled [106Ru(phen)3]2+ showed that the intact complex cation was the bioactive unit, that this complex was not metabolized in vivo and did not accumulate in any organ, and that nearly all the complex was recovered in the urine.(13) Despite this lack of reactivity in vivo, these RPC’s were shown to possess biological activity both in vitro and in vivo.(13–16)
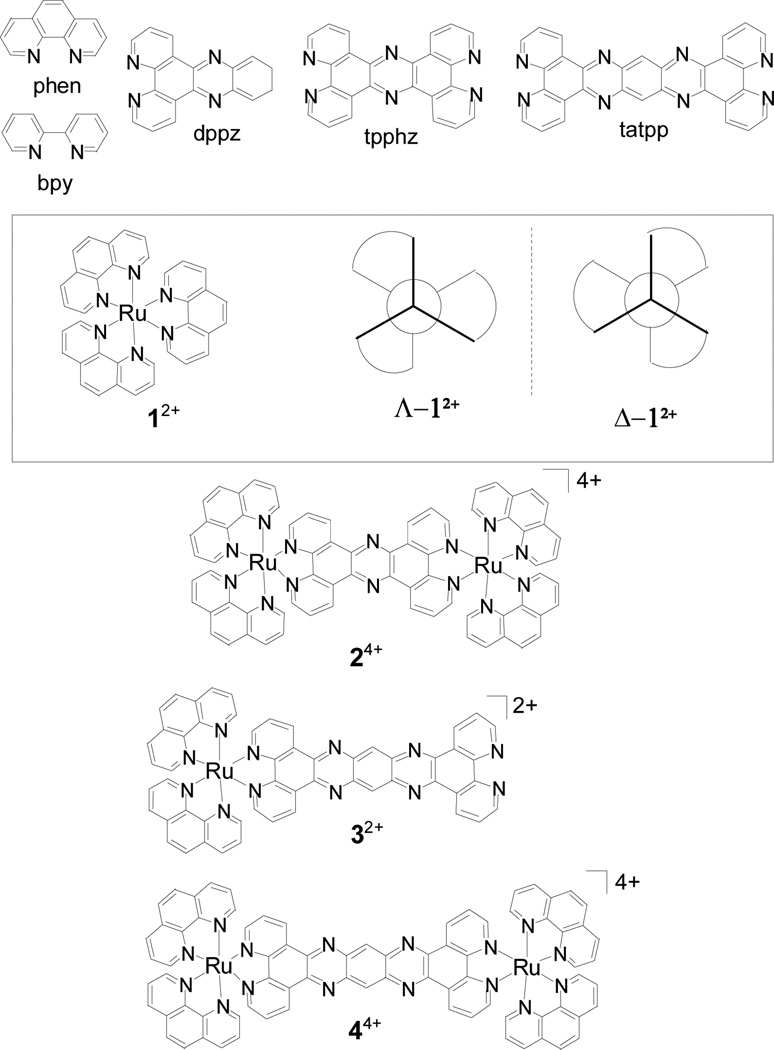
Fig. 1. Ruthenium-polypyridyl complexes (RPCs) and associated ligand structures.
The chemical structure of the important ligands and RPCs, 12+, 24+, 32+, and 44+, in this study are shown. Chloride counterions are not shown. All of the complexes are chiral due to the three-fold helical chirality at the ruthenium centers, shown schematically for RPC 12+. These RPCs are isolated as a racemic mixture for the monomers (12+ and 32+) or as diastereotopic mixture of stereoisomers (ΔΔ, ΛΛ and ΔΛ) for the dinuclear complex (24+ and 44+). In certain cases, the RPCs were prepared in diastereomerically and enantiomerically pure forms, i.e., Δ-12+ and Λ-12+, Δ-32+ and Λ-32+, ΔΔ-44+, ΛΛ-44+ and ΔΛ-44+, where the descriptor Δ or Λ is used to indicate right or left-handed chirality, respectively, at the ruthenium centers.
RPC 12+ is modestly cytotoxic (IC50 ~ 90 µM) with enhanced cytoxicity generally observed by increasing the lipophilicity of the complex.(17) The 3,4,7,8-tetramethyl-1,10-pheanthroline derivative of 12+ was among the most cytotoxic of the derivatives found and was shown to inhibit the growth of dispersed tumor cells (Landshultz ascites) in mice.(14) Since these early studies, there have been numerous studies exploring the DNA-binding activity of RPCs (18) and to a lesser extent the cytotoxicity of RPCs,(19–23) however, the anti-cancer activity of these complexes in vivo has not been extensively studied. The paucity of follow up studies is due, in part, to their observed neurotoxicity, as 12+ has been shown to be a competitive inhibitor of acetyl cholinesterase (AChE) both in vitro and in vivo.(13, 24, 25) As a consequence, the minimum lethal doses of 12+ (iodo salt) in mice is 12 µmol/kg when administered by i.p. injection. A study of the peak ruthenium blood concentrations showed that RPCs able to diffuse quickly through tissue and enter the bloodstream were more neurotoxic than those slower to do so and that by increasing the complex lipophilicity it was possible to ameliorate the neurotoxicity.(13) Interestingly, 12+ is non-toxic when administered orally.(11)
It has been possible to track the intracellular distribution of many of these RPC’s due to their inherent luminescence with both the nucleus and mitochondria often showing significant accumulation.(19, 22, 26–28) As many RPCs have been shown to bind DNA, it is often assumed that this is the biological target,(29) although the mitochondria(19, 25, 30) and the cell-cell interface (20, 31) are also potential targets. In general, these RPCs bind to duplex DNA via electrostatic and intercalative modes but usually they do not damage DNA unless an external factor, such a light irradiation, is introduced.(32, 33)
We have shown that the dinuclear tatpp complex, [(phen)2Ru(tatpp)Ru(phen)2]Cl4 ([4]Cl4), (Fig. 1) is an effective DNA cleaving agent upon in situ reduction by glutathione (GSH) and that the cleavage activity is potentiated upon lowering the oxygen concentration.(34) We also demonstrated that the reduced forms of 44+ are competent for DNA cleavage under anaerobic conditions without GSH present. Under identical reducing conditions, the closely related RPCs 12+, [(phen)2Ru(dppz)]2+ and [(phen)2Ru(tpphz)Ru(phen)2]4+ (24+)(see Figure 1 for the structures of dppz, tpphz, and 24+) are neither reduced nor do they exhibit any DNA cleavage activity. We postulated that the redox-activity of the RPC, 44+, was responsible for the DNA cleavage activity and that the cleavage activity may be useful therapeutically. Moreover, the inverse sensitivity to the dioxygen concentration could enhance the cleavage activity in hypoxic tumors. Finally, we reasoned that the increased size and lipophilicity of 44+ and the related mononuclear complex, [(phen)2Ru(tatpp)]Cl2 (32+) may ameliorate any neurotoxic side effects of these complexes in vivo. Herein, we evaluate 32+ and 44+ as potential anti-cancer agents in a pre-clinical study.
MATERIALS AND METHODS
Abbreviations, Stereochemistry, and Nomenclature
The ruthenium complexes, [1]Cl2, [2]Cl4, [3]Cl2, and [4]Cl4, are all water soluble salts composed of the complex cations, 12+, 24+, 32+, and 44+ and chloride anions. These complex cations, collectively identified as RPCs, are composed of Ru(II) ions bound to 1,10-phenanthroline (phen), tpphz, and/or (tatpp) polypyridyl ligands (Fig. 1). If the counterion is not explicitly listed, it can be assumed to be chloride. All of the complexes are chiral (Δ or Λ) due to the three-fold helical chirality at the ruthenium centers where the descriptor Δ or Λ is used to indicate right or left-handed chirality, respectively, at the ruthenium centers. Typically, these complex cations are isolated as a racemic mixture for the monomers (12+ and 32+) or as diastereotopic mixtures (ΔΔ, ΛΛ and ΔΛ) for the dinuclear complex (24+ and 44+). The complex cations are chemically robust and stable to decomposition and racemization in aqueous solution over extended periods of time (i.e. months). Because of this stability, it is possible to prepare and isolate these complexes in diastereomerically and enantiomerically pure forms, i.e., Δ-12+ and Λ-12+, Δ-32+ and Λ-32+, ΔΔ-44+, ΛΛ-44+ and ΔΛ-44+. In the following studies, we typically used the racemic or diastereotopic mixture initially, and only used enantiopure substances where specifically indicated with the appropriate chiral descriptor. If no descriptor is given, then the racemate was used for 12+ and 32+ and the diastereotopic mixture was used for 24+ and 44+.
Reagents
Reagents for cell culture were purchased from Life Sciences Technologies, Inc., Grand Island, NY. Supercoiled plasmid pUC 18 DNA was purchased from Bayou Biolabs, New England. Agarose, ethidium bromide, glutathione (GSH), dimethyl sulfoxide (DMSO), 2,2,6,6-tetramethylpiperidine-1-oxyl (TEMPO), trizma base and horseradish peroxidase (HRP)-conjugated secondary antibodies were from Sigma Aldrich. Fluorescein terminal deoxynucleotidyl transferase (TdT)–mediated dUTP nick end labeling (TUNEL) apoptosis assay kit was from Promega (Madison, WI). pAkt (S473) antibody was procured from Upstate Cell Signaling (Lake Placid, NY). pPI3K (Y458), pEGFR (Y1068), and cleaved PARP antibodies were from Cell Signaling Technologies (Danvers, MA). Antibodies against Bcl2, Bax, cdk4, cyclin B1, pRb (S780), caspase, and GAPDH were obtained from Santa Cruz Biotechnology (Columbus, OH). CasPACE FITC-VAD-FMK in situ marker, apoptosis detection system was purchased from Promega Inc. (Madison, WI). Mounting medium Fluro-gel II containing DAPI was procured from Electron Microscopy Sciences, (Hatfield, PA, USA). The complexes, [1]Cl2(35) [2]Cl4(36) [(phen)2Ru(tpphz)]Cl2(36, 37), [3]Cl2 (38), [4]Cl4 (39), and select enantiomers, Δ-12+ and Λ-12+,(35) Δ-32+ and Λ-32+,(38) and diastereomers, ΔΔ-34+, ΛΛ-34+ and ΔΛ-34+,(40) were prepared previously described. Enantiopurity was ascertained using chiral HPLC as described in ref (35) and was 97% or greater for all complexes.
DNA Cleavage Assay
DNA cleavage was carried out as previously described.(34) Briefly, RPC was added to a reaction mixture containing 7 mM sodium phosphate (pH 7.0) and pUC18 DNA (0.1 µg/µL). The reaction was quenched by addition of 2 µL sodium acetate (pH 5.2) and 80 µL ethanol and incubated at −20 °C to complete precipitation. The DNA was pelleted, air-dried, resuspended in 15 mM Tris-HCl pH 8.0, 0.3 mM EDTA, 3% glycerol and 0.1% w/v bromophenol blue, and subjected to horizontal slab agarose (1%) gel at 80 V for 90 min with the gel immersed in TAE/0.2 mM ethidium bromide (0.2 mM). Results were visualized and recorded by using a UVPGDS 8000 gel analysis system. For studies under anaerobic conditions, all reagent solutions were degassed using N2. DNA stock was degassed using five freeze-pump-thaw cycles under N2. Degassed reagents were taken into a N2 glove box and all the solutions were prepared inside it to minimize further contamination with oxygen. DMSO and TEMPO were added inside the glove box where the assays were completed and reactions quenched by precipitating the DNA using 2 µL of degassed sodium acetate at pH 5.2 and 80 µL degassed ethanol under N2.
Cell Lines and Cultures
Human non small cell lung cancer (NSCLC) cell lines H358 (bronchioalveolar), H226 (squamous cell carcinoma), human aortic vascular smooth muscle cells (HAVSMC), and human umbilical vascular endothelial cells (HUVEC) were obtained and maintained as previously described.(41)
Cytotoxicity Assays
Cell viability and density was measured by counting trypan-blue dye excluding cells in a hemocytometer, and 2 × 104 viable cells in respective full culture media were placed into each well of a 96-well plate and allowed to grow at 37 °C for 24 h prior to addition of RPCs. After 96-h incubation, MTT assay was carried out as previously described.(41) Both Δ-[3]Cl2 and ΔΔ-[4]Cl4 were accepted by the Developmental Therapeutics Program (DTP) of the NCI for the NCI-60 cell line one-dose screen and were assigned registration numbers NSC 747949 and 747950, respectively. Only [Δ-3]Cl2 (NSC 747949) had sufficient activity in the single dose screen to proceed to the 5 dose screen. However, because the NCI-60 screen is not done under hypoxic conditions, thus may not truly reflect in-vivo activity, we included ΔΔ-[4]Cl4 in present studies.
Non-Hypoxic/Hypoxic cytotoxicity
H358 cells were plated in 60mm × 15mm cell dishes with equal cell densities respectively. 10 dishes were used per compound being tested; 5 dishes for normoxic and 5 dishes for hypoxic. Before assay each 5 set dishes were washed with PBS and media changed with 3 ml aliquot of compound solution and put into incubators (normoxic was monitored at 5% CO2 and 18% O2 and hypoxic was monitored at 5% CO2 and 1% O2) accordingly. Cells were left in both incubator conditions for 24 h and every 4 h cells were checked for viability. Cytotoxicity of cells was determined by cell count with hemocytometer w/Trypan blue staining.
In situ caspase-3 assay for Apoptosis
Normal (HUVEC) and lung cancer (H358) cells (0.1×106) were plated on glass cover slip in tissue culture treated 12 well plates and incubated with either 10 µM of Δ-32+ or ΔΔ-44+ for 12 h at 37 °C. Apoptotic cells were detected by staining with 5 µM Caspase FITC-VAD-FMK (Promega) in situ marker for 30 min in the dark. The slides were rinsed with PBS and fixed with 4% paraformaldehyde for 30 min, and mounted in a medium containing DAPI (1.5 µg/ml). Images were taken on Olympus Provis AX70 fluorescence microscope. Photographs taken at identical exposure at 40 × magnification are presented.
Effect of Ru-compounds on apoptosis by TUNEL assay
Normal (HUVEC) and lung cancer (H358) cells were grown on cover-slips. For Ru-compounds treatment, cells were incubated with 10 µM either Δ-32+ or ΔΔ-44+ before TUNEL assay using Promega fluorescence detection kit according to the protocol provided by the manufacturer. Slides were analyzed by fluorescence microscope (Olympus Provis AX70). Photographs taken at identical exposure at 40 × magnification are presented. Apoptotic cells showed green fluorescence.
Animal Studies
All animal experiments were carried out in accordance with a protocol approved by the Institutional Animal Care and Use Committee. Hsd: Athymic nude nu/nu mice were obtained from Harlan, Indianapolis, IN. C57 BL/6 mice were obtained from Harlan (Indianapolis, Indiana). The maximum tolerable dose (MTD, mg complex/kg mouse) was determined in 10 weeks old male mice (3 per group) treated with 100 µL (i.p. injection) of RPC at 6 concentrations ranging from 1 to 50 mg/mL. MTD was defined as the dose level causing no deaths. For lung cancer xenograft studies, H358 NSCLC cells (1 × 106/ µL PBS) were injected into the flanks of 10 wk nu/nu mice, Harlan (Indianapolis, Indiana), followed 14 days later by i.p. injection of RPC in 100 µL PBS. Animals were examined daily for signs of distress and tumors were measured in two dimensions using Vernier calipers.(42) For statistical analysis, the mean tumor volume data versus time for the two RPC treated groups were combined to a single group with n = 6 and a control group. A repeated measures ANOVA analysis of the treated and control group data was obtained using the SAS® programmer PROC MIXED program.
RESULTS
Cytotoxicity
The IC50 data from initial screening studies of antineoplastic activity of a series of RPCs were performed in two NSCLC cell lines, H358 and H226 (Table 1). The tatpp containing complexes 32+ and 44+ were significantly more cytotoxic (IC50s ~12–16 µM) than the tpphz complexes, 24+ and [(phen)2Ru(tpphz)]4+ (~45 µM), or the homoleptic complexes phen 12+ (90 µM) and [Ru(bpy)3]2+ (200 µM) for both cell lines. Stereoisomers with the Δ configuration were more cytotoxic than those with the Λ configuration. The ΔΔ enantiomer of 44+ and the Δ enantiomer for 32+ were the most cytotoxic with IC50 values approximately one-half that of their mirror images. The IC50 value for racemic 32+ falls between the values of the two enantiomers suggesting that the toxicity of the racemate is simply the average of the two enantiomers. For the stereochemically more complicated dimers, the mixture of ΔΔ-44+, ΛΛ-44+, and ΔΛ-44+ (mix-44+), the IC50 also appears to be a weighted average of the IC50’s for individual diastereomers. Interestingly, the IC50 for meso-diastereomer, ΔΛ-44+, was not significantly different from the ΛΛ–diastereomer, indicating that both chiral centers must be Δ to achieve a significant biological effect.
Table 1.
IC50 of Ru (II) polypyridyl complexes (H358 and H226)
| Complex | Abbr. | IC50 (µM) H358 |
IC50 (µM) H226 |
|---|---|---|---|
| rac-[Ru(phen)3]2+ | 12+ | 86.7 ± 4.1 | 92.8 ± 5.7 |
| Δ-[Ru(phen)3]2+ | Δ-12+ | 64.8 ± 4.2 | 93.6 ± 6.3 |
| Λ-[Ru(phen)3]2+ | Λ-12+ | 61.4 ± 4.7 | 116 ± 6.9 |
| rac-[Ru(bpy)3]2+ | 209 ± 16 | 198 ± 15 | |
| rac-[(phen)2Ru(tatpp)]2+ | 32+ | 13.2 ± 1.8 | 12.5 ± 1.9 |
| Δ-[(phen)2Ru(tatpp)]2+ | Δ-32+ | 8.8 ± 1.0 | 6.7 ± 1.0 |
| Λ-[(phen)2Ru(tatpp)]2+ | Λ-32+ | 13.8 ± 1.5 | 13.0 ± 2.0 |
| mix-[(phen)2Ru(tatpp)Ru(phen)2]4+ | 44+ | 15.2 ± 1.8 | 16.2 ± 1.9 |
| ΔΔ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΔΔ-44+ | 9.5 ± 1.2 | 9.3 ± 0.9 |
| ΛΛ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΛΛ-44+ | 16.7 ± 1.0 | 17.3 ± 1.5 |
| ΔΛ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΔΛ-44+ | 15.3 ± 1.5 | 18.6 ± 1.5 |
| mix-[(phen)2Ru(tpphz)Ru(phen)2]4+ | 24+ | 41.8 ± 2.7 | 51.1 ± 3.4 |
| rac-[(phen)2Ru(tpphz)]2+ | 44.0 ± 3.0 | 49.2 ± 3.1 | |
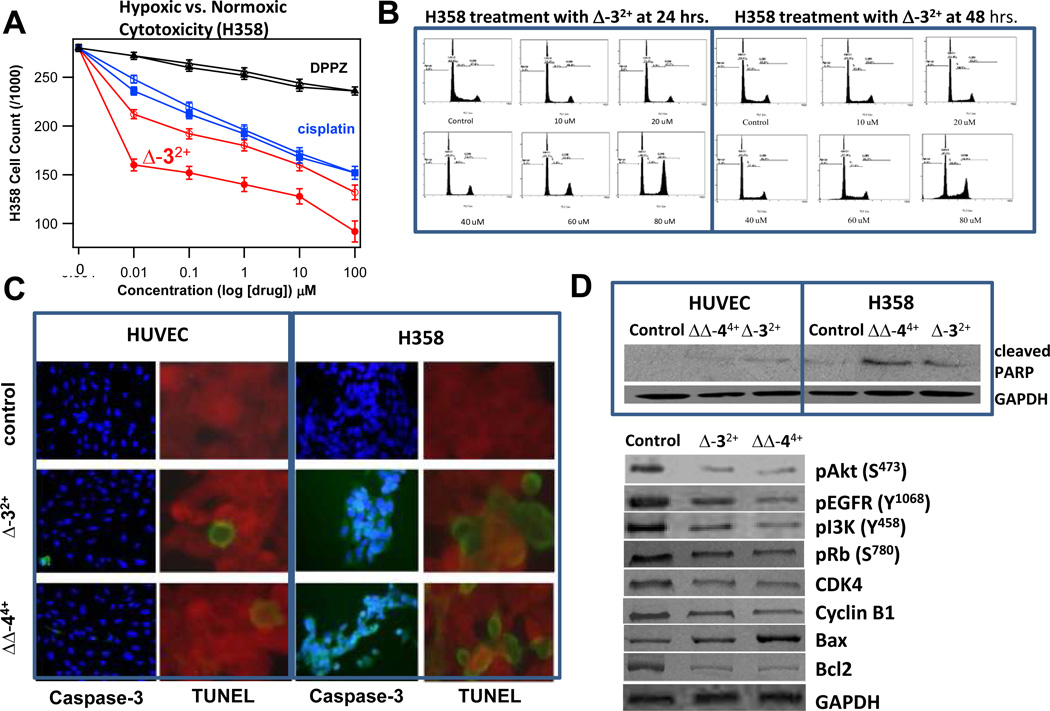
As shown in Figure 2A, the cytotoxicity of Δ-32+ is enhanced when the H358 cells were incubated under hypoxic conditions (1 %O2) relative to normoxic conditions. This figure also includes the data from two controls, cisplatin and [Ru(phen)2(dppz)]2+, neither of which showed significant potentiation under hypoxic conditions as expected. Significantly, Δ-32+ and ΔΔ-44+ are significantly more cytotoxic towards malignant cell lines as the IC50’s for normal HUVEC and HUVSMC cells are around 100 µM, an approximate 10-fold difference in cytotoxicity. This selectivity is also observed in the Caspase-3 and TUNEL assays, (Fig. 2C) and appears to be unique to the tatpp-based complexes. Dwyer and co-workers found little selectivity in the cytotoxicity of RPCs such as 12+ or its tetramethylphenanthroline derivative towards malignant, adult, embryonic, epithielial, and fibroblastic cells.(11)
Fig. 2. Hypoxia-Selective, anti-proliferative and pro-apoptotic effect of ruthenium compounds in human NSCLC.
H358m cell count after incubation with Δ-32+, cisplatin, and [Ru(phen)2(dppz)]2+ (DPPZ) for 24 h in hypoxic (1 % O2) and non-hypoxic incubators (filled and open symbols, respectively). Cells were checked every 4 h to ensure viability. Complex concentrations were made prior to inoculation to ensure consistent time. Cell viability was counted using a hemocytometer w/ Trypan blue staining. 5 dishes were used per set of different concentrations from 0.01 – 100 µM in the cell medium and were put in respectively in 25 min increments to allow cell counting with respect to 24 h mark for each cell set. (panel A). Inhibitory effect of ruthenium compounds Δ-32+ and ΔΔ-44+ on cell cycle distribution was determined by FACS analysis (panel B). For in situ caspase-cleavage assay, normal (HUVEC) and lung cancer (H358) cells (0.1×106) were grown on glass cover-slips and treated with either 10 µM of Δ-32+ and ΔΔ-44+ for 12 h and caspase-cleavage was measured using caspase FITC-VAD-FMK kit. Activated caspase-positive cells appeared fluorescing green and pink when co-stained with DAPI. For detection of late event of apoptosis, TUNEL apoptotic assay was performed. Green fluorescence represents apoptotic cells (panel C). The effect of Δ-32+ and ΔΔ-44+ on PARP-cleavage in H358 cells and on apoptotic, proliferative, and tumor suppressor proteins was assayed. H358 cells, control, and treated (10 µM either Δ-32+ and ΔΔ-44+ for 24 h), were lysed and analyzed by Western blot by using specific antibodies for each protein. Membranes were stripped and re-probed for GAPDH as a loading control (panel D).
DNA Cleavage by Δ-32+ and ΔΔ-44+ under aerobic and anaerobic conditions
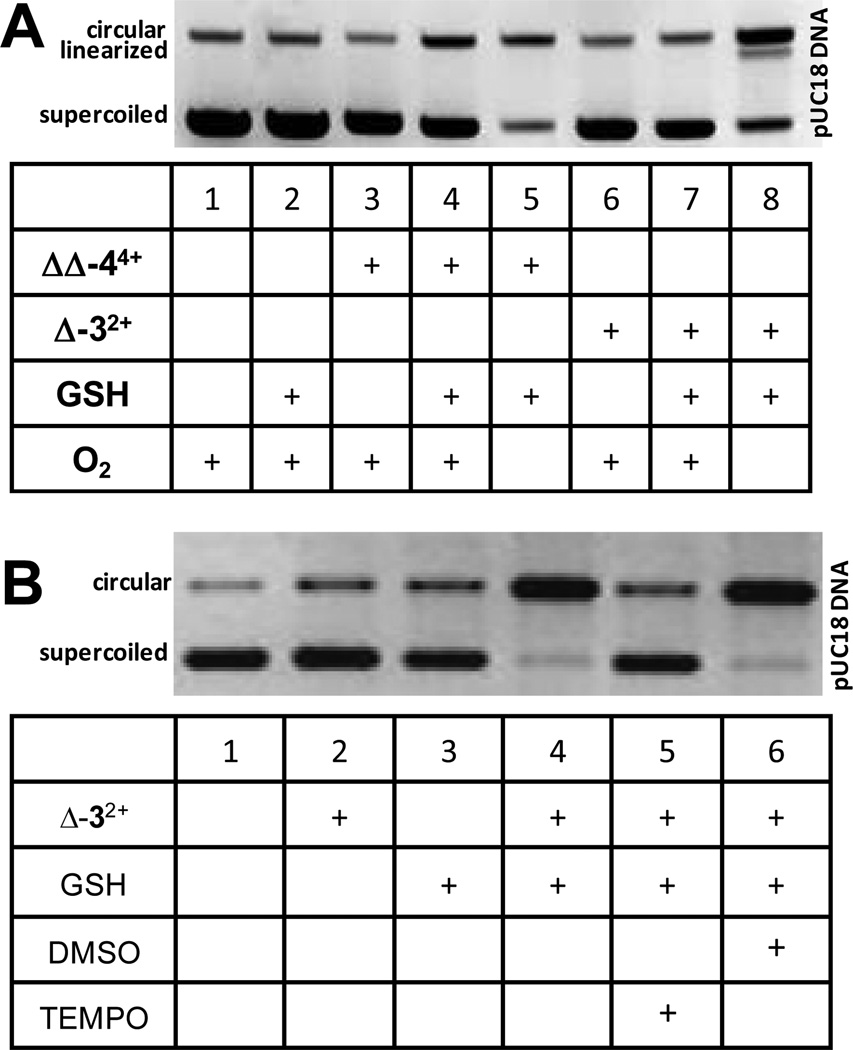
Since the above studies indicated that Δ-32+ and ΔΔ-44+ were the most active compounds, sufficient amounts of these were synthesized, purified, and authenticated for further studies. We have previously shown that the reduced form of the di-ruthenium compound 44+ shows hypoxia sensitive DNA cleavage and that this reduced complex is readily formed via addition of common agents such GSH or ascorbic acid.(34) The data presented here in Figure 2 confirmed similar activity of the mono-ruthenium complex, Δ-32+. Supercoiled plasmid DNA was incubated with Δ-32+ and ΔΔ-44+ in the presence or absence of GSH and O2. In this assay, the conversion of supercoiled plasmid DNA to the circular form is used to monitor single-strand cleavage events using agarose gel electrophoresis. As can be seen in Figure 3A, RPCs Δ-32+ and ΔΔ-44+ only show DNA cleavage when GSH is present (lanes 4, 5, 7, 8) and this cleavage is appreciably enhanced when O2 is absent (lanes 5 and 8, respectively). As shown in Figure 3B, RPC Δ-32+ was also incubated with DNA in the presence of GSH and the oxygen radical scavenger DMSO (43) and the carbon and metal centered radical scavenger TEMPO (43) under anaerobic conditions. As seen in Lane 4 (Fig 3B) there is an extensive amount of DNA cleavage upon treatnment with Δ-32+ and GSH under anerobic conditions. Addition of DMSO did not affect this cleavage activity (lane 6) whereas addition of TEMPO (lane 5) appreciably attenuated the cleavage, supporting a carbon-based radical intermediate, and ruling out any significant role for reactive oxygen species. The activity is indeed unique in comparison with transition metal-mediated DNA cleavage by cisplatin, which is inhibited by GSH.(44) Having demonstrated DNA-cleaving activity, both compounds were submitted to the DTP for NCI-60 panel screening.
Fig. 3. GSH-stimulated hypoxic DNA Cleavage by RPCs.
DNA cleavage assay showing conversion of supercoiled pUC18 plasmid DNA to circular DNA upon treatment with GSH and Δ-32+ or ΔΔ-44+ under aerobic and anaerobic (3 ppm O2) conditions (panel A) DNA cleavage of GSH and Δ-32+ under anaerobic conditions in the presence of DMSO (oxygen radical scavinger) and TEMPO (carbon or metal-based radical scavinger) (panel B). In each figure, the upper band is the circular (cleaved) plasmid and the lower band is the (uncleaved) supercoiled plasmid.
Spectrum of Activity and Predicted Mechanism of Action
The NCI-60 panel single dose cytotoxicity (10 µM) screen for compounds Δ-32+ and ΔΔ-44+ performed by the DTP revealed that RPC Δ-32+ is more cytotoxic than ΔΔ-44+ (mean growth percentages of 42 % and 80 %, respectively for the whole panel). RPC Δ-32+ was subsequently selected for the 5-dose screen. RPC Δ-32+ showed some its best activity against leukemia, colon cancer, lung cancer and melanoma cell lines. Interestingly, the 5-dose assay predicted a unique and complex mechanism (Table 2), a cross between the antimitotic agent paclitaxel (corr. 0.909), the RNA/DNA antimetabolite pyrazofurin (corr. 0.666), the Topo II inhibitor, dibenzyldaunomycin (corr. 0.515) and multiple alkylating agents (corr. 0.4 – 0.5). It is useful to note that there are no alkylating moieties in the complex and no chemical evidence of its behavior as an alkylating agent. These observations are consistent flow-cytometry studies showing a marked G2-M block (see Fig. 2B). Similar degree of specific G2-block was seen with paclitaxel, an agent that does not directly cause DNA-fragmentation, but does activate intrinsic apoptosis through bcl2. Some similarity in mechanism to other tubulin-binding drugs that also activate bcl2 agents that cause single (most alkylating agents) or double-stranded DNA breaks (radiation, intercalating agents) also typically cause G2 arrest. These findings concur with the known subcellular localization of these compounds in the nucleus and mitochondria, and with previous studies of RPC 32+ and 44+ that binds DNA tightly (Kb ~ 108 M−1, in vitro) via an intercalative mode, (45, 46) typical for RPCs with large planar aromatic rings.(18) Indeed, analysis of the whole cell (wc) and nuclear fractions (nf) of RPC treated (5 µM) H-358 cells for uptake of ruthenium by graphite furnace atomic absorption spectroscopy, a showed early concentration spikes in the nf. After 1 h incubation, cells revealed [Ru] of ~7 µg RPC/mg wc and ~27 µg RPC/mg nf for both Δ-32+ and ΔΔ-44+. The ruthenium concentration in the whole cells and nucleus level off and were essentially equal at 30 µg RPC/mg for Δ-32+ and 18 µg RPC/mg for ΔΔ-44+ after 24 h and remain this way for 72 h.
Table 2.
DTP COMPARE Analysis1: Mechanism of Action of RPC Δ-32+ (NSC 747949)
| Class | Drug | Correlation |
|---|---|---|
| Antimitotic Agent | Paclitaxel | 0.91 |
| Vincristine | 0.55 | |
| Echinomycin | 0.49 | |
| RNA/DNA Antimetabolite | Pyrazofurin | 0.67 |
| 5-FUDR | 0.53 | |
| 5-fluorouracil | 0.53 | |
| Ftoafur | 0.52 | |
| Thioguanine | 0.41 | |
| Alkylating Agent | Cyclodisone | 0.62 |
| Piperazine alkylator | 0.57 | |
| Mitomycin C | 0.52 | |
| Porfiromycin | 0.51 | |
| CCNU | 0.50 | |
| Cisplatin | 0.47 | |
| Chlorambucil | 0.43 | |
| Thio-tepa | 0.43 | |
| Tetraplatin | 0.42 | |
| Chlorozotocin | 0.40 | |
| Topo II | "N, N-dibenzyldaunomycin” | 0.52 |
The DTP Compare Analysis uses cytotoxicity data for the test-drug across the NCI-60 spectrum of cell lines to determine is a correlation coefficient of measurement of the similarity in cytotoxic mechanism between the test subject drug, and known cytotoxic drugs with known mechanisms of action. The correlation coefficient ranging from 0 to 1 reflects the degree of similarity. Thus the above findings indicate maximum similarity of mechanism to paclitaxel and significant similarity to several drugs across the antimetabolite and alkylating agent class.
Spectrum of Activity and Mechanism of Cell Death
The activity predicted by the NCI-60 panel towards NSCLC was confirmed in further studies showing significant activity in NSCLC (see Fig. 2). Cell-cycle analysis indicated that RPC Δ-32+ caused a profound G2 block, characteristic of DNA fragmenting agents such as radiation and anthracyclines, but also paclitaxel, which does not directly cause DNA fragmentation. Caspase-3 activation, PARP-cleavage, and TUNEL positivity in IHC was observed upon treatment of H358 with RPCs confirming an increase in the percentage of cells in early and late apoptosis. In contrast, similar treatment of HUVEC or HUVSMC cells with RPCs Δ-32+ or ΔΔ-44+ did not show apoptosis, consistent with the lesser degree of cytotoxicity observed against these cell lines. Simultaneous cytochrome-c release and caspase-8 activation in H358 cells indicated that both intrinsic and extrinsic pathways are activated. Treatment with either Δ-32+ or ΔΔ-44+ caused a generalized decrease in kinase signaling (Fig. 2D), with reduction in pAKT, pEGFR, PI3K, pRb, CDK4, cyclinB1, and bcl2. Bax was significantly increased by ΔΔ-44+, but less so by Δ-32+, perhaps a reason for its greater efficacy.
Animal Toxicity in Mouse
MTD determinations were performed with 12+, 24+, 32+, 44+, and the individual stereoisomers of 44+. As seen in Table 3, it was clear that the compounds containing the tatpp ligand, 32+ and 44+, were significantly less toxic relative to the others, with the tetracationic complex 24+ exhibiting the greatest cytotoxicity. The single ruthenium-tatpp complex 32+ (MTD 65 µmol/Kg), was the least toxic but comparable with the MTD found for the diastereotopic mixture of the di-ruthenium complex 44+ (MTD 43 µmol/Kg), it was equitoxic with the ΔΔ-44+ enantiomer (64 µmol/Kg). Interestingly, the toxicity does not seem to correlate with the complex overall charge or size. Complexes 24+ and 44+ differ little in charge or size, but exhibit significantly different toxicities (MTD 2.5 vs. 43 µmol/Kg, respectively). Among the stereoisomers of 44+, the ΔΔ-enantiomer exhibited the least cytotoxicity and was equitoxic with 32+. From these data and the cytotoxcity studies, It is clear that there is some benefit in working with ΔΔ-44+ and Δ-32+ both in terms of efficacy and in lessened animal toxicity, which is consistent with earlier studies on Δ-12+ and Λ-12+.(13) Mice receiving lethal doses were observed to display lethargy, hind-limb paralysis, and respiratory distress shortly after ip injection. These symptoms are associated with neurotoxicity which has been reported in earlier studies of 12+. (13) Mice showing no abnormal symptoms 15 min after ip injection were generally fine thereafter.
Table 3.
MTDs of Ru (II) polypyridyl complexes in C57 BL/6 mice
| Complex (chloride salt) | Abbr. | Maximum Tolerable Dose MTD µmol complex /kg mouse (mg complex /kg mouse) |
|---|---|---|
| rac-[Ru(phen)3]2+ | 12+ | 9.1 (6.6) |
| rac-[(phen)2Ru(tatpp)]2+ | 32+ | 65 (67) |
| mix-[(phen)2Ru(tatpp)Ru(phen)2]4+ | 44+ | 43 (66) |
| ΔΔ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΔΔ-44+ | 64 (100) |
| ΛΛ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΛΛ-44+ | 43 (66) |
| ΔΛ-[(phen)2Ru(tatpp)Ru(phen)2]4+ | ΔΛ-44+ | 43 (66) |
| mix-[(phen)2Ru(tpphz)Ru(phen)2]4+ | 24+ | 2.5 (3.3) |
Growth Inhibition of H358 NSCLC in a Nude Mouse Xenograft Model
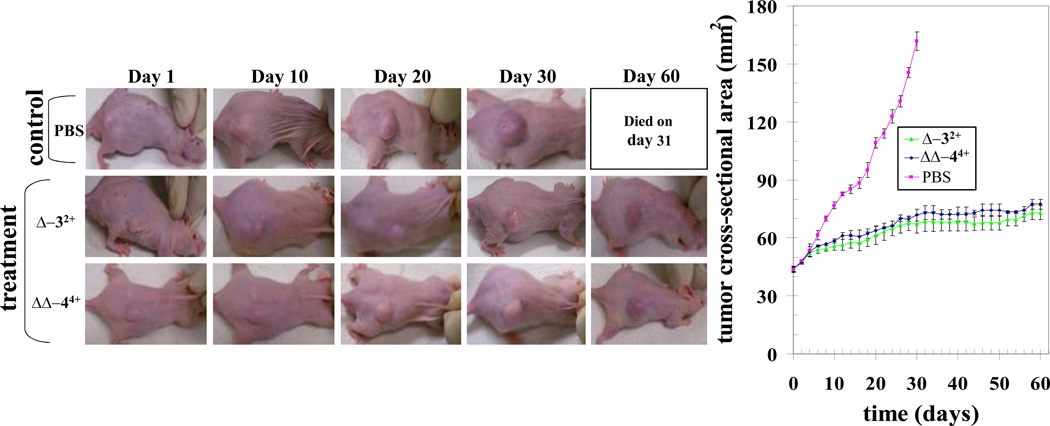
Tumor-bearing animals with established s.c. implanted H358 tumors ~40 mm2 were treated on days 0, 15, and 30 by i.p. injection containing 1 mg (~50% of MTD) RPC (Δ-3Cl2 or ΔΔ-4Cl4) in 100 µL PBS or PBS alone. As seen in Figure 4, rapid and near linear growth in tumor volume was seen in the control group. The treated groups initially showed similar tumor growth to the control but after day 6, tumor growth slowed and only gradually increased over the remaining course of the experiment. Notably, the two treated groups show nearly identical behavior with the Δ-32+ treated group showing slightly less tumor growth than the ΔΔ-44+ group. For statistical purposes, the two treated groups were combined (n = 6) and using a mixed linear model to fit the data, the equality of the group means was tested using a repeated measures ANOVA analysis. Results indicate that the main group effect, the main time effect, and their interaction term are all very significant (P-values <0.00001). If we consider the divergence of the combined treated (n = 6) and control (n = 3) groups by day 14, one day before the second drug dose was administered, the MTV of the treated group is 35 % that of the control group. By 32 days, all control animals had died or were sacrificed for tumors > 150 mm2. Tumors in the treated group were only 20 % the size of the control group at that time (P <0.0001). The last treatment was given on day 30 and, as can be seen in Figure 4, no further tumor growth was seen in either individual treated group over the next 30 day period. Survival was also improved, with 0 alive at 32 days for control, 2/3 and 2/3 alive at 60 days with both Δ-42+ and ΔΔ-34+, respectively. Ruthenium treated mice had weight gain comparable to non-tumor bearing controls (Fig. 4).
Fig. 4. Effect of ruthenium compounds on H358 human lung cancer nude mice xenograft model.
Hsd: Athymic nude nu/nu mice were obtained from Harlan, Indianapolis, IN. All animal experiments were carried out in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC). Nine 10-week-old nude mice were divided into three groups of 3 animals (treated with PBS (pink), ruthenium compound Δ-3Cl2 (green) and ΔΔ-4Cl4 (blue) (70 mg / kg b.w.). All 9 animals were injected with 1 × 106 human lung cancer cells (H358) suspensions in 100 µl of PBS, subcutaneously into one flank of each nu/nu nude mouse. At the same time, animals were randomized treatment groups as indicated in the figure. Treatment was administered when the tumor surface area exceeded ~ 42 mm2 (day 24). Treatment consisted of Δ-32+ (2 mg) and ΔΔ-44+ (2 mg) in 200 µl PBS. Control groups were treated with 200 µl PBS. Animals were examined daily for signs of tumor growth. Tumors were measured in two dimensions using calipers. Photographs of animals were taken at day 1, day 10, day 20, day 30, and day 60 after treatment, are shown for all groups.
DISCUSSION
These studies are the first demonstration of in-vivo anti-cancer activity of systemically administered RPCs (lacking labile ligands) in NSCLC. It is clear that RPC’s containing the tatpp ligand (32+ and 44+) outperform the other RPCs (12+, 24+, [Ru(phen)2(dppz)]2+ and [Ru(phen)2(tpphz)]2+) in terms of efficacy and selectivity for inducing apoptosis in cultured NSCLC cell lines, causing a specific G2M arrest, and triggering apoptosis through both intrinsic and extrinsic pathways. Most significantly, they demonstrate statistically significant tumor growth repression (P < 0.0001) in vivo and a doubling of survival time, as shown in Figure 4.
By far, the most important feature discovered was the enhanced biological activity associated with use of tatpp ligand (found only in RPC’s 32+ and 44+). Although tatpp is insoluble alone, coordination to Ru(II) leads to stable, soluble complexes, like 32+ and 44+, in which tatpp-based reductive processes are observed at potentials positive of −0.25 V (vs. NHE at pH 7.0)(47, 48). In contrast, most RPC’s like 12+ and 24+ are significantly harder to reduce and usually require reduction potentials negative of −0.70 V or more.(18, 33) This low reduction potential means that RPCs are readily reduced by common cellular reductants, and we have shown that these reductions are centered on the tatpp ligand, not the Ru(II) ion, with concomitant formation of a carbon radical species. Because RPCs 32+ and 44+ as well as related complexes 24+, [Ru(phen)2(dppz)]2+, and [Ru(phen)2(tpphz)]2+, are known to bind DNA via intercalation,(18) the tatpp carbon radical in the reduced species is in intimate contact with the DNA. We hypothesize that H atom abstraction from the deoxyribose unit as the likely mechanism of DNA damage. Dioxygen can react with this radical species to reform the tatpp complex(47) and in this manner attenuate the DNA cleavage process. As the tatpp complex can be reduced again, the steady-state concentration of the key carbon-radical species is dependent on the local concentrations of O2 and cellular reducing agents. Ruthenium atomic absorption analysis shows ruthenium in the nuclear fractions of whole H358 cells incubated with Δ-32+ and ΔΔ-44+, indicating that the RPC are making it to the nucleus. The enhanced cytotoxicity seen for Δ-32+ in treated H358 cells under hypoxia (Fig 2A.) suggests this carbon-radical mediated cleavage mechanism is active in live cells and potentially in vivo.
RPCs 32+ and 44+ show good selectivity for malignant cells and one possible explanation is that these RPCs exhibit different transport mechanisms in cells compared to the simpler RPCs, such as 12+, 24+, and [(phen)2Ru(dppz)]2+, which are generally shown to enter cells via passive diffusion.(27, 28) Recently, Thomas and coworkers showed that the structurally related tpphz dimers, 24+ and [(bpy)2Ru(tpphz)Ru(bpy)2]4+ are actively transported into MCF-7 cells in a non-endocytotic but temperature-dependent manner.(22) If 32+ and 44+ similarly undergo active transport, the differences in the transport pathways in malignant cells versus normal cells could explain the selectivity. Not only are the tpphz dimers actively transported, they were also observed to localize in the nucleus as demonstrated by fluorescence and co-localization with another nuclear stain, DAPI. Complexes 32+ and 44+ are not luminescent but given the observed detection of ruthenium in the nucleus and the similarity of their structure to [(bpy)2Ru(tpphz)Ru(bpy)2]4+, it is reasonable that they exhibit the same transport and localization properties as the tpphz RPCs.(22) The important difference between these two classes of RPCs (tatpp- vs tpphz-based RPCs) is that the latter are not easily reduced, do not show any DNA cleavage activity, and are considerably less cytotoxic which only further emphasizes the importance and role of the redox-active ligand in the observed biological activity.
The analysis of the cytotoxicity of Δ-32+ and ΔΔ-44+ (NSC 747949 and 747950, respectively) in the NCI-60 panel revealed unique patterns of cytotoxicity and the former had sufficient activity to proceed to multidose screening. The predicted mechanism of action by the NCI panel was found to be consistent with the known nucleotide and mitochondrial binding activity and DNA cleaving activity of this compound. Because an anaerobic environment is not utilized in the NCI-60 panel, mechanism of action is incompletely assessed, however, our cytotoxicity studies under hypoxic conditions reveal enhanced activity for Δ-32+ in H358 cells relative to those under normoxic conditions (Fig. 2A). It is clear that these molecules bind DNA and mitochondria (19, 25, 30) strongly, cleave DNA, cause a specific G2M arrest, and trigger apoptosis through both intrinsic and extrinsic pathways. Another possibility is the involvement of receptor-mediated death pathways where the tumor factor necrosis (TNF) family of death receptors activate the initiator caspase-8 which can activate caspase-3 without involve the mitochondrial-mediated apoptosis.
In summary, we have systematically optimized structural features of RPCs, selecting features that optimize cancer-selective apoptosis in cultured NSCLC cell lines and on the basis of animal toxicity, and have demonstrated unique chemical mechanism of reductive DNA cleavage through a carbon-centered radical mechanism. Due to the ease with which the tatpp ligand can be reduced in situ, RPCs 32+ and 44+, display a unique and potentially hypoxia-selective ability to bind and cleave DNA through a carbon-centered radical mechanism. Combined these data show that RPCs, and specifically RPCs with bioreducible ligands, represent a pharmacologically favorable class of compounds with a natural selectivity for cancer cells and promising hypoxia selective activity.
Acknowledgments
Grant Support: The Robert A. Welch Foundation Grant Y-1301 to F. M. MacDonnell and the NIH-NCI for grants 2R15CA113747 to F. M. MacDonnell, and CA77495 to S. Awasthi are acknowledged for their support.
Footnotes
Conflicts of Interest: None
REFERENCES
- 1.Lippert B, editor. Cisplatin Chemistry and Biochemistry of a Leading Anticancer Drug. Weinheim: Wiler-VCH; 1999. [Google Scholar]
- 2.Clarke MJ. Ruthenium metallopharmaceuticals. Coord Chem Rev. 2003;236:209–233. [Google Scholar]
- 3.Jakupec MA, Galanski M, Arion VB, Hartinger CG, Keppler BK. Antitumour metal compounds: more than theme and variations. Dalton Transactions. 2008:183–194. doi: 10.1039/b712656p. [DOI] [PubMed] [Google Scholar]
- 4.Allardyce CS, Dyson PJ. Ruthenium in medicine: Current clinical uses and future prospects. Platinum Metals Review. 2001;45:62–69. [Google Scholar]
- 5.Bergamo A, Gava B, Alessio E, Mestroni G, Serli B, Cocchietto M, et al. Ruthenium-based NAMI-A type complexes with in vivo selective metastasis reduction and in vitro invasion inhibition unrelated to cell cytotoxicity. Int J Oncol. 2002;21:1331–1338. [PubMed] [Google Scholar]
- 6.Depenbrock H, Schmelcher S, Peter R, Keppler BK, Weirich G, Block T, et al. Preclinical activity of trans-indazolium [tetrachlorobisindazoleruthenate(III)] (NSC 666158; IndCR; KP 1019) against tumour colony-forming units and haematopoietic progenitor cells. European Journal of Cancer. 1997;33:2404–2410. doi: 10.1016/s0959-8049(97)00277-3. [DOI] [PubMed] [Google Scholar]
- 7.Lentz F, Drescher A, Lindauer A, Henke M, Hilger RA, Hartinger CG, et al. Pharmacokinetics of a novel anticancer ruthenium complex (KP1019, FFC14A) in a phase I dose-escalation study. Anti-Cancer Drugs. 2009;20:97–103. doi: 10.1097/CAD.0b013e328322fbc5. [DOI] [PubMed] [Google Scholar]
- 8.Hartinger CG, Jakupec MA, Zorbas-Seifried S, Groessl M, Egger A, Berger W, et al. KP1019, A New Redox-Active Anticancer Agent – Preclinical Development and Results of a Clinical Phase I Study in Tumor Patients. Chemistry & Biodiversity. 2008;5:2140–2155. doi: 10.1002/cbdv.200890195. [DOI] [PubMed] [Google Scholar]
- 9.Morris RE, Aird RE, Murdoch PdS, Chen H, Cummings J, Hughes ND, et al. Inhibition of Cancer Cell Growth by Ruthenium(II) Arene Complexes. J Med Chem. 2001;44:3616–3621. doi: 10.1021/jm010051m. [DOI] [PubMed] [Google Scholar]
- 10.Meng X, Leyva ML, Jenny M, Gross I, Benosman S, Fricker B, et al. A Ruthenium-Containing Organometallic Compound Reduces Tumor Growth through Induction of the Endoplasmic Reticulum Stress Gene CHOP. Cancer Research. 2009;69:5458–5466. doi: 10.1158/0008-5472.CAN-08-4408. [DOI] [PubMed] [Google Scholar]
- 11.Shulman A, Dwyer FP. Metal Chelates in Biological Systems. In: Dwyer FP, Mellor DP, editors. Chelating Agents and Metal Chelates. New York: Academic Press; 1964. pp. 383–439. [Google Scholar]
- 12.Beccari E. Pharmacological studies on the ferrous tri-2,2'-bipyridyl complex. IV. Relation between concentration in the blood and action on the central nervous system. Boll - Soc Ital Biol Sper. 1941;16:216–218. [Google Scholar]
- 13.Koch JH, Rogers WP, Dwyer FP, Gyarfas EC. The metabolic fate of tris-1,10-phenanthroline ruthenium-106 (II) perchlorate, a compound with anticholinesterase and curare-like activity. Australian J Biol Sci. 1957;10:342–350. [Google Scholar]
- 14.Dwyer FP, Mayhew E, Roe EMF, Shulman A. Inhibition of Landschuetz ascites tumor growth by metal chelates derived from 3,4,7,8 - tetramethyl-- 1,10 - phenanthroline. Br J Cancer. 1965;19:195–199. doi: 10.1038/bjc.1965.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch JH, Gyarfas EC, Dwyer FP. Biological activity of complexions. Mechanism of inhibition of acetylcholinesterase. Aust J Biol Sci. 1956;9:371–381. [Google Scholar]
- 16.Dwyer FP, Gyarfas EC, Rogers WP, Koch JH. Biological activity of complex ions. Nature (London, U K) 1952;170:190–191. doi: 10.1038/170190a0. [DOI] [PubMed] [Google Scholar]
- 17.White DO, Harris AW, Cheyne IM, Shew M. Actions of metal chelates of substituted 1,10-phenanthrolines on viruses and cells. 3. Actions on cultured cells. Austral J Exp Biol Med Sci. 1969;47:81–89. doi: 10.1038/icb.1969.7. [DOI] [PubMed] [Google Scholar]
- 18.Augustyn KE, Pierre VC, Barton JK, editors. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons, Inc; 2009. Metallointercalators as probes of DNA recognition and reactions. [Google Scholar]
- 19.Pisani MJ, Weber DK, Heimann K, Collins JG, Keene FR. Selective mitochondrial accumulation of cytotoxic dinuclear polypyridyl ruthenium(II) complexes. Metallomics. 2010;2:393–396. doi: 10.1039/c004922k. [DOI] [PubMed] [Google Scholar]
- 20.Schatzschneider U, Niesel J, Ott I, Gust R, Alborzinia H, Wölfl S. Cellular Uptake, Cytotoxicity, and Metabolic Profiling of Human Cancer Cells Treated with Ruthenium(II) Polypyridyl Complexes [Ru(bpy)2(N-N)]Cl2 with N-N=bpy, phen, dpq, dppz, and dppn. ChemMedChem. 2008;3:1104–1109. doi: 10.1002/cmdc.200800039. [DOI] [PubMed] [Google Scholar]
- 21.Pascu G, Hotze A, Sanchez-Cano C, Kariuki B, Hannon M. Dinuclear Ruthenium(II) Triple-Stranded Helicates: Luminescent Supramolecular Cylinders That Bind and Coil DNA and Exhibit Activity against Cancer Cell Lines. Angew Chem Int Ed. 2007;46:4374–4378. doi: 10.1002/anie.200700656. [DOI] [PubMed] [Google Scholar]
- 22.Gill MR, Garcia-Lara J, Foster SJ, Smythe C, Battaglia G, Thomas JA. A ruthenium(II) polypyridyl complex for direct imaging of DNA structure in living cells. Nat Chem. 2009;1:662–667. doi: 10.1038/nchem.406. [DOI] [PubMed] [Google Scholar]
- 23.Novakova O, Kasparkova J, Vrana O, van Vliet PM, Reedijk J, Brabec V. Correlation between Cytotoxicity and DNA Binding of Polypyridyl Ruthenium Complexes. Biochemistry. 1995;34:12369–12378. doi: 10.1021/bi00038a034. [DOI] [PubMed] [Google Scholar]
- 24.Dwyer FP, Gyarfas EC, Wright RD, Shulman A. Effect of inorganic complex ions on transmission at a neuromuscular junction. Nature. 1957;179:425–426. doi: 10.1038/179425a0. [DOI] [PubMed] [Google Scholar]
- 25.Koch JH, Gallagher CH. Effect of Some Neuromuscular Blocking Agents on Mitochondrial Enzyme Systems. Nature. 1959;184:1039–1041. doi: 10.1038/1841039a0. [DOI] [PubMed] [Google Scholar]
- 26.Mayhew E, Roe EM, Shulman A. Microscopical observations on the effects of phenanthroline chelates on Landschütz ascites tumour cells. J R Microsc Soc. 1965;84:475–483. doi: 10.1111/j.1365-2818.1965.tb02148.x. [DOI] [PubMed] [Google Scholar]
- 27.Puckett CA, Barton JK. Methods to Explore Cellular Uptake of Ruthenium Complexes. J Am Chem Soc. 2007;129:46–47. doi: 10.1021/ja0677564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puckett CA, Barton JK. Mechanism of Cellular Uptake of a Ruthenium Polypyridyl Complex. Biochemistry. 2008;47:11711–11716. doi: 10.1021/bi800856t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brabec V, Novakova O. DNA binding mode of ruthenium complexes and relationship to tumor cell toxicity. Drug Resist Updates. 2006;9:111–122. doi: 10.1016/j.drup.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Tan C, Lai S, Wu S, Hu S, Zhou L, Chen Y, et al. Nuclear Permeable Ruthenium(II) Î2-Carboline Complexes Induce Autophagy To Antagonize Mitochondrial-Mediated Apoptosis. J Med Chem. 2010;53:7613–7624. doi: 10.1021/jm1009296. [DOI] [PubMed] [Google Scholar]
- 31.Zava O, Zakeeruddin SM, Danelon C, Vogel H, Gratzel M, Dyson PJ. A Cytotoxic Ruthenium Tris(Bipyridyl) Complex that Accumulates at Plasma Membranes. ChemBioChem. 2009;10:1796–1800. doi: 10.1002/cbic.200900013. [DOI] [PubMed] [Google Scholar]
- 32.Sun Y, Joyce LE, Dickson NM, Turro C. Efficient DNA photocleavage by [Ru(bpy)2(dppn)]2+ with visible light. Chem Commun. 2010;46:2426–2428. doi: 10.1039/b925574e. [DOI] [PubMed] [Google Scholar]
- 33.Moucheron C, De MAK, Kelly JM. Photophysics and photochemistry of metal polypyridyl and related complexes with nucleic acids. Struct Bonding (Berlin) 1998;92:163–216. [Google Scholar]
- 34.Janaratne TK, Yadav A, Ongeri F, MacDonnell FM. Preferential DNA Cleavage under Anaerobic Conditions by a DNA-Binding Ruthenium Dimer. Inorg Chem. 2007;46:3420–3422. doi: 10.1021/ic0619714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun P, Krishnan A, Yadav A, Singh S, MacDonnell FM, Armstrong DW. Enantiomeric Separations of Ruthenium(II) Polypyridyl Complexes Using High-Performance Liquid Chromatography (HPLC) with Cyclodextrin Chiral Stationary Phases (CSPs) Inorg Chem. 2007;46:10312–10320. doi: 10.1021/ic701023x. [DOI] [PubMed] [Google Scholar]
- 36.MacDonnell FM, Bodige S. Efficient Stereospecific Syntheses of Chiral Ruthenium Dimers. Inorg Chem. 1996;35:5758–5759. [Google Scholar]
- 37.Bolger J, Gourdon A, Ishow E, Launay J-P. Mononuclear and Binuclear Tetrapyrido[3,2-a:2',3'-c:3'',2''-h:2''',3'''-j]phenazine (tpphz) Ruthenium and Osmium Complexes. Inorg Chem. 1996;35:2937–2944. [Google Scholar]
- 38.Wärnmark K, Thomas JA, Heyke O, Lehn J-M. Stereoisomerically Controlled Inorganic Architectures: Synthesis of Enantio- and Diastereo-merically Pure Ruthenium-Palladium Molecular Rods from Enantiopure Building Blocks. Chem Commun. 1996:701–702. [Google Scholar]
- 39.Kim M-J, Konduri R, Ye H, MacDonnell FM, Puntoriero F, Serroni S, et al. Dinuclear ruthenium(II) polypyridyl complexes containing large, redox-active, aromatic bridging ligands: synthesis, characterization, and intramolecular quenching of MLCT excited states. Inorg Chem. 2002;41:2471–2476. doi: 10.1021/ic011028u. [DOI] [PubMed] [Google Scholar]
- 40.Kim M-J. Chiral Metallodendrimers and oligomers Containing Ru(II) Polypyridyl Complexes [Ph. D.] Arlington, TX: University of Texas at Arlington; 2000. [Google Scholar]
- 41.Singhal SS, Singhal J, Yadav S, Dwivedi S, Boor PJ, Awasthi YC, et al. Cancer Res. 2007;67:4382–4389. doi: 10.1158/0008-5472.CAN-06-4124. [DOI] [PubMed] [Google Scholar]
- 42.Singhal SS, Singhal J, Yadav S, Dwivedi S, Boor PJ, Awasthi YC, et al. Regression of lung and colon cancer xenografts by depleting or inhibiting RLIP76 (ral-binding protein 1) Cancer Res. 2007;67:4382–4389. doi: 10.1158/0008-5472.CAN-06-4124. [DOI] [PubMed] [Google Scholar]
- 43.Mohler DL, Downs JR, Hurley-Predecki AL, Sallman JR, Gannett PM, Shi X. DNA Cleavage by the Photolysis of Cyclopentadienyl Metal Complexes: Mechanistic Studies and Sequence Selectivity of Strand Scission by CpW(CO)3CH3. J Org Chem. 2005;70:9093–9102. doi: 10.1021/jo050338h. [DOI] [PubMed] [Google Scholar]
- 44.Pratviel G, Bernadou J, Meunier B. DNA and RNA cleavage by metal complexes. Adv Inorg Chem. 1998;45:251–312. [Google Scholar]
- 45.Janaratne TK. Investigation of Ruthenium (II) Polypyridyl Dimers as Potential Chemotherepeutic Agents [Ph. D. thesis] Arlington, TX: University of Texas at Arlington; 2006. [Google Scholar]
- 46.Rajput C, Rutkaite R, Swanson L, Haq I, Thomas JA. Dinuclear monointercalating RuII complexes that display high affinity binding to duplex and quadruplex DNA. Chem Eur J. 2006;12:4611–4619. doi: 10.1002/chem.200501349. [DOI] [PubMed] [Google Scholar]
- 47.de Tacconi NR, Lezna RO, Chitakunye R, MacDonnell FM. Electroreduction of the Ruthenium Complex [(bpy)2Ru(tatpp)]Cl2 in Water: Insights on the Mechanism of Multielectron Reduction and Protonation of the Tatpp Acceptor Ligand as a Function of pH. Inorg Chem. 2008;47:8847–8858. doi: 10.1021/ic8009157. [DOI] [PubMed] [Google Scholar]
- 48.de Tacconi NR, Lezna RO, Konduri R, Ongeri F, Rajeshwar K, MacDonnell FM. Influence of pH on the photochemical and electrochemical reduction of the dinuclear ruthenium complex, [(phen)2Ru(tatpp)Ru(phen)2]Cl4, in water: Proton-coupled sequential and concerted multi-electron reduction. Chem Eur J. 2005;11:4327–4339. doi: 10.1002/chem.200401287. [DOI] [PubMed] [Google Scholar]